Summary
Members of sirtuin family regulate multiple critical biological processes, yet their role in carcinogenesis remains controversial. To investigate the physiological functions of SIRT2 in development and tumorigenesis, we disrupted Sirt2 in mice. We demonstrated that SIRT2 regulates the anaphase-promoting complex/cyclosome activity through deacetylation of its co-activators, APCCDH1 and CDC20. SIRT2 deficiency caused increased levels of mitotic regulators, including Aurora-A and -B that direct centrosome amplification, aneuploidy, and mitotic cell death. Sirt2-deficient mice develop gender-specific tumorigenesis, with females primarily developing mammary tumors, and males developing more hepatocellular carcinoma (HCC). Human breast cancers and HCC samples exhibited reduced SIRT2 levels compared with normal tissues. These data demonstrate that SIRT2 is a tumor suppressor through its role in regulating mitosis and genome integrity.
Keywords: APC/C-CDH1, APC/C-CDC20, Aurora-A, mitosis, centrosome
INTRODUCTION
In yeast, silence information regulator 2 (Sir2), a histone deacetylase, acts as a chromatin silencer to regulate gene expression, DNA recombination, genomic stability, and aging (Guarente and Kenyon, 2000). In mammals, sirtuins constitute a gene family of 7 (SIRT1-7) NAD+-dependent type III histone and protein deacetylases that share homology with Sir2 (Finkel et al., 2009; Li and Kazgan, 2011; Saunders and Verdin, 2007). It has been shown that SIRT1, 6, and 7 primarily localize to the nucleus, while SIRT3, 4, and 5 are present in mitochondria (Haigis and Sinclair, 2010; Li and Kazgan, 2011; Saunders and Verdin, 2007). SIRT2 is predominantly localized in the cytoplasm where it co-localizes with and deacetylates microtubules (North et al., 2003). During mitosis, SIRT2 is localized to the chromosome and serves as a histone deacetylase with a preference for histone H4 lysine 16 (H4K16Ac), and may regulate chromosomal condensation during mitosis (Inoue et al., 2007; Vaquero et al., 2006). SIRT2 is also associated with mitotic structures, including the centrosome, mitotic spindle, and midbody during mitosis, presumably to ensure normal cell division (North and Verdin, 2007).
Progression of mitosis is also regulated by the anaphase-promoting complex/cyclosome (APC/C), a multi-subunit member of the RING finger family of ubiquitin ligases. APC/C is composed of many different subunits, including APC1–8, APC9–11, and CDC26 (Peters, 2006; Pines, 2009). APC/C recognizes its substrates through two adaptor proteins, CDH1 and CDC20, which serve as co-activators for APC/C through binding to substrates at different phases of mitosis (Peters, 2006; Pines, 2009). In mammalian cells, CDC20 activates APC/C in early mitosis until anaphase, while CDH1 acts in late mitosis and during G1 phase (Pines, 2006). APC/C mediates ubiquitination of many protein substrates that have distinct functions during mitosis, including Aurora-A and -B, cyclins-A and -B, survivin, Plk1, Nek2A, and securin (Li and Zhang, 2009). Although both SIRT2 and APC/C play important functions during mitosis, the relationship between these proteins is unknown.
Currently, all sirtuins, except for SIRT2, have been knocked out in mice by gene targeting (Ahn et al., 2008; Cheng et al., 2003; Haigis et al., 2006; Jacobs et al., 2008; Lombard et al., 2007; McBurney et al., 2003; Mostoslavsky et al., 2006; Nakagawa et al., 2009; Vakhrusheva et al., 2008; Wang et al., 2008a). Studies of these mutant mice have provided useful information regarding sirtuin function in many important processes, including cell fate determination, DNA damage repair, neuronal protection, adaptation to calorie restriction, organ metabolism and function, age-related diseases, and tumorigenesis, although much of the information has come from studies of SIRT1 (Ahn et al., 2008; Deng, 2009; Finkel et al., 2009; Jacobs et al., 2008; Kim et al., 2010; Saunders and Verdin, 2007; Wang et al., 2008a; Wang et al., 2008b). To study the physiological function of SIRT2, we have disrupted the Sirt2 gene in mice and reported our findings below.
RESULTS
Impaired Mitotic Function due to SIRT2 Deficiency
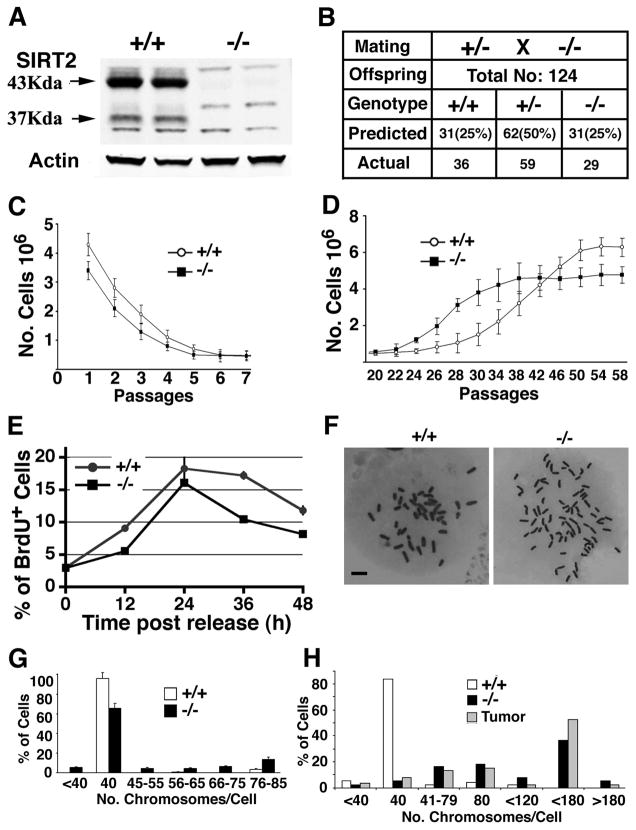
The Sirt2 gene was disrupted by deleting exons 5 through 8, which encodes the entire catalytic domain (Figure S1A–1C). Western blot analysis revealed that there was no truncated protein in embryos homozygous for the mutation (Figure 1A), suggesting the creation of a candidate null mutation of SIRT2. Despite expression of SIRT2 in multiple tissues (Figure S1D), Sirt2−/− mice presented at weaning in a Mendelian ratio (Figure 1B) and developed normally (Figures S1E and S1F). Histopathological analysis of multiple organs also did not reveal obvious abnormalities (data not shown). These data indicate that SIRT2 is not essential for embryonic viability and postnatal development.
Figure 1. SIRT2 Deficiency Reduces Proliferation of MEFs but Does Not Affect Animal Survival.
(A) Western blot analysis of proteins extracted from adult Sirt2+/+ (+/+) and Sirt2−/− (−/−) mice. WT mice have two forms of SIRT2, 43 KD and 37KD, and both are absent in mutant mice.
(B) Number of predicted and actual offspring from interbreeding of heterozygous mice.
(C–D) Growth curve of Sirt2+/+ and Sirt2−/− MEFs from passages 1 to 7 (C), and 20–58 (D) (MEFs cells from at least 5 pairs of embryos were analyzed in this experiment).
(E) BrdU incorporation of MEFs released from serum starvation.
(F–H) Chromosome spread (F) and summary of chromosome number from wild-type and SIRT2−/− MEFs at P2 (G), and at P35 (H) (three pairs at each passage). The chromosome number of three primary tumors from mammary gland (2) and liver (1) was also included in (H). Data are presented as average ± SD. Scale bars = 10 μm in (F)
(See also Figure S1)
To provide a comprehensive analysis of potential abnormalities associated with SIRT2 deficiency, we assessed the growth properties of mouse embryonic fibroblasts (MEFs). The SIRT2 mutant MEFs proliferated significantly more slowly than did wild-type (WT) controls, and stopped growing at passage P5, one passage earlier than WT MEFs (Figure 1C). Our analysis of P2 MEFs revealed that the reduced proliferation of mutant MEFs was associated with decreased BrdU incorporation into DNA (Figure 1E). Next, we performed chromosome spreads from 3 pairs of mutant and WT MEFs. Our data showed that about 35% of mutant cells were aneuploid, with chromosome numbers ranging from more than 40 to about 80 per cell, while less than 5% of wild-type cells were aneuploid (Figure 1F and 1G). These observations suggest that loss of SIRT2 resulted in reduced cell proliferation that is associated with increased genetic instability.
