Abstract
We describe a general computational approach to site-specific resonance assignments in multidimensional NMR studies of uniformly 15N,13C-labeled biopolymers, based on a simple Monte Carlo/simulated annealing (MCSA) algorithm contained in the program MCASSIGN2. Input to MCASSIGN2 includes lists of multidimensional signals in the NMR spectra with their possible residue-type assignments (which need not be unique), the biopolymer sequence, and a table that describes the connections that relate one signal list to another. As output, MCASSIGN2 produces a high-scoring sequential assignment of the multidimensional signals, using a score function that rewards good connections (i.e., agreement between relevant sets of chemical shifts in different signal lists) and penalizes bad connections, unassigned signals, and assignment gaps. Examination of a set of high-scoring assignments from a large number of independent runs allows one to determine whether a unique assignment exists for the entire sequence or parts thereof. We demonstrate the MCSA algorithm using two-dimensional (2D) and three-dimensional (3D) solid state NMR spectra of several model protein samples (α-spectrin SH3 domain and protein G/B1 microcrystals, HET-s218–289 fibrils), obtained with magic-angle spinning and standard polarization transfer techniques. The MCSA algorithm and MCASSIGN2 program can accommodate arbitrary combinations of NMR spectra with arbitrary dimensionality, and can therefore be applied in many areas of solid state and solution NMR.
Keywords: Sequential assignment, Solid state NMR, Magic-angle spinning, Multidimensional spectroscopy
Introduction
Assignment of chemical shifts to specific sites is generally a preprequisite for nuclear magnetic resonance (NMR) studies of the structures, dynamics, interactions, or other properties of biopolymers such as proteins and nucleic acids. The assignment process can be difficult and error-prone for a variety of reasons, including limited spectral resolution, low signal-to-noise in the NMR spectra, and uncertainty regarding the identity of the segments of the biopolymer sequence that contribute to the NMR spectra. This is particularly true in solid state NMR studies of uniformly 15N,13C-labeled proteins with magic-angle spinning (MAS), which are the primary motivation for the work described below. However, similar difficulties may arise in other types of experiments.
The standard approach to the assignment problem in solid state MAS NMR is to record several multidimensional spectra that correlate 15N and/or 13C chemical shifts of residue k with the chemical shifts of residues k − 1 and k + 1, and to determine the assignments by tracing pathways of connections among crosspeaks in the spectra in a systematic, sequential manner, either by hand or with the assistance of computer-based tools. When the connections among crosspeaks are ambiguous (e.g., due to insufficient resolution), such a systematic approach becomes dangerous, as numerous candidates for sequential assignment pathways may exist, all of which appear to be consistent with the available data, or nearly so. Automated assignment algorithms for solution NMR spectroscopy have been proposed by several groups (Bailey-Kellogg et al. 2000; Bartels et al. 1997; Buchler et al. 1997; Hitchens et al. 2003; Hyberts and Wagner 2003; Lemak et al. 2008; Leutner et al. 1998; Li and Sanctuary 1997; Lukin et al. 1997; Moseley et al. 2001; Nelson et al. 1991).
As an alternative to manual assignment procedures in solid state MAS NMR of proteins, we recently introduced a computational approach that employs a simple Monte Carlo/simulated annealing (MCSA) algorithm to search for chemical shift assignments (Tycko and Hu 2010). Advantages of the MCSA approach include its simplicity, its objectivity, and its ability to identify all assignments that satisfy the experimental constraints in a reproducible and explicit manner. Our initial implementation and demonstration of the MCSA algorithm (contained in the program MCASSIGN1) was limited to the analysis of 2D NCACX and NCOCX spectra, which are frequently used for assignments in protein systems with relatively low molecular weight and good spectral resolution (Debelouchina et al. 2010; Franks et al. 2005; Helmus et al. 2008; Igumenova et al. 2004; McDermott et al. 2000; Pauli et al. 2001; Petkova et al. 2003; Siemer et al. 2006b; Tycko et al.2010). In this paper, we describe and demonstrate a generalized MCSA algorithm (contained in the program MCASSIGN2) that is capable of analyzing arbitrary combinations of spectra with arbitrary dimensions, involving signals from arbitrary combinations of nuclei. We expect the generalized algorithm to have broad applicability in solid state MAS NMR studies of proteins. The same algorithm is also applicable to nucleic acids and other heteropolymers, to solid state NMR of oriented systems (Ishii and Tycko 2000; Sinha et al. 2007), and to solution NMR.
Generalized MCSA algorithm
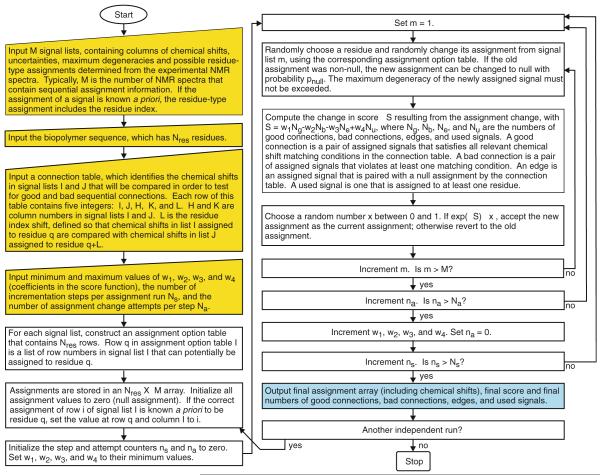
The algorithm is summarized in a flow chart in Fig. 1. Fortran95 source code for MCASSIGN2, which implements this algorithm, is included in Supporting Information, as are the input files used in the demonstrations discussed below. MCASSIGN2, examples of input files, and instructions are also available upon request from the authors (address robertty@mail.nih.gov). The following points supplement the information in Fig. 1:
Signal lists can come from any NMR spectra that provide constraints on resonance assignments through agreement among chemical shifts in pairs of spectra. Each row of a signal list is considered to be one signal that can be assigned to one residue (or more than one residue, if the degeneracy exceeds one). Roughly speaking, signal lists are crosspeak lists, except that the number of chemical shift columns in a signal list does not necessarily equal the dimensionality of the NMR spectrum. For example, if one row of a 2D spectrum contains two or more crosspeaks that definitely arise from the same residue, the number of chemical shift columns in the corresponding signal list can be greater than two. Similarly, if one plane of a 3D spectrum contains multiple crosspeaks that definitely arise from the same residue, the number of chemical shift columns can be greater than three.
Biopolymer sequences and possible residue-type assignments are given in one-letter codes. If the assignment of a signal is known a priori, the possible residue-type assignment in the signal list is a single letter followed by the residue index (e.g., V32 for valine at residue 32). Otherwise, the possible residue-type assignments are strings of one-letter codes (e.g., EQKRH for glutamate, glutamine, lysine, arginine, and histidine).
Any relevant information can be used for residue-type assignments. In the examples below, residue-type assignments come from 13C chemical shifts, using the known shift ranges for Cα and sidechain carbon sites for the various residues. If the signal lists themselves include many sidechain chemical shifts, then residue-type assignments can be based entirely on information in the signal lists. Alternatively, chemical shifts in the signal lists can be compared with intra-residue crosspeak patterns in other spectra that contain the full sidechain signals, for example a 2D 13C–13C spectrum as in Fig. 4c, in order to determine possible residue-type assignments. Similarly, once chemical shifts that are present in the signal lists are assigned to specific residues by MCASSIGN2, additional sidechain chemical shift assignments can be made through comparisons with other spectra, such as a 2D 13C–13C spectrum.
The degeneracy of a signal in a signal list is the maximum number of residues to which it can be assigned. Degeneracies greater than one can be used if it appears that a signal may contain unresolved contributions from more than one residue. If the corresponding crosspeaks are broad, the chemical shift uncertainties should include the full crosspeak widths. Alternatively, several copies of the signal, each with a degeneracy of one, could be included in the signal list.
The connection table specifies the pairs of chemical shifts from pairs of signal lists that should agree if a sequential assignment is correct. The values in the connection table depend on the identities of chemical shift columns in the signal lists and on the types of NMR spectra that are being analyzed. “Residue index shift” values determine whether the chemical shifts that are being compared are shifts that have been assigned to the same residue (index shift = 0) or to residues whose indices differ by the specified integer. Residue index shift values can be −1, 0, and +1.
Residue index shift values depend on the way in which signals are assigned to residues. For example, if a signal contains chemical shifts of residues q and q’, either chemical shift can be used as the basis for assignment (i.e., the signal can be assigned either to residue q or to residue q’). The two possibilities lead to different residue index shift values, but either choice is acceptable as long as consistency is maintained. In the demonstrations discussed below, assignments are based on 13Cα chemical shifts because these shifts were measured in all of the NMR experiments and appear in all of the signal lists. Alternatively, backbone amide 15N chemical shifts could have been used as the basis for assigning signals to specific residues, but then the residue index shifts in the connection tables would change (see below).
When a connection between two signals involves more than one pair of chemical shifts (i.e., when several chemical shifts in a pair of multidimensional signals should agree when the sequential assignment is correct), the connection table contains more than one row for that connection. The connection is considered to be “good” when all chemical shift comparisons involving the same pair of residues and the same pair of signal lists produce a positive result, meaning that the chemical shift difference Δ satisfies the condition that , where δi and δj are the relevant chemical shift uncertainties in the two signal lists. When one or more of the chemical shift comparisons involving the same pair of residues and the same pair of signal lists produces a negative result, the connection is considered to be “bad”. If a chemical shift comparison in the connection table can not be made in a specific instance because one of the relevant chemical shift values is missing from the signal list (e.g., if the connection table specifies a column that contains β-carbon chemical shifts, but the signal under consideration arises from a glycine residue or from a non-glycine residue whose β-carbon signal is too weak to detect), the comparison is ignored.
A null assignment means that a given residue has no signal assigned to it from a given signal list. When certain segments of the biopolymer sequence do not contribute to the NMR signals, the final, correct assignment should include null assignments for those segments. An assignment “edge” is a connection that can not be tested for agreement of chemical shifts because one of the relevant residues has a null assignment (and the other residue has a non-null assignment). Edges are penalized in the score function (if w3 > 0) in order to favor assignments in which the signals are concentrated into relatively few segments, rather than being scattered over the biopolymer sequence with many intervening gaps.
The definition of the score function can easily be changed without changing the rest of the algorithm. The values of w1, w2, w3, and w4 can be tailored to specific applications. In particular, penalties for assignment edges and unused signals can be eliminated by setting w3 = w4 = 0.