To study SIRT2 function further, we immortalized SIRT2 mutant and WT MEFs, using a 3T3 protocol. SIRT2 mutant cells escaped senescence at P22 and increased their proliferation and reached maximum growth at P38, while WT cells escaped senescence and reached maximum growth approximately 4 passages later, although they gained faster growth starting from P45 (Figure 1D). Meanwhile, our study revealed that SIRT2−/− MEFs exhibited loss of contact inhibition, as reflected by the formation of foci in cultured monolayer cells (Figure S1H) and increased soft agar colony formation (Figure S1I), suggesting that some of the mutant cells may become malignantly transformed. To investigate this hypothesis, the immortalized MEFs were inoculated into nude mice. All mice that received an allograft of SIRT2 mutant cells developed tumors, while none were observed in those inoculated with WT MEFs (Figure S1J). More profound genetic instability was found in immortalized SIRT2−/− MEFs and tumors (Figure 1H), suggesting that tumorigenesis of SIRT2−/− MEFs could result from increased genetic instability.
SIRT2 Deficiency Causes Tumorigenesis
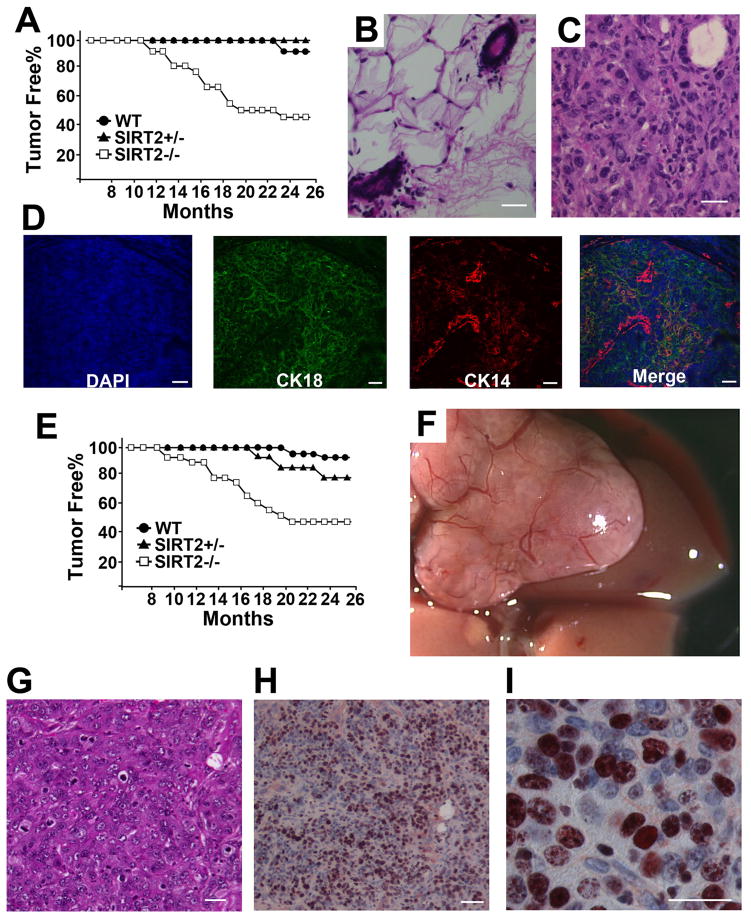
The association of SIRT2 loss with spontaneous malignant transformation of mutant MEFs suggests that SIRT2 might serve as a tumor suppressor. To provide in vivo evidence, a cohort of mice was monitored for possible tumor formation, and it was observed that SIRT2 mutant mice exhibited a gender-specific spectrum of tumorigenesis. SIRT2 mutant females started to develop tumors, primarily in the mammary glands, at about 10 months of age, with cancer incidence reaching about 60% by 24 months (Figure 2A, and Table S1). The mammary tumors were poorly differentiated, with obvious nuclear polymorphisms (Figure 2C). Notably, many cells were positive for both basal and luminal markers, suggesting they share a common origin (Figure 2D). Hyperplasia in the mutant glands was also detected prior to mammary tumor development (Figures S2A and S2B).
Figure 2. Absence of SIRT2 Results in Tumor Formation in Multiple Organs.
(A) Kaplan–Meier survival curve showing cancer incidence in SIRT2−/− (n = 26), SIRT2+/− (n = 13), and WT (n = 22) female mice.
(B–C) Histological sections of a mammary gland from wild-type mice (B) and an adenocarcinoma (C) from SIRT2 mutant mice. Scale bars = 10 μm
(D) Immunofluorescent staining of an adenocarcinoma using basal (CK14) and luminal (K18) markers. Scale bars = 10 μm
(E) Kaplan–Meier survival curve showing cancer incidence in SIRT2−/− (n = 19), SIRT2+/− (n = 10), and WT (n = 20) male mice.
(F,G) Whole-mount view (F) and histological section (G) of a HCC from liver of SIRT2 mutant mice. Scale bars = 10 μm in (G)
(H,I) Proliferation assay using Ki67 staining; 20 × (H) and 63 × (I) magnifications. Scale bars = 10 μm.
SIRT2 mutant male mice also developed cancers in multiple organs, starting from 8 months of age, with the cancer incidence reaching about 60% by 20 months of age (Figures 2E, and Table S1). Of 19 SIRT2−/− mice studied, five (26%) developed from the liver (Fig. 2F), two (11%) from the lung and one each from pancreas, stomach, duodenum and prostate, respectively (Figures S2D–S2G). Our analysis of liver cancers revealed that they are hepatocellular carcinoma (HCC) with extensive cellular proliferation (Figures 2G–2I).
SIRT2 Deficiency Caused Centrosome Amplification Associated with Increased Expression of Aurora-A
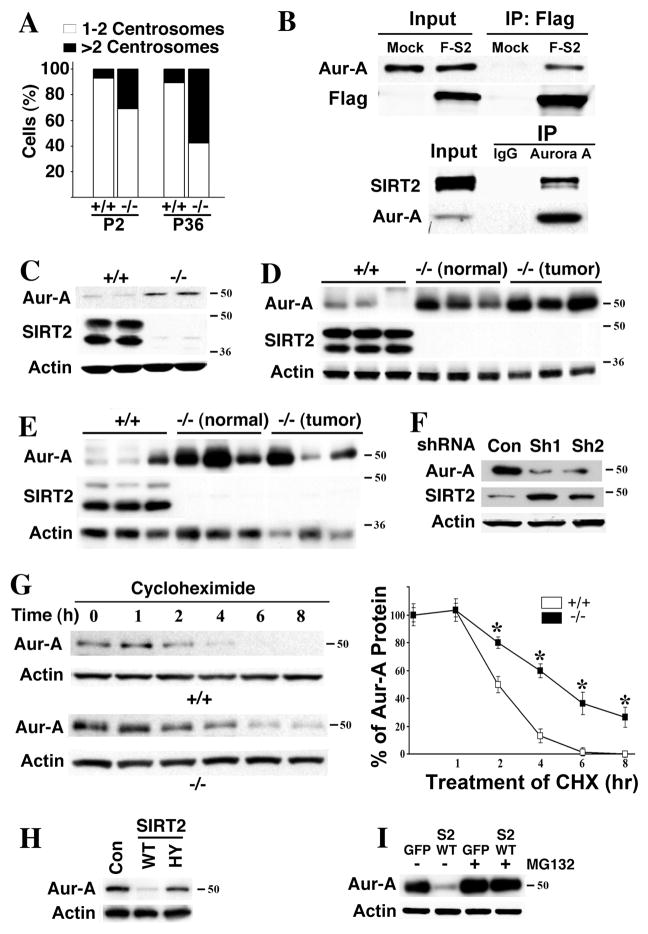
The presence of a substantial percentage of aneuploid cells suggests that the fidelity of chromosome segregation is compromised. It was shown that SIRT2 is localized on the centrosome (North and Verdin, 2007), and our data confirmed this (Figure S3A). Because aberrant replication of centrosomes can lead to aneuploidy (Wang et al., 2006; Xu et al., 1999), we next investigated whether loss of SIRT2 affects centrosome duplication. Using an antibody to γ-tubulin, we compared the centrosome number of Sirt2−/− and wild-type MEFs at P2. Our data showed that 31% (55/181) of Sirt2−/− cells contained 3 or more centrosomes, while only 7% (15/212) of control MEFs contained more than 2 centrosomes (Figure 3A–Figure S3B). Profound centrosome amplification was also observed in immortalized Sirt2−/− MEFs (Figure 3A). Similar data was obtained by using an antibody to pericentrin (data not shown).