Any final assignment with Nb = 0 and with no unused signals should be considered fully consistent with available data. There may be multiple consistent assignments, with scores that vary due to variations in Ng and Ne. If the scores vary greatly, it may be justifiable to retain only the highest-scoring assignments. Some runs may produce final assignments with Nb ≠ 0. Such assignments are inconsistent with available data.
Execution of the generalized MCSA algorithm is rapid on modern computers. Typical execution times in the examples discussed below were 50 min for 100 independent runs with Ns = 20 and Na = 106, executed on a 3.0 GHz AMD Opteron 8222 processor.
Fig. 1.
Flow chart describing the generalized MCSA algorithm within MCASSIGN2. Yellow boxes describe input files that are provided by the user. Blue box describes the output. Other boxes describe operations that are performed within MCASSIGN2
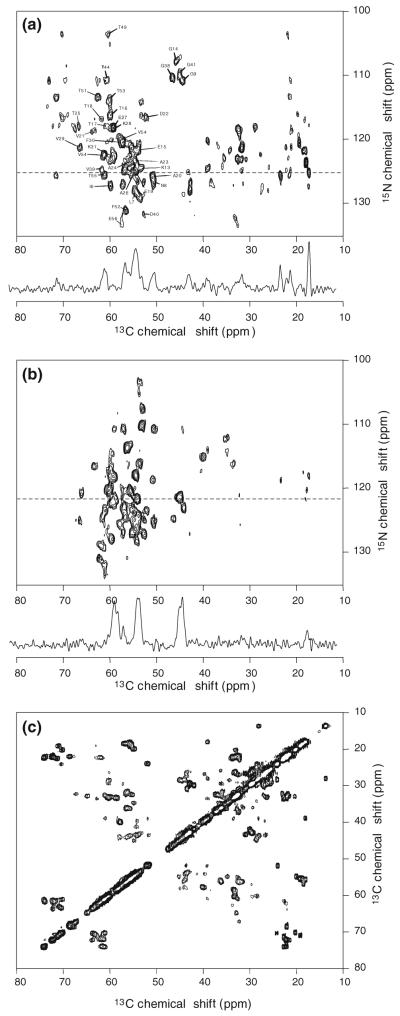
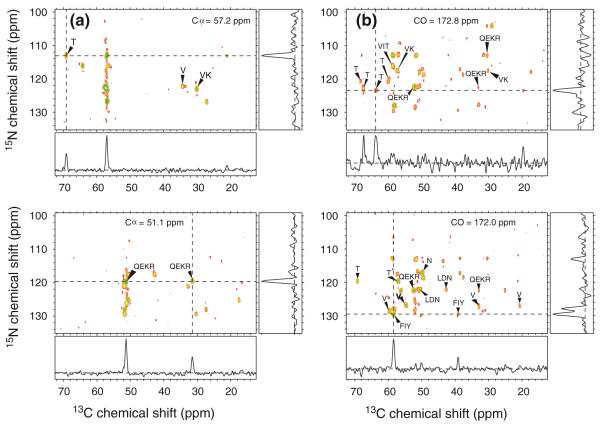
Fig. 4.
2D solid state NMR spectra of uniformly 15N,13C-labeled protein G/B1 microcrystals, obtained at 9.4 T. a 2D NCACX spectrum, showing a subset of the intra-residue 15N–13Cα crosspeak assignments determined by MCASSIGN2. The 1D slice, taken at the dashed line in the 2D spectrum, illustrates the signal-to-noise ratio and linewidths. b 2D NCOCX spectrum with a representative 1D slice. c Aliphatic region of the 2D 13C–13C spectrum
Results
Microcrystalline α-spectrin SH3
Uniformly 15N,13C-labeled α-spectrin SH3 was produced and crystallized essentially as described by Pauli et al. (2000) Microcrystalline samples were packed in a 1.8 mm MAS NMR probe from the group of Dr. Ago Samoson (NMR Institute, Tallinn University of Technology, Estonia) for measurements at 17.6 T and in a 3.2 mm Varian MAS rotor for measurements at 9.4 T. Protein quantities were 5 mg at both fields.
At 17.6 T, 3D NCACX and NCOCX spectra were acquired with MAS at 17.00 kHz, using 6.0 ms cross-polarization periods for 15N–13C polarization transfer after t1 (13C carrier frequency at 56 ppm for NCACX and 175 ppm for NCOCX; 39 kHz 15N radio-frequency (rf) amplitude; 20–25 kHz ramped 13C rf amplitude) and 20 ms rf-assisted diffusion (RAD) (Morcombe et al. 2004; Takegoshi et al. 2001) periods for 13C–13C polarization transfer after t2. A 3D CONCA spectrum was recorded with 6.0 ms 13C–15N and 15N–13C cross-polarization periods after t1 and t2 (13C carrier frequencies at 175 and 56 ppm, respectively). 1H decoupling fields were 96 kHz, with two-pulse phase modulation (TPPM) (Bennett et al. 1995) during t1, t2, and t3. 48 complex t1 and 26 complex t2 values were recorded, with 235.3 μs increments. Total measurement times for NCACX and NCOCX spectra were 17 h, with a 1.5 s recycle delay. The total measurement time for the CONCA spectrum was 3.7 days, with a 2.0 s recycle delay. Conditions for measurements at 9.4 T were similar, except that the MAS frequency was 9.0 kHz. Spectra were processed with NMRPipe (Delaglio et al. 1995) and analyzed with Sparky (available at http://www.cgl.ucsf.edu/home/sparky/).