Figure 3. Loss of SIRT2 Causes Centrosome Amplification That Is Associated with Increased Expression of Aurora-A.
(A) SIRT2 deficiency results in centrosome amplification in MEFs. Centrosome number of 181 and 175 SIRT2−/− cells were counted at passage (P) 2 and P36, while 212 and 120 wild-type cells were counted at P2 and P36, respectively. Percentage of cells with normal number (1–2/cell) and abnormal number (>2/cells) of centrosomes were shown.
(B) Interaction between SIRT2 and Aurora-A (Aur-A). 293T cells were transiently transfected with flag-SIRT2 (F-S2) expression vector or mock vector (pCMV5), lysed with IP buffer, and lysates were immunoprecipitated with either anti-Flag conjugated beads (upper panel) or antibody to endogenous Aurora-A (lower panel). Blots were immunoblotted with either anti-SIRT2, Flag, or Aurora-A antibodies. 5% of input was used in all panels.
(C–E) SIRT2 deficiency results in an increase of Aurora-A protein in MEFs (C), mammary tissues (“normal”, D) and mammary tumors (D), and liver and liver tumors (E).
(F) shRNA-mediated acute knockdown of SIRT2 increases Aurora-A in HepG2 cells. Two lentiviral-based shRNA constructs against different region of SIRT2 (Sh1 and Sh2) and control (pLKO.1-Luc) are used.
(G) Aurora-A protein is more stable in SIRT2−/− MEFs than in SIRT2 WT MEFs at G1 phase. Cells were kept in serum free medium for 72 hours, re-plated in 15% FBS DMEM for 4 hours, and then treated with 10 μg/ml of cycloheximide (CHX) at the indicated time points before subjected to Western Blot. The experiments were repeated three times and band intensities were measured using Image Lab Version 3.0. Statistic analysis was provided on the right panel. * represents student ρ value <0.005.
(H–I) Ectopic overexpression of SIRT2-WT, but not SIRT2-HY reduces Aurora-A in SIRT2−/− MEFs (Con: pcDNA3.1 vector only) (H), but this effect is blocked in the presence of 10 μM of MG132 for 24 hours (I).
(See also Figure S3)
Aurora-A plays an essential role in centrosome replication (Cowley et al., 2009). We previously found that overexpression of Aurora-A causes centrosome amplification and mammary tumor formation in mice (Wang et al., 2006). These data prompted us to investigate the potential relationship between SIRT2 and Aurora-A. We observed that SIRT2 and Aurora-A interact with each other by reciprocal immunoprecipitation (Figure 3B), and they co-localize to the centrosome (Figure S3C). We had also detected significantly higher levels of Aurora-A in SIRT2 mutant MEFs (Figure 3C), mammary tissues (Figure 3D), and liver (Figure 3E), compared with controls. High levels of Aurora-A were also detected in the mammary and liver tumors from SIRT2 mutant mice (Figures 3D and 3E).
SIRT2 Interacts with and Degrades Aurora-A, Although It Does Not Deacetylate Aurora-A
To assess whether increased expression of Aurora-A is a direct consequence of SIRT2 deficiency, we performed shRNA-mediated knockdown in HepG2 cells. We found that shRNA-mediated acute knockdown of SIRT2 increased Aurora-A protein (Figure 3F) but did not affect transcriptional levels of Aurora-A (data not shown), suggesting that the absence of SIRT2 stabilized Aurora-A at the post-transcriptional level. To verify this, we treated MEFs with cycloheximide to block new protein synthesis and then measured Aurora-A levels at either G1 phase or unsynchronized cell population. The level of Aurora-A was reduced to about half after 2 hours of treatment, and was almost completely diminished after 6 hours in wild-type cells (Figure 3G). In contrast, the level of Aurora-A was reduced to about half after 4 hours and about 30% protein was maintained up to 8 hours of treatment in SIRT2−/− cells (Figure 3G). Similar observation was also made in the unsynchronized cell populations (Figure S3D).
Because SIRT2 is a well-known deacetylase and it interacts with Aurora-A, it is conceivable that SIRT2 deacetylates Aurora-A, which contributes to Aurora-A’s degradation. However, we were not able to detect acetylation of Aurora-A (data not shown), suggesting that Aurora-A is not a direct target of SIRT2 deacetylase activity, despite their interaction with each other. Next, we investigated whether SIRT2 deacetylase activity is involved in regulating Aurora-A stability. We transfected SIRT2−/− MEFs with WT SIRT2 (SIRT2-WT) or a deacetylation mutant SIRT2 (SIRT2-HY) and measured Aurora-A levels in these cells. Overexpression of SIRT2-WT, but not SIRT2-HY, significantly reduced levels of Aurora-A (Figure 3H). This data suggests that although SIRT2 does not deacetylate Aurora-A, its deacetylase activity is involved in regulating Aurora-A stability, perhaps through deacetylating some other unidentified factors. We also found that degradation of Aurora-A caused by SIRT2 overexpression can be blocked by treatment with the proteosome inhibitor MG132 (Figure 3I), suggesting that Aurora-A may be ubiquitinated before its degradation.
SIRT2 Forms a Protein Complex with Anaphase-promoting Complex/Cyclosome (APC/C)
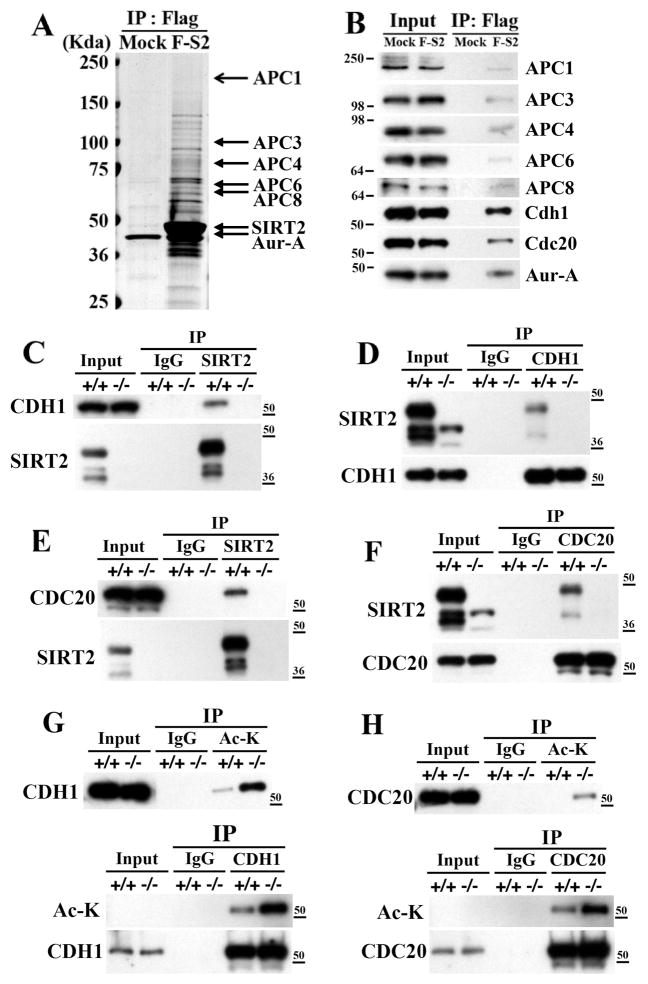
To investigate the underlying mechanism how SIRT2 deficiency affect Aurora-A stability, we performed a proteomic study to identify interaction proteins and potential targets of SIRT2 deacetylase activity. After expressing a Flag-SIRT2 construct in HeLa cells, followed by a pull-down using an antibody to Flag, we identified several distinct protein bands on the SDS-PAGE gel that were present in Flag-SIRT2 transfected, but not in the vector transfected cells (Figure 4A). Analysis of these bands using mass spectrometry identified many proteins, including several components of APC/C and Aurora-A (Figure 4A). The interaction of these proteins with Flag-SIRT2 was confirmed by immunoblots with individual antibodies after pull-down with Flag-SIRT2 (Figures 4A and 4B).