Representative 2D planes from the 3D spectra acquired at 17.6 T are shown in Fig. 2. Multidimensional signal lists from these spectra are shown in Table S1. Signals from 3D NCACX and NCOCX spectra contain up to five chemical shifts. Signals from the 3D CONCA spectrum contain only the chemical shifts of directly-bonded 13CO, 15N, and 13Cα sites, as no period for 13C–13C polarization transfer before t3 was included. 51, 49, and 49 signals were identified in the 3D NCACX, NCOCX, and CONCA spectra, respectively. Unique residue-type assignments were obtained for Ile, Gly, Ala, Ser, Thr, Pro, and certain other signals. No degeneracy values greater than one were required.
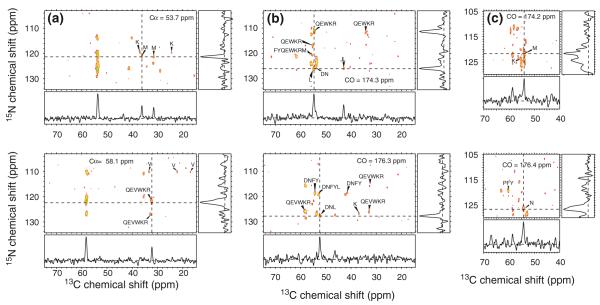
Fig. 2.
Representative 2D planes from 3D solid state NMR spectra of uniformly 15N,13C-labeled α-spectrin SH3 microcrystals, obtained at 17.6 T. a Two planes from the 3D NCACX spectrum, taken at the indicated chemical shifts in the second dimension. b Two planes from the 3D NCOCX spectrum, taken at the indicated chemical shifts in the second dimension. c Two planes from the 3D NCOCX spectrum, taken at the indicated chemical shifts in the first dimension. 1D slices, taken at the dashed lines in each 2D plane, illustrate the signal-to-noise ratios and linewidths. Residue-type assignments for selected crosspeaks are indicated
90 MCASSIGN2 runs with Na = 106 and Ns = 20 and with w1, w2, w3, and w4 incremented from 0 to 10, 20, 3, and 1, respectively resulted in 90 assignments with Nb = 0 and Nu = 148. Unique chemical shift assignments for all backbone and some sidechain sites were obtained for residues 7–20, 22–45, and 49–61 of α-spectrin SH3, as shown in Table S2. These assignments are in good agreement with assignments reported by Pauli et al. (2001), with the root-mean-squared deviation (rmsd) of 15N shifts being 0.93 ppm (after adding 0.13 ppm to values in Table S2 to correct for possible differences in referencing) and the rmsd of 13Cα and 13CO shifts being 0.36 and 0.44 ppm, respectively.
Representative 2D planes from the 3D spectra acquired at 9.4 T are shown in Fig. 3. Multidimensional signal lists from these spectra are shown in Table S3. Only 46, 44, and 37 signals were identified in the 3D NCACX, NCOCX, and CONCA spectra, respectively, reflecting the poorer resolution in these spectra compared with their higher-field counterparts. Degeneracy values of 2 were used for signals that appeared to contain contributions from more than one residue. Residue-type assignments were aided by examination of a 2D 13C–13C spectrum (not shown). 100 MCASSIGN2 runs with Na = 106 and Ns = 20 and with w1, w2, w3, and w4 incremented from 0 to 10, 25, 4, and 1, respectively resulted in 98 assignments with Nb = 0 and Nu = 131–134. Unique chemical shift assignments for all backbone and some sidechain sites were obtained for residues 8–19, 22–40, 49–52, and 54–60, as shown in Table S4. These assignments agree well with the higher-field assignments in Table S2. Thus, the poorer resolution at the lower field does not prevent useful results from being obtained, although the number of unique assignments is reduced.
Fig. 3.
Same as Fig. 2, but showing data obtained at 9.4 T
Note that the connection tables for 3D spectra of α-spectrin SH3 (see Tables S1 and S3) assume that the assignment of multidimensional signals to individual residues is based on the 13Cα chemical shifts. Thus, a signal in the 3D NCOCX spectrum that contains 13C chemical shifts of residue q and the backbone amide 15N chemical shift of residue q + 1 is assigned to residue q; a signal in the 3D CONCA spectrum that contains the backbone 13CO chemical shift of residue q − 1, the backbone 15N chemical shift of residue q, and the 13Cα chemical shift of residue q is assigned to residue q; a signal in the 3D NCACX spectrum that contains 15N and 13C chemical shifts of residue q is also assigned to residue q. Consequently, when comparing signals in NCACX and NCOCX spectra, the residue index shift for backbone 15N chemical shifts is −1. When comparing signals in NCACX and CONCA spectra, the residue index shift for backbone 13CO chemical shifts is +1. When comparing signals in NCOCX and CONCA spectra, the residue index shifts for backbone 13CO and backbone 15N shifts are +1. All other residue index shifts are 0.