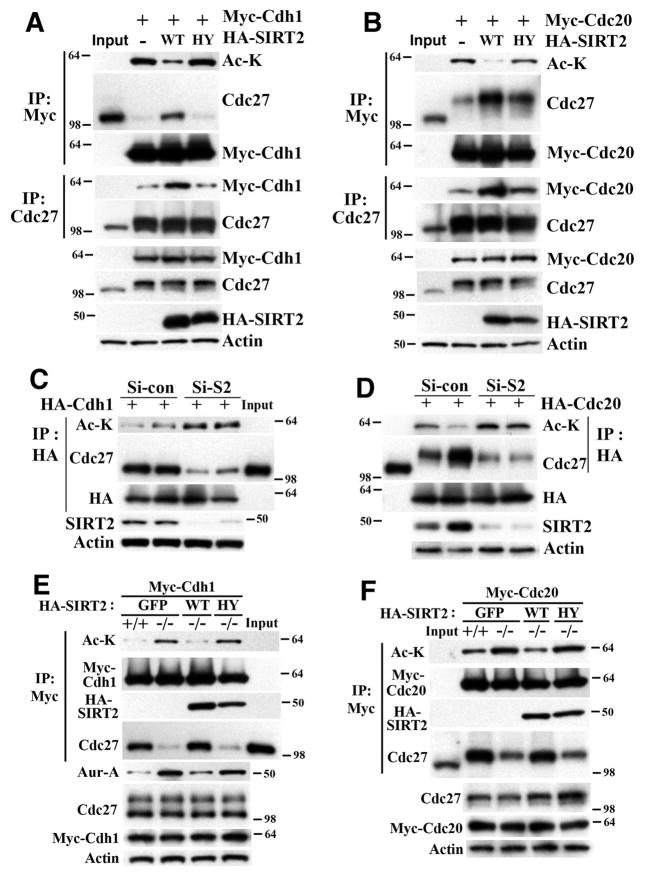
Figure 4. SIRT2 Interacts with APC/C and Deacetylates CDH1 and CDC20.
(A) Mass spectrometry analysis of Flag-SIRT2-interactingproteins after transfection of Flag-SIRT2 into HeLa cells. Lysates were immunoprecipitated with anti-Flag-agarose beads and eluted using Flag peptide, resolved by SDS-PAGE gel, and subjected to mass spectrometry analysis.
(B) Immunoblots of Flag-SIRT2 interacting proteins.
(C–F) Reciprocal co-immunoprecipitation of endogenous SIRT2 and CDH1 (C,D) or CDC20 (E,F) from immortalized SIRT2+/+ and SIRT2−/− MEF cells. After synchronization using serum starvation for 72 hr, cells were released into normal media for 16 hr and harvested for co-immunoprecipitation. The lysates were immunoprecipitated with anti-SIRT2, -CDH1, or CDC20 antibodies, respectively, and IPed samples were analyzed by immunoblotting with anti-SIRT2, -CDH1, or –CDC20, respectively.
(G,H) In vivo deacetylation analysis of endogenous CDH1 (G) and –CDC20 (H) from liver of SIRT2 WT and KO mice 48 hours after partial hepatectomy.
(See also Figure S4)
CDH1 and CDC20 are essential E3-ligases of the APC/C complex, which serve as co-activators for APC/C and have substrate specificity for different APC/C substrates, including Aurora-A (Peters, 2006; Pines, 2009). Given our earlier observation that SIRT2 overexpression mediated Aurora-A degradation is blocked by proteosome inhibitor MG132 (Figure 3I), we hypothesized that SIRT1 might modulate Aurora-A stability through interacting with CDH1 and CDC20. Therefore, we further studied interactions between SIRT2 with CDH1 and CDC20. Using reciprocal immunoprecipitation, we confirmed reciprocal interactions of endogenous SIRT2 with CDH1 (Figure 4C,D) and CDC20 (Figure 4E,F) in wild type MEFs, but not in SIRT2−/− MEFs (Figures 4C-F). Similar interactions between endogenous or ectopically overexpressed SIRT2 with CDH1 and CDC20 were also observed in HeLa cells (data not shown).
SIRT2 Deacetylates CDH1 and CDC20 and Enhances Their Binding with APC/C
Next, we tested whether SIRT2 could deacetylate CDH1 and CDC20. We first obtained acetylated CDH1 and CDC20 from 293T cells (Figure S4A, lanes 1 and 4) and then performed an in vitro deacetylation assay by adding purified SIRT2-WT (Figure S4A, lanes 2 and 5) or SIRT2-HY (Figure S4A, lanes 3 and 6) proteins, which showed that SIRT2-WT, but not SIRT2-HY was able to deacetylate both proteins. A similar deacetylation pattern was observed when 293T cells were transfected with a SIRT2-WT construct, but not a SIRT2-HY construct (Figures S4B and S4C). Furthermore, the acetylation of CDH1 (Figure S4D) or CDC20 (Figure S4E) proteins was much higher in SIRT2−/− than WT MEFs. shRNA specific for CDH1 (Fig. S4D) or CDC20 (Figure S4E) reduced their levels of acetylation. Altogether, this data shows that SIRT2 is a potent deacetylase of CDH1 and CDC20. To provide validation at the organism level, we examined the acetylation status of endogenous CDH1 and CDC20 from normal mouse liver or mouse liver 48 hours after partial hepatectomy (hepatectomy is used to induce hepatocyte proliferation and liver regeneration). Our data indicated that in the absence of SIRT2, there was markedly increased acetylated CDH1 in both the normal liver and the liver 48 hours after partial hepatectomy (Figure 4G and data not shown). Our data indicated that CDC20 was not detected in the normal liver but strongly induced 48 hours after partial hepatectomy (data not shown), which is consistent to an observation that CDC20 is only present in the mitotic cells (Gieffers et al., 1999). Absence of SIRT2 significantly increased acetylated CDC20 in the hepatectomized lever compared with hepatectomiced liver of wild type mice (Figure 4H).
CDH1 and CDC20 activate APC/C by binding to CDC27 (Peters, 2006; Pines, 2009). Therefore, we determined whether deacetylation of CDH1 and CDC20 by SIRT2 could affect their interaction with CDC27. In HeLa cells, we found that expression of SIRT2-WT, but not SIRT2-HY, reduced the acetylated form of CDH1 (Figure 5A) and CDC20 (Figure 5B), and increased their interaction with CDC27 (Figures 5A and 5B). Conversely, siRNA-mediated knockdown of SIRT2 increased acetylation of CDH1 and CDC20 and decreased their interaction with CDC27 (Figures 5C and 5D). Furthermore, we showed that SIRT2−/− MEFs displayed hyperacetylated CDH1 (Figure 5E) and CDC20 (Figure 5F) that was accompanied by decreased interaction with CDC27, and these phenotypes could be reversed by re-expression of SIRT2-WT, but not SIRT2-HY (Figures 5E and 5F). Because the interaction of CDH1 and CDC20 with CDC27 plays an important role in activation of APC/C (Peters, 2006; Pines, 2009), these data suggest that SIRT2 deficiency impairs APC/C activity through causing hyperacetylation of CDH1 and CDC20, which fail to interact with CDC27.
Figure 5. SIRT2 Regulates the Function of CDH1 and CDC20 through Deacetylation.
(A) Overexpression of WT but not deacetylase mutant (HY) SIRT2 decreases CDH1 acetylation and increases its interaction with CDC27 in HeLa cells. Ac-K: pan-acetyl lysine antibody.