If assignments of multidimensional signals to individual residues were based on backbone 15N chemical shifts, then (for example) the residue index shift values would be 0 for backbone 15N chemical shifts and +1 for all 13C chemical shifts when signals in NCACX and NCOCX spectra were compared.
Microcrystalline protein G/B1
Uniformly 15N,13C-labeled protein G/B1 was produced essentially as described by Franks et al. (2005) After dialysis against 50 mM potassium phosphate buffer at a protein concentration of approximately 10 mg/ml, the protein solution was mixed into a three-fold excess of methyl-2,4-pentanediol/isopropyl alcohol (2:1 volume ratio of the alcohols) in three steps, with thorough vortexing after each step. The mixture was incubated at room temperature for 2 days to allow formation of microcrystals with a needle-like appearance. Microcrystals (5 mg) were pelleted by centrifugation at 2,000×g for 10 min and packed into a 3.2 mm Varian MAS rotor.
2D 13C–13C, NCACX, and NCOCX spectra of uniformly 15N,13C-labeled protein G/B1 were acquired at 9.4 T, using a 3.2 mm Varian MAS NMR probe and a Varian Infinity spectrometer console. The spectra are shown in Fig. 4. The 2D 13C–13C spectrum (Fig. 4c) was obtained with MAS at 20.00 kHz, using a finite-pulse radio-frequency-driven recoupling (fpRFDR) sequence (Bennett et al. 1998; Ishii 2001) with 15.0 μs 13C π pulses during the 2.4 ms exchange period and 110 kHz 1H decoupling fields with TPPM during t1 and t2. 2D NCACX and NCOCX spectra (Fig. 4a, b) were acquired with MAS at 9.00 kHz, using a 4.5 ms cross-polarization period with 20 kHz 15N and 29 kHz 13C rf fields for 15N–13C polarization transfer and a 2.8 ms fpRFDR period with 25.0 μs 13C π pulses for 13C–13C polarization transfer between the t1 and t2 periods. During 15N–13C cross-polarization, the 13C rf carrier frequency was set to the middle of the 13Cα or the 13CO chemical shift range (for NCACX or NCOCX, respectively); a linear amplitude ramp of roughly 5% was applied to the 13C rf field. 275 complex t1 points were acquired for the 2D NCACX and NCOCX spectra, with a 72.8 μs increment. Total measurement times were 20 h, with a 2.0 s recycle delay.
Multidimensional signal lists extracted from the 2D NCACX and NCOCX spectra are shown in Table S5. Residue-type assignments were determined by comparisons of the 2D NCACX and NCOCX spectra with the 2D 13C–13C spectrum in Fig. 4c, taking into account the known 13C chemical shift ranges of each residue type. Signals from Thr, Gly, Ala, and Val had unique residue types. Signals from Ile6 and Met1 were assigned site-specifically in the signal lists because the protein G/B1 sequence contains only one Ile and one Met residue. All other signals had ambiguous residue-type assignments. Note that signals in Table S5 contain up to four NMR frequencies, even though the spectra were two-dimensional. This is because it was possible to associate up to three 13C shifts with each 15N shift, by examining one-dimensional slices parallel to the 13C axis. Sidechain signals beyond Cβ were not observed for most residues in the 2D NCACX and NCOCX spectra. Therefore, these signals are not included in Table S5 or in the final assignments in Table S6.
100 MCASSIGN2 runs were performed with Na = 106 and Ns = 30 and with w1, w2, w3, and w4 incremented from initial values of 0 to final values of 20, 40, 7, and 1, respectively. These runs resulted in 100 assignments with Nb = 0 and Nu = 96–100. Unique chemical shift assignments were obtained for backbone sites of residues 1, 6–9, 13–31, 38–41, 44, 49, and 52–56, as shown in Table S6. Compared with chemical shift assignments reported by Franks et al. (2005), rmsd values for 15N, 13Cα, and 13CO shifts are 1.08, 0.41, and 0.37 ppm, respectively (after subtracting 0.98 ppm from the 15N shifts in Table S6 to correct for an apparent difference in 15N shift referencing). Imperfect agreement is attributable in part to the relatively low resolution and signal-to-noise of the spectra in Fig. 4, but our protein G/B1 crystals are also different from the crystals studied by Franks et al. Schmidt et al. have demonstrated that crystal polymorphism leads to variations in chemical shifts for protein G/B1 (Schmidt et al. 2007).
We note that Moseley et al. have recently described automated solid state NMR assignments for microcrystalline protein G/B1 (Moseley et al. 2010), based on an adaptation of the AutoAssign program (Moseley et al. 2001).