(B) Overexpression of WT but not deacetylase mutant (HY) SIRT2 decreases CDC20 acetylation and increases its interaction with phospho-CDC27 in HeLa cells.
(C–D) Knockdown of SIRT2 by siRNA increases acetylation of CDH1 (C) and CDC20 (D) and decreases interaction with CDC27 or with phospho-CDC27, respectively, in HeLa cells. Si-S2 (siRNA against Sirt2), Si-Con (scramble siRNA).
(E) SIRT2 deficiency increases acetylation of CDH1 and decreases its interaction with CDC27, which is accompanied by an increase in Aurora-A in immortalized Sirt2 +/+ and −/− MEF cells.
(F) SIRT2 deficiency increases acetylation of CDC20 and decreases its interaction with CDC27 in immortalized SIRT2 WT and KO MEF cells. In all above panels, 24 hours after transfection, cells were synchronized by double thymidine block (HeLa cells) or serum starvation for 48 hr (MEF cells), released into regular media for 12 hr or 18 hr, respectively, Input: lysates from non-transfected and unsynchronized cells (5% in relation to IP samples) were used as control to distinguish phospho-CDC27 (migration slower) and non-phospho-CDC27.
(See also Figure S5)
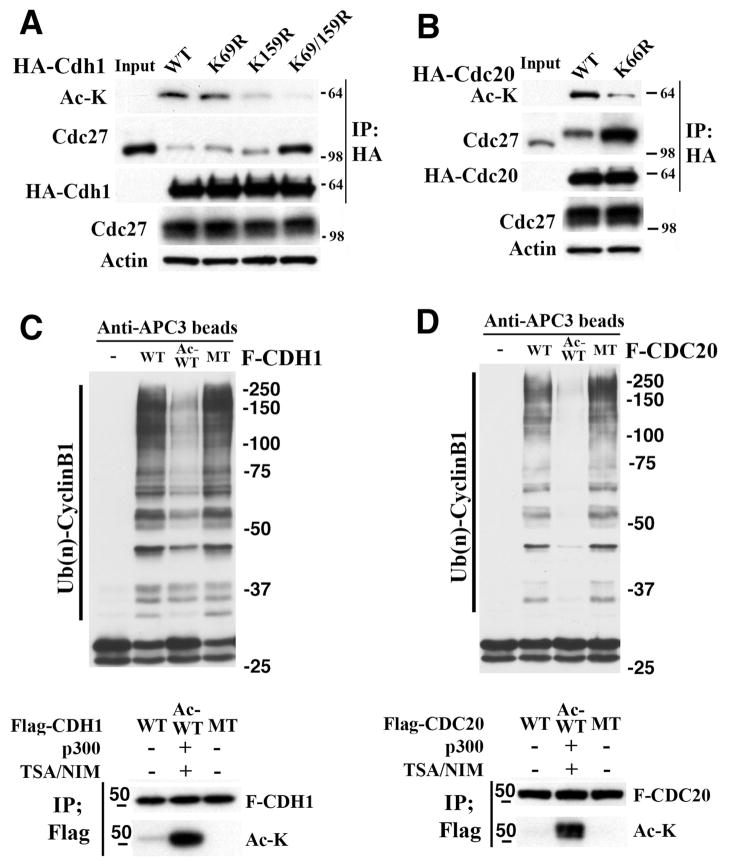
Next, we used mass spectrometry to identify amino acids in these proteins that may be deacetylated by SIRT2. We detected two potential lysine acetylation sites in CDH1 (i.e., K69 and K159) (Figures S5A and S5B) and one in CDC20 (K66) (Figure S5C). A single mutation of K69 or K159 to arginine (R) in CDH1 reduced acetylation levels, while the combined mutations of both sites diminished its acetylation (Figure 5G). On the other hand, mutation of K66 to R66 in CDC20 partially reduced its acetylation level (Figure 6A), suggesting that other unidentified site(s) could also be acetylated. We further demonstrated that mutations of the acetylation sites, K69R and K159R of CDH1 (Figure 6A), and K66R of CDC20 (Figure 6B), reduced the acetylation of these proteins and enhanced their interaction with CDC27, which is consistent with our earlier finding that the deacetylated form of these proteins interact with CDC27.
Figure 6. Analysis of acetylation sites in CDH1 and CDC20 and their effect on APC/C activity.
(A,B) Mutations of Cdh1 (K69R, K159R, and K69/159R) (A) and CDC20 (K66R) (B), which mimic the deacetylation of lysine residue, reduces their interaction with CDC27, as revealed by IP against HA-Cdh1 (A) and HA-Cdc20 (B) followed by Western blot against Cdc27.
(C,D) in vitro APC/C activity assay using Cyclin B1 ubiquitination as reporter in the presence or absence of acetylated or non-acetylated forms of CDH1 (C) and CDC20 (D) proteins purified from 293T cells and HeLa cells, respectively. Lane 1, reaction mix without adding CDH1 or CDC20 proteins. Lane 2, non-acetylated forms of Flag tagged wild type (WT) CDH1 or CDC20. Line 3, acetylated forms of Flag tagged wild type (Ac-WT) CDH1 or CDC20. Lane 4, Flag tagged CDH1-K69/159R or CDC20-K66R (MT). Lower panels show status of acetylation of CDH1 and CDC20.
Because mutation of K69R and K159R of CDH1 nearly completely reduces the acetylated form of CDH1 and markedly increases its interaction with CDC27 (Figure 6A), we hypothesized that the non-acetylated form of CDH1 should have much higher ability to activate APC/C compared with the acetylated form. To investigate this, we isolated the Flag-CDH1-WT (non-acetylated form), Flag-CDH1-WT (acetylated form), and Flag-CDH1-K69/159R (which cannot be acetylated) from 293 cells, and performed in vitro APC/C activity assay using Cyclin B1 ubiquitination as reporter. Our data indicated that the acetylated CDH1 has significantly lower APC/C activity than non-acetylated CDH1 and the CDH1-K69/159R, while no significant difference is observed between the non-acetylated CDH1 and the CDH1-K69/159R mutated CDH1 (Figure 6C).
SIRT2 Deficiency Impairs APC/C Activity, Leading to Increased Levels of Mitotic Regulators
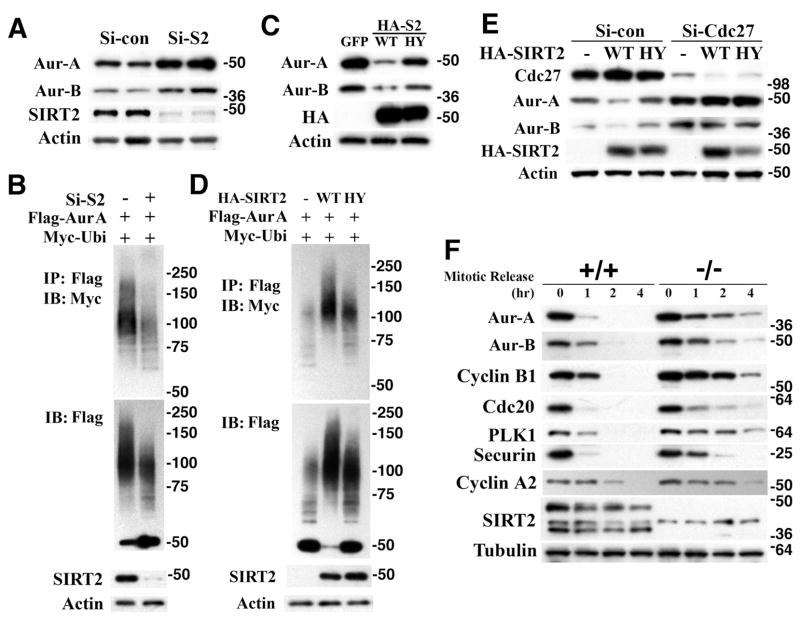
In light of this finding, we believe that the increased level of Aurora-A in SIRT2 mutant cells is due to impaired APC/C activity. To provide evidence for this notion, we examined whether SIRT2 could affect Aurora-A ubiquitination, since APC/C has E3 ubiquitin ligase activity. Our data revealed that knockdown of SIRT2 caused upregulation of Aurora-A that is associated with decreased ubiquitination (Figures 7A and 7B). Conversely, overexpression of SIRT2-WT, but not SIRT2-HY, decreased the protein level of Aurora-A through increased ubiquitination (Figures 7C and 7D). We also confirmed that the effect of SIRT2 on Aurora-A stability requires CDC27, since SIRT2 cannot reduce the protein level of Aurora-A in CDC27 knockdown HeLa cells (Figure 7E). These data provide strong evidence that degradation of Aurora-A by SIRT2 is mediated by APC/C. Since APC/C is responsible for degrading multiple proteins during mitosis, we hypothesized that SIRT2 deficiency might cause a broader alteration in protein abundance. Therefore, we performed further analysis and detected increased levels of multiple mitotic regulators, including Aurora-A, Aurora-B, Plk1, securin, and cyclin A2 in SIRT2−/− cells (Figure 7F). As a validation for these changes, we detected similar results for Aurora-B as compared with Aurora-A, i.e., siRNA against SIRT2 increased levels of Aurora-B (Figure 7A), while overexpression of SIRT2 decreased it (Figure 7C), and such changes were blocked when CDC27 was knocked down (Figure 7E). Consistent with the altered expression of multiple mitotic regulators, SIRT2−/− cells displayed abnormalities in mitosis besides the centrosome amplification, including cell death during mitosis (Figure S6B,D,E), failure to complete cytokinesis (Figure S6C), and arrest at metaphase (Figure S6D), some of which were not observed in control cells (Figures S6A and S6F).