HET-s218–289 fibrils
A single 3D 15N–13C–13C spectrum was acquired at 17.6 T, using the same uniformly 15N,13C-labeled HET-s218–289 fibril sample as in our earlier work (Tycko and Hu 2010), a 1.8 mm MAS NMR probe from the group of Dr. Ago Samoson (NMR Institute, Tallinn University of Technology, Estonia), and a Varian Infinity spectrometer console. The MAS frequency was 17.00 kHz. After 1H–15N cross-polarization and a t1 evolution period, non-selective 15N–13C cross-polarization was performed with 15N and 13C rf fields of 41 and 58 kHz, respectively, with a ±5% amplitude ramp on the 13C rf field. The 15N–13C cross-polarization time was 5.0 ms, and the 13C carrier frequency was 111.9 ppm. With these conditions, the net efficiencies for creation of 13CO and 13Cα spin polarization were roughly 22 and 29% relative to a simple 1H–13C cross-polarization experiment, as determined from signal areas. After the t2 evolution period, a 2.824 ms fpRFDR mixing period was used for 13C–13C polarization transfer before the t3 period, with 20 μs 13C π pulses and 85.3 ppm carrier frequency. 1H decoupling fields were 110 kHz, with TPPM during t1, t2, and t3. Maximum values of t1, t2, and t3 were 8.845, 2.594, and 8.163 ms, respectively, with increments of 327.6, 27.3, and 27.3 μs. The total experiment time was 96 h, with a 1.0 s recycle delay.
The 3D 15N–13C–13C spectrum was split into separate 3D NCACX and NCOCX spectra, based on the 13C chemical shifts in t2 (which determine whether one-bond 15N–13C polarization transfer occurs within residue q or from residue q to residue q − 1). Representative 2D planes from the 3D spectra are shown in Fig. 5. MCASSIGN2 input files, contain 51 NCACX signals and 49 NCOCX signals, were derived from the 3D spectra (see Table S7). 100 independent runs with Na = 106 and Ns = 20 and with w1, w2, w3, and w4 incremented from 0 to 10, 20, 3, and 1, respectively, resulted in 100 assignments with Nb = 0 and Nu = 95–99. In the 74 highest-scoring runs, unique chemical shift assignments were obtained for nearly all backbone and Cβ sites of residues 226–248 and 259–282 (see Table S8). These shifts are in good agreement with shifts for HET-s218–289 determined originally by Siemer et al. through manual analysis of multiple 2D and 3D spectra (Siemer et al. 2006b), and with our previous results from analysis of 2D NCACX and NCOCX spectra with the MCASSIGN1 program (Tycko and Hu 2010). Sidechain signals beyond Cb were not observed for most residues in the 3D NCACX and NCOCX spectra. Therefore, these signals are not included in Tables S7 and S8.
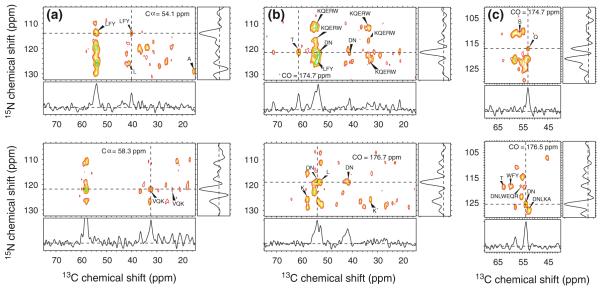
Fig. 5.
Representative 2D planes from the 3D 15N–13C–13C solid state NMR spectra of uniformly 15N,13C-labeled HET-s218–289 fibrils, obtained at 17.6 T. a Two planes from the NCACX region of the 3D spectrum, taken at the indicated chemical shifts in the second dimension. b Two planes from the NCOCX region of the 3D spectrum, taken at the indicated chemical shifts in the second dimension. 1D slices, taken at the dashed lines in each 2D plane, illustrate the signal-to-noise ratios and linewidths. Residue-type assignments for selected crosspeaks are indicated
As originally demonstrated by Meier and coworkers (Siemer et al. 2006a, b; Wasmer et al. 2008), fibrillar HET-s218–289 contains dynamically disordered segments at the N- and C-termini and between the two immobilized segments that form the β-helical core structure. Siemer et al. obtained assignments for residues 226–248 and 262–282 in the immobilized segments (Siemer et al. 2006b); in our previous work, we obtained unambiguous assignments for residues 226–248 and 260–283 (Tycko and Hu 2010). Van Melckebeke et al. subsequently extended the assignments to residues 222–249 and 260–287 by analyzing relatively weak signals from partially mobile residues (Van Melckebeke et al. 2010). Variations in the lengths of the assigned segments are attributable to variations in signal-to-noise in the solid state NMR spectra and possibly to variations in sample temperatures and hydration levels. Residues 226–244 and 260–280 appear most highly ordered in the molecular structure determined by Meier and coworkers (PDB file 2KJ3).
Discussion
Results for the model systems presented above demonstrate the ability of the generalized MCSA algorithm to handle a variety of 2D and 3D NMR data and to find unique assignments for most signals when the data are of moderate quality or better. In applications to data sets with relatively low spectral resolution or signal-to-noise, the MCAS-SIGN2 program typically finds multiple sets of chemical shift assignments that are consistent with information in the input files (i.e., multiple assignments with no bad connections and few unassigned signals). Examination of the output from MCASSIGN2 may then help the investigator design additional experiments (e.g., additional NMR measurements or additional samples with different isotopic labeling patterns) that distinguish among the possible assignments. It may also be productive to re-examine the original NMR spectra, to determine whether residue-type assignments in signal lists can be further restricted or whether the uncertainties in chemical shifts can be reduced. If information in the input files accurately represents the NMR data and no additional experiments are possible, the existence of multiple sets of chemical shift assignments reflects inherent limitations of the NMR measurements. Even when unique assignments do not exist for the entire biopolymer sequence, certain segments may have unique assignments, certain segments may always have non-null assignments, and certain segments may always have null assignments. Identification of segments that definitely contribute or definitely do not contribute to the NMR data can have useful structural and dynamical implications (Helmus et al. 2008, 2010; Siemer et al. 2006a, b; Tycko et al. 2010; Wasmer et al. 2008).