Figure 7. SIRT2 Regulates Aurora-A Levels Mediated by APC/C.
(A) Knockdown of SIRT2 by siRNA against Sirt2 (Si-S2), but not scramble siRNA (Si-Con), in HeLa cells increases expression level of Aurora-A and Aurora-B (Aur-B) protein.
(B) Increased level of Aurora-A is correlated with reduced Aurora-A. After co-transfection with indicated plasmids and Si-S2) into HeLa cells for 24 hrs, cells were treated with 10 μM of MG132 for 4 hrs. Lysates were immunoprecipitated with Flag antibody conjugated-beads, and immunoblotted with indicated antibodies.
(C) Overexpression of HA-tagged SIRT2-WT, but not SIRT2-HY (HA-S2), reduces expression levels of Aurora-A and Aurora-B compared with GFP transfected HeLa cells.
(D) Reduces level of Aurora-A is associated with increased ubiquitination. After transfection with indicated plasmids for 24 hrs, cells were treated with 10 μM of MG132 for 4 hrs. Lysates were immunoprecipitated with Flag antibody conjugated-beads, and immunoblotted with indicated antibodies.
(E) Effect of SIRT2 on the protein level of Aurora-A and -B was blocked by CDC27 knockdown. Lysates were analyzed by Western blot 48 hours after co-transfection with indicated plasmids, siRNA against Cdc27 (Si-Cdc27) or scramble siRNA (Si-Con) into HeLa cells.
(F) Western blot analysis of protein lysates in primary SIRT2 +/+ and −/− MEFs (Passage 2) after mitotic release for indicated time points. Cells were synchronized by serum starvation for 72 hr, replaced with normal media containing nocodazole for 18 hr, and then mitotic cells collected were replated and harvested at indicated time points for Western blot analysis.
(See also Figure S6)
Expression of SIRT2 in Human Tumor Samples
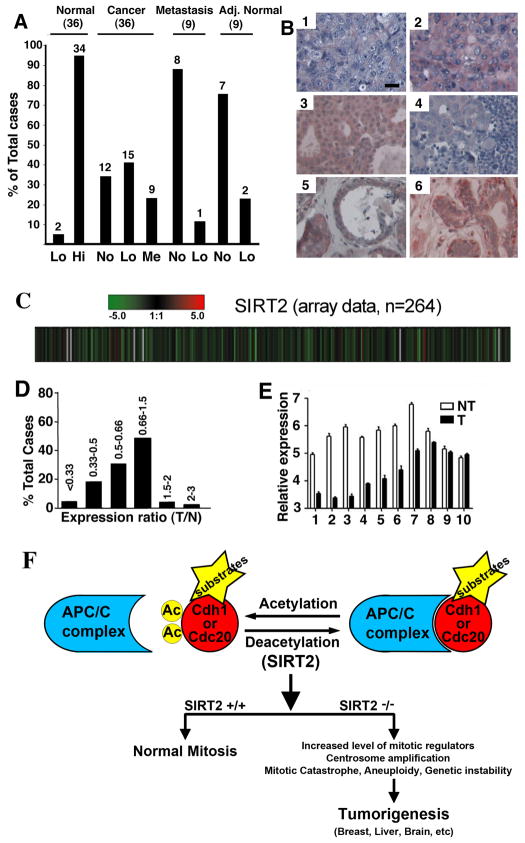
Our data so far indicate that SIRT2 acts as a tumor suppressor in mice. To investigate whether SIRT2 may have a similar function in humans, we performed a tissue array to compare SIRT2 protein levels between 36 pairs of breast cancers and cancer-adjacent normal breast tissues, as well as 18 metastatic cancers. There were significantly higher levels of SIRT2 in all normal breast tissues as compared with cancer tissues (Figures 8A-B). We also analyzed SIRT2 expression levels from microarray data containing 264 HCC samples and found that many HCCs showed lower levels of SIRT2 than normal liver (Figure 8C). Expression of SIRT2 was reduced by 2- to 3-fold in 44 (16.77%) tumors and by more than 3-fold in 8 (3.03%) tumors, while only 3 (1.14%) exhibited increased SIRT2 levels by 2- to 3-fold. Expression of SIRT2 was also reduced by 1.5- to 2-fold in 77 (29.17%) tumors, and increased by 1.5- to2-fold in 7 (2.65%) tumors, and had no significant changes (varying from −1.5 to +1.5) in the remaining 125 (47.35%) tumors (Figure 8D). To provide validation of the microarray data, we picked 10 HCC samples that showed reduced levels of SIRT2 and performed real-time RT-PCR. There was a reduction of SIRT2 in 8 samples, as compared with their normal controls (Figure 7E). This observation is consistent with our view that SIRT2 may play a role in human HCC development. Oncomine analysis also showed reduced SIRT2 mRNA expression in anaplastic oligodendroglioma, glioblastoma, clear cell renal carcinoma, and prostate carcinoma, as compared with normal tissue (Figure S7).
Figure 8. SIRT2 Gene Expression in Human Cancers.
(A) Comparison of SIRT2 protein levels revealed by tissue array in 36 pairs of primary breast cancers and adjacent normal breast tissues, as well as 9 pairs of lymph node metastatic infiltrating ductal carcinomas and cancer adjacent normal lymph node tissues (adj. normal). Levels of SIRT2 staining were classified as high (Hi), medium (Me), low (Lo), and negative (No).
(B) Examples of immunohistochemical images of 3 primary cancers (1: No; 2: Lo, 3: Me), 1 lymph node metastatic infiltrating ductal carcinoma (4: No), and 2 adjacent normal breast tissues (5: Lo; 6: Hi). Scale bars = 10 μm.
(C–D) SIRT2 expression levels from microarray data of 264 HCC samples (C), presented as raw log2 ratio (tumor/normal: T/N) (D). Red represents high ratio of T/N and green represents low ratio of T/N.
(E) Real-time RT-PCR of 10 pairs of samples was also presented. Data shown is average ±SD.
(F) A model illustrating actions of SIRT2 in regulating APC/C activity through deacetylation of CDH1 and CDC20, which is critical for maintaining normal mitosis. SIRT2 deficiency impairs APC/C activity, leading to mitotic catastrophe, genetic instability, and tumorigenesis.
(See also Figure S7)
DISCUSSION
In this study, we analyzed the physiological function of SIRT2 in mutant mice generated by gene targeting. We show that SIRT2-deficient cells displayed centrosome amplification and cell death during mitosis that was accompanied by genetic instability. Initially, SIRT2-deficient MEFs exhibited reduced proliferation; however, they gradually gained a faster growth rate and became malignantly transformed after immortalization, suggesting that the absence of SIRT2 eventually triggers tumorigenesis. Consistently, many aged Sirt2−/− mice developed tumors in multiple tissues. These data yield important information suggesting an essential role of SIRT2 in maintaining genetic stability and repressing tumor formation.
SIRT2 Functions as a Tumor Suppressor
Previous investigations indicate that two members of the sirtuin family, SIRT1 and SIRT3, have tumor suppressor function (Bell et al, 2011; Deng, 2009; Finley et al. 2011; Kim et al., 2010; Tao et al. 2010; Wang et al., 2008a; Wang et al., 2008b). Here our data suggest that another member of this gene family, SIRT2, also acts as a tumor suppressor. Current information regarding a role of SIRT2 in tumorigenesis is scarce and conflicting. It was suggested that SIRT2 might promote tumor formation because SIRT2 deacetylates and inhibits the activity of p53 (Jin et al., 2008; Peck et al., 2010). Consistently, dual inhibitors of SIRT1 and SIRT2 induce apoptosis in tumor cell lines (Peck et al., 2010; Zhang et al., 2009) and inhibit growth of Burkitt lymphoma xenografts (Heltweg et al., 2006), while an inhibitor for SIRT1 alone did not have such an anti-tumor effect (Peck et al., 2010) Moreover, a greater correlation between tumor tissue and the expression levels of SIRT2 (6 of 11) than SIRT1 (1 of 11) was also reported (Ouaissi et al., 2008). On the other hand, it was shown that SIRT2 expression is downregulated in human gliomas, and ectopic expression of SIRT2 in glioma cell lines led to a remarkable reduction of colony formation ability (Hiratsuka et al., 2003). We also detected reduced expression of SIRT2 in human breast cancer and HCC, compared to normal tissue (Figure 8A-E). An analysis of Oncomine datasets also revealed reduced SIRT2 mRNA expression in anaplastic oligodendroglioma, glioblastoma, clear cell renal carcinoma, and prostate carcinoma, as compared with normal tissue (Figure S7). These data suggest that SIRT2 inhibits tumor formation, rather than promotes it. Importantly, we show that SIRT2 mutant animals developed tumors, providing strong genetic evidence for a tumor suppressor function.
SIRT2 Is a Positive Regulator of APC/C Activity
Cells lacking Sirt2, both in vitro and in vivo, displayed widespread genetic instability and abnormal mitosis both of which can serve as causes for tumorigenesis suggesting a possible underlying mechanisms for tumorigenesis associated with SIRT2 deficiency. In this regard, it has been shown that SIRT2 deacetylates α-tubulin and overexpression of SIRT2 blocks chromosome condensation in response to mitotic stress (Inoue et al., 2007). SIRT2 downregulation was also found to confer resistance to microtubule inhibitors by prolonging chronic mitotic arrest (Inoue et al., 2009). However, our data showed that while ectopic overexpression of SIRT2 reduces α-tubulin acetylation in cultured cells, there were no acetylation changes in SIRT2 mutant cells and tissues (data not shown), suggesting that SIRT2-mediated tubulin deacetylation does not play a obvious role in promoting genetic instability and tumorigenesis in the SIRT2 mutant mice. Instead, our analysis indicates that hyperacetylation of CDH1 and CDC20 reduces their interaction with CDC27, which decreases APC/C activity; and in contrast, deacetylation of CDH1 and CDC20 by SIRT2 enhances the interaction of these co-activators with CDC27, leading to activation of APC/C. Thus, this data indicates that SIRT2 is a positive regulator for APC/C activity.
Several lines of evidence suggest that APC/C-CDH1 is involved in tumorigenesis. First, several APC/C-CDH1 substrates, including cyclins, Aurora-A, and Plk1, are overexpressed in human cancers and appear to promote tumor growth (Saeki et al., 2009; Schmit et al., 2009). Second, although it is unclear whether CDH deficiency could cause tumor formation due to middle gestation lethality of CDH1−/− embryos (Garcia-Higuera et al., 2008; Li et al., 2008), CDH1-homozyous cells displayed profound genetic instability and the heterozygous mice exhibited increased susceptibility to spontaneous tumors (Garcia-Higuera et al., 2008). These data suggest that CDH1 contributes to the maintenance of genomic stability and acts as a haplo-insufficient tumor suppressor. Third, it is well established that cancer cells preferentially use glycolysis to generate energy to continue proliferation in the aerobic conditions that is referred to as the “Warburg effect” (Warburg, 1956). A recent study revealed that the proliferative response, regardless of whether it occurs in normal or neoplastic cells, is dependent on a decrease in the activity of CDH1 that activates both proliferation and glycolysis. Thus, CDH1 activity may account, at least in part, for the Warburg effect that may link glycolysis to cell proliferation (Almeida et al. 2010).
SIRT2 Maintains the Integrity of Mitosis through Regulating APC/C Activity
Our results also showed that Sirt2 deficiency results in genomic instability that is associated with centrosome amplification, mitotic cell death in early passage MEFs, and tumorigenesis at later stages. As such, it is proposed that these abnormalities can be caused by a combined effect of altered expression of mitotic regulators that are regulated by APC/C. While it is impossible to investigate the actual influence of SIRT2 on each of these regulators, or the impact of them on the SIRT2-dependent phenotypes observed in this study, we first chose to present Aurora-A as a mechanistic example. In this regard, considerable data has implicated Aurora-A in genetic instability and cancer. First, Aurora-A is localized at the centrosome and is required for centrosome maturation and separation (Anand et al., 2003; Giet et al., 2005), and ectopic overexpression of Aurora-A impairs the spindle assembly checkpoint and results in aneuploidy (Stenoien et al., 2003; Zhou et al., 1998). Second, the absence of Aurora-A in mice causes mitotic arrest and monopolar spindle formation (Cowley et al., 2009) and its overexpression in mammary epithelium results in centrosome amplification and murine mammary tumor formation (Wang et al., 2006). Third, in addition to its effect on spindle assembly checkpoint and centrosome duplication, Aurora-A overexpression could also activate the Akt-mTOR pathway that is involved in a transformed phenotype (Taga et al., 2009). Indeed, overexpression of Aurora-A is frequently detected in several different types of cancers, including breast malignancy (Giet et al., 2005; Hu et al., 2005; Saeki et al., 2009; Tanaka et al. 2005; Zhou et al., 1998). A recent study also provided data that strongly associate the high Aurora-A expression with decreased survival (P = 0.0005) of breast cancer patients (Nadler et al. 2008). Our observation that mammary cancer is the predominant form of cancers in SIRT2−/− females is consistent with the high level of Aurora-A in these mice. In addition, García-Higuera et al. (2008) showed that Cdh1+/− female mice developed higher incidence of mammary tumor than other types of tumors. The authors believed that the accumulation of genomic instability might specifically promote the development of epithelial tumours that typically require a genomic instability component (García-Higuera et al. 2008).
On the other hand, our data reveal about 26% (5/19) of SIRT2−/− males developed HCC, whereas it only occurred in 4% (1/26) SIRT2−/− females. This observation is consistent with the fact that human HCC occurs at a much higher rate in males than females (Ruggieri et al. 2010) although it may suggest that the Aurora-A overexpression does not have a major impact on HCC formation, or alternatively, its effect is compromised due to the presence of some protective factors in females. While the role of Aurora-A needs to be further investigated, previous studies have implicated some other APC/C targets to HCC, such as Aurora B (Lin et al. 2010) and L2DTL (Pan et al. 2006). A recent study carried on Drosophila indicated that irradiation induced hormetic effect was only revealed in Sirt2 mutant males but not in females (Moskalev et al. 2011). Thus, the gender difference in cellular stress resistance and tumorigenesis associated with SIRT2 mutation should be interesting topics and deserve further investigation in the future.
Of note, the APC/C and most of its main regulators are essential for the development, as knockouts of any of these proteins cause embryonic lethality or early postnatal death (Wirth et al., 2004; Lee et al., 2006), while SIRT2−/− mice are born at normal mendelian ratio. We believe this is mainly because SIRT2 deficiency leads to a reduced, but not complete knockout of APC/C, activity, which is sufficient to support their development although the aged mutant mice displayed spontaneous tumorigenesis. In this regards, it was shown that mice heterozygous for CDH1 (Garcia-Higuera et al., 2008), CDC20 (Li et al., 2009), MAD1 (Iwanaga et al., 2007), and MAD2 (Michel et al., 2001) are developmentally normal, but suffered from tumorigenesis at later stages. Thus, reduced APC/C activity in SIRT2 mutant mice, although does not impair development of the mutant mice, eventually results in tumor formation.
In summary, we demonstrated that SIRT2 regulates APC/C activity through deacetylating its co-activators, CDH1 and CDC20. Loss of SIRT2 consequently causes increased levels of many mitosis regulators that may contribute to centrosome amplification, aneuploidy, mitotic cell death, and most importantly spontaneous tumor formation (Figure 8F). In addition, SIRT2 expression is reduced in several human malignancies including breast, liver, brain, kidney, and prostate cancers. Thus, these results identify SIRT2 as a fidelity or tumor suppressor gene and uncover an essential role for SIRT2 in maintaining the integrity of mitosis through positively regulating APC/C activity, a dysfunction of which leads to genetic instability and tumorigenesis.
EXPERIMENTAL PROCEDURES
Mating and Genotyping Mice
Chimeric mice, obtained by injecting the targeted Sirt2+/− ES cells into blastocysts, were mated with NIH Black Swiss or C57B6 females to screen for germline transmission. Male mice bearing germline transmission were mated with female FVB EII-Cre mice (Lakso et al., 1996) to generate complete deletion of Sirt2 exons 5–8. The animals were genotyped using either southern blot or PCR with the following primers: 1: 5′ gccttagctacatagaaggc 3′. Primers 2: 5′ gaatgacctacaatgggcca 3′ and 3: 5′ gtgtagccctggctcttcta 3′. Primers 1, 2, and 3 are located within introns 4, 7, and 8, respectively. Primers 2 and 3 amplify the wild-type allele (200 bp) and floxed allele (255 bp). The combination of primer 1 and 3 amplifies the deleted allele (350 bp). All experiments were approved by the Animal Care and Use Committee of the National Institute of Diabetes, Digestive and Kidney Diseases (ACUC, NIDDK).
Clinical Specimens
The tissue array of breast cancer samples was purchased from US Biomax (Cat. BR1002). All tissues were collected with the donor being informed completely and with their consent and the samples were subsequently de-identified prior to analysis. Immunohistochemical staining against SIRT2 (Cat. S8447, Sigma) was carried out with a HistoMouse-SP (AEC) kit (Cat. 95-9544, Zymed). cDNA microarray analysis from 264 hepatic cell carcinomas was as previously described (Yamashita et al., 2008). The validation of microarray data was carried out by qRT-PCR with the following primers: SIRT2-RT-F1:
ccggcctctatgacaaccta, SIRT2-RT-R1: ggagtagccccttgtccttc
18S-F1: agtccctgccctttgtacaca, 18S-R1: cgatccgagggcctcacta
Use of human tissues was approved by the NIH Office of Human Subjects Research.
Purification of SIRT2-associated Proteins
HeLa cells, transiently transfected with Flag-SIRT2 or pCMV vector by Fugene 6 (Roche Applied Science; Indianapolis, IN), were lysed with IP buffer (10 mM Hepes, Ph 7.9, 180 mM KCl, 1.5 mM MgCl2, 0.1% NP-40, 1 mM EDTA, 0.1 mM PMSF), including protease inhibitors. Total cell extracts were incubated with anti-Flag M2 agarose (Sigma; St. Louis, MO) for 12 hr at 4°C. After washing five times with IP buffer, bound proteins were eluted using 0.25 mg/ml Flag peptide (Sigma), concentrated using a microcon column (Millipore), resolved by 4–12% SDS-PAGE, stained with Coomassie blue, and analyzed via in-gel digestion followed by liquid chromatography-mass spectrometry.
Growth Curve, Chromosome Spread and Mitotic Exit Analysis
For growth properties, primary MEFs were plated at a density of 5 × 105 cells per 100-mm dish, and the number of cells counted using the classical 3T3 protocol. Chromosome spread was conducted as previously described (Deng and Xu, 2004). Briefly, cells in an exponentially growing plate are treated for 1 hour with colcemid at a final concentration of 0.01 μg/ml. Cells are then treated with 0.56% KCl and fixed with ice cold fixative for three times. Gently suspend cells in the fixative and drop a small quantity of the suspension using a pasteur pipette on glass slides. After air dry, the slides are stained with Giemsa (based on manufacture’s instructions). For mitotic exit analysis, primary MEFs were synchronized, using the double thymidine block method, followed by treatment with nocodazole, and harvesting of mitotic cells. The mitotic cells were washed three times with PBS, incubated with standard DMEM containing 15% FBS, and harvested at indicated time points. The harvested samples were analyzed using Western blot analysis.
Immunohistochemical Staining
For immunohistochemical staining, tissues were fixed in 10% formalin, blocked in paraffin, sectioned, stained with hematoxylin and eosin, and analyzed by light microscopy. Detection of primary antibodies was carried out using the Zymed Histomouse SP Kit (Invitrogen) following the manufacturer’s instructions.
Supplementary Material
Significance.
Although the connection between chromatin maintenance, carcinogenesis, and aging is well established, the underlying mechanism remains elusive. During mitosis, SIRT2 is relocalized from the cytoplasm to the nucleus and associates with chromosome and mitotic structures. Nonetheless, the function of SIRT2 in the nucleus and its deacetylation targets are unclear. We hypothesized that SIRT2 is a fidelity protein that ensures normal mitotic progression and genetic stability. This work shows that SIRT2 plays a critical role in maintaining the mitosis through modulating the activity of APC/C via deacetylation of the co-activator proteins CDH1 and CDC20 and regulating their interaction with CDC27. This finding identifies SIRT2 as a positive regulator of APC/C activity ensuring normal mitotic progression that is critical for maintaining genome integrity and suppressing tumorigenesis.
Highlights.
Hyperacetylation of adaptor protein CDH1 and CDC20 inhibits APC/C activity
SIRT2 positive regulates APC/C activity through deacetylation of CDH1 and CDC20
Expression of SIRT2 is reduced in human breast and liver cancers.
SIRT2 deficiency in mice causes mitotic cell death, genetic instability, and tumorigenesis.
Acknowledgments
We thank members of Dr. Deng’s lab for their critical discussion of this work. This research was supported (in part) by the Intramural Research Program of the NIDDK, NCI, and CCR, NIH. DG is supported by 1R01CA152601-01 and 1R01CA152799-01 from the NCI, BC093803 from the DOD, and SPORE P50CA98131. This project was also supported with federal funds from the National Cancer Institute, National Institutes of Health, DHHS, in part under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organization imply endorsement by the U.S. government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A, Bolanos JP, Moncada S. E3 ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc Natl Acad Sci U S A. 2010;107:738–741. doi: 10.1073/pnas.0913668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley DO, Rivera-Perez JA, Schliekelman M, He YJ, Oliver TG, Lu L, O’Quinn R, Salmon ED, Magnuson T, Van Dyke T. Aurora-A kinase is essential for bipolar spindle formation and early development. Mol Cell Biol. 2009;29:1059–1071. doi: 10.1128/MCB.01062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CX, Xu X. Generation and analysis of Brca1 conditional knockout mice. Methods Mol Biol. 2004;280:185–200. doi: 10.1385/1-59259-788-2:185. [DOI] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19:416–28. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- Gieffers C, Peters BG, Kramer ER, Dotti CG, Peters JM. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci U S A. 1999;96:11317–22. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa A, et al. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun. 2003;309:558–566. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- Hu W, Kavanagh JJ, Deaver M, Johnston DA, Freedman RS, Verschraegen CF, Sen S. Frequent overexpression of STK15/Aurora-A/BTAK and chromosomal instability in tumorigenic cell cultures derived from human ovarian cancer. Oncol Res. 2005;15:49–57. doi: 10.3727/096504005775082101. [DOI] [PubMed] [Google Scholar]
- Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, Nakano S, Katoh M, Ito H, Oshimura M. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2007;26:945–957. doi: 10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- Inoue T, Nakayama Y, Yamada H, Li YC, Yamaguchi S, Osaki