Obviously, the generalized MCSA algorithm does not fully automate the chemical shift assignment process. Measurement of crosspeak positions and chemical shift uncertainties and determination of possible residue-type assignments are still done manually. We believe that manual approaches to these phases of the assignment process are best, particularly for solid state NMR data, because there are large variations in the quality of the data and in the nature of spectroscopic clues that can be used to determine residue-type assignments. It is not straightforward to develop a general algorithm that can handle these variations as well as does the human mind. On the other hand, once crosspeaks have been measured and possible residue-type assignments have been determined, exhaustive exploration of the possible site-specific assignments is best done by a computer.
Since the MCASSIGN2 program assumes that all information in the input files is equally correct, one must be careful not to include speculative information in the input files. For example, residue-type assignment lists should not be unjustifiably restrictive; sidechain chemical shifts that are not unambiguously associated with backbone shifts should not be included; uncertainties in chemical shift values should not be underestimated (or grossly overestimated).
Important advantages of the MCSA algorithm include the facts that the algorithm is well-defined and that all assumptions regarding the relevant properties of the NMR data are contained explicitly in the input files. Therefore, results obtained by one laboratory can be evaluated and reproduced by investigators in another laboratory in ways that are impossible when purely manual assignment procedures (which generally are not documented or described in detail) are used to make assignments. Even within one laboratory, reproducibility of the assignment process can be important.
Although the generalized MCSA algorithm was developed for applications in solid state NMR of uniformly 15N,13C-labeled proteins with MAS, the same algorithm has broader applicability. In principle, it may be applied to data from nucleic acids or other heteropolymers and to data from samples with other isotopic labeling patterns, provided the data place constraints on sequential assignments in a manner that can be described by a connection table. The generalized MCSA algorithm is also applicable in solid state NMR studies of oriented systems without MAS if multidimensional techniques that correlate anisotropic chemical shifts of sequential pairs of residues are employed (Ishii and Tycko 2000; Sinha et al. 2007). Finally, the same algorithm may be useful in solution NMR studies of proteins, nucleic acids, and other heteropolymers.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases within the National Institutes of Health. Computational facilities of the Helix Systems at the National Institutes of Health were used in this work.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10858-011-9517-1) contains supplementary material, which is available to authorized users.
References
- Bailey-Kellogg C, Widge A, Kelley JJ, Berardi MJ, Bushweller JH, Donald BR. The NOESY jigsaw: automated protein secondary structure and main-chain assignment from sparse, unassigned NMR data. J Comput Biol. 2000;7:537–558. doi: 10.1089/106652700750050934. [DOI] [PubMed] [Google Scholar]
- Bartels C, Guntert P, Billeter M, Wuthrich K. Garant: a general algorithm for resonance assignment of multidimensional nuclear magnetic resonance spectra. J Comput Chem. 1997;18:139–149. [Google Scholar]
- Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- Bennett AE, Rienstra CM, Griffiths JM, Zhen WG, Lansbury PT, Griffin RG. Homonuclear radio frequency-driven recoupling in rotating solids. J Chem Phys. 1998;108:9463–9479. [Google Scholar]
- Buchler NEG, Zuiderweg ERP, Wang H, Goldstein RA. Protein heteronuclear NMR assignments using mean-field simulated annealing. J Magn Reson. 1997;125:34–42. doi: 10.1006/jmre.1997.1106. [DOI] [PubMed] [Google Scholar]
- Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG. Magic-angle spinning NMR analysis of β2-microglobulin amyloid fibrils in two distinct morphologies. J Am Chem Soc. 2010;132:10414–10423. doi: 10.1021/ja102775u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe: a multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM. Magic-angle spinning solid state NMR spectroscopy of the β1 immunoglobulin binding domain of protein G (GB1): 15N and 13 chemical shift assignments and conformational analysis. J Am Chem Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP. Molecular conformation and dynamics of the Y145Stop variant of human prion protein. Proc Natl Acad Sci USA. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP. Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid state nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2010;132:2393–2403. doi: 10.1021/ja909827v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchens TK, Lukin JA, Zhan YP, McCallum SA, Rule GS. Monte: an automated Monte Carlo based approach to nuclear magnetic resonance assignment of proteins. J Biomol NMR. 2003;25:1–9. doi: 10.1023/a:1021975923026. [DOI] [PubMed] [Google Scholar]
- Hyberts SG, Wagner G. IBIS: a tool for automated sequential assignment of protein spectra from triple resonance experiments. J Biomol NMR. 2003;26:335–344. doi: 10.1023/a:1024078926886. [DOI] [PubMed] [Google Scholar]
- Igumenova TI, Wand AJ, McDermott AE. Assignment of the backbone resonances for microcrystalline ubiquitin. J Am Chem Soc. 2004;126:5323–5331. doi: 10.1021/ja030546w. [DOI] [PubMed] [Google Scholar]
- Ishii Y. 13C–13C dipolar recoupling under very fast magic angle spinning in solid state nuclear magnetic resonance: applications to distance measurements, spectral assignments, and high-throughput secondary-structure determination. J Chem Phys. 2001;114:8473–8483. [Google Scholar]
- Ishii Y, Tycko R. Multidimensional heteronuclear correlation spectroscopy of a uniformly 15N- and 13C-labeled peptide crystal: toward spectral resolution, assignment, and structure determination of oriented molecules in solid state NMR. J Am Chem Soc. 2000;122:1443–1455. [Google Scholar]
- Lemak A, Steren CA, Arrowsmith CH, Llinas M. Sequence specific resonance assignment via multicanonical Monte Carlo search using an Abacus approach. J Biomol NMR. 2008;41:29–41. doi: 10.1007/s10858-008-9238-2. [DOI] [PubMed] [Google Scholar]
- Leutner M, Gschwind RM, Liermann J, Schwarz C, Gemmecker G, Kessler H. Automated backbone assignment of labeled proteins using the threshold accepting algorithm. J Biomol NMR. 1998;11:31–43. doi: 10.1023/a:1008298226961. [DOI] [PubMed] [Google Scholar]
- Li KB, Sanctuary BC. Automated resonance assignment of proteins using heteronuclear 3D NMR. 2. Side chain and sequence-specific assignment. J Chem Inf Comput Sci. 1997;37:467–477. doi: 10.1021/ci960372k. [DOI] [PubMed] [Google Scholar]
- Lukin JA, Gove AP, Talukdar SN, Ho C. Automated probabilistic method for assigning backbone resonances of 13C, 15N-labeled proteins. J Biomol NMR. 1997;9:151–166. doi: 10.1023/a:1018602220061. [DOI] [PubMed] [Google Scholar]
- McDermott A, Polenova T, Bockmann A, Zilm KW, Paulsen EK, Martin RW, Montelione GT. Partial NMR assignments for uniformly 13C, 15N-enriched BPTI in the solid state. J Biomol NMR. 2000;16:209–219. doi: 10.1023/a:1008391625633. [DOI] [PubMed] [Google Scholar]
- Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- Moseley HNB, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- Moseley HNB, Sperling LJ, Rienstra CM. Automated protein resonance assignments of magic angle spinning solid state NMR spectra of β1 immunoglobulin binding domain of protein G (GB1) J Biomol NMR. 2010;48:123–128. doi: 10.1007/s10858-010-9448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SJ, Schneider DM, Wand AJ. Implementation of the main chain directed assignment strategy: computer-assisted approach. Biophys J. 1991;59:1113–1122. doi: 10.1016/S0006-3495(91)82326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli J, van Rossum B, Forster H, de Groot HJM, Oschkinat H. Sample optimization and identification of signal patterns of amino acid side chains in 2D RFDR spectra of the α-spectrin SH3 domain. J Magn Reson. 2000;143:411–416. doi: 10.1006/jmre.2000.2029. [DOI] [PubMed] [Google Scholar]
- Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H. Backbone and side-chain 13C and 15N signal assignments of the α-spectrin SH3 domain by magic-angle spinning solid state NMR at 17.6 Tesla. ChemBioChem. 2001;2:272–281. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Petkova AT, Baldus M, Belenky M, Hong M, Griffin RG, Herzfeld J. Backbone and side chain assignment strategies for multiply labeled membrane peptides and proteins in the solid state. J Magn Reson. 2003;160:1–12. doi: 10.1016/s1090-7807(02)00137-4. [DOI] [PubMed] [Google Scholar]
- Schmidt HLF, Sperling LJ, Gao YG, Wylie BJ, Boettcher JM, Wilson SR, Rienstra CA. Crystal polymorphism of protein GB1 examined by solid state NMR spectroscopy and x-ray diffraction. J Phys Chem B. 2007;111:14362–14369. doi: 10.1021/jp075531p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, Riek R, Meier BH. Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc. 2006a;128:13224–13228. doi: 10.1021/ja063639x. [DOI] [PubMed] [Google Scholar]
- Siemer AB, Ritter C, Steinmetz MO, Ernst M, Riek R, Meier BH. 13C, 15N resonance assignment of parts of the HET-s prion protein in its amyloid form. J Biomol NMR. 2006b;34:75–87. doi: 10.1007/s10858-005-5582-7. [DOI] [PubMed] [Google Scholar]
- Sinha N, Grant CV, Park SH, Brown JM, Opella SJ. Triple resonance experiments for aligned sample solid state NMR of 13C and 15N labeled proteins. J Magn Reson. 2007;186:51–64. doi: 10.1016/j.jmr.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- Tycko R, Hu KN. A Monte Carlo/simulated annealing algorithm for sequential resonance assignment in solid state NMR of uniformly labeled proteins with magic-angle spinning. J Magn Reson. 2010;205:304–314. doi: 10.1016/j.jmr.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R, Savtchenko R, Ostapchenko VG, Makarava N, Baskakov IV. The α-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel β-sheet structure in PrP fibrils: evidence from solid state nuclear magnetic resonance. Biochemistry. 2010;49:9488–9497. doi: 10.1021/bi1013134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melckebeke H, Wasmer C, Lange A, Eiso AB, Loquet A, Böckmann A, Meier BH. Atomic-resolution three-dimensional structure of HET-s(218–289) amyloid fibrils by solid state NMR spectroscopy. J Am Chem Soc. 2010;132:13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a β-solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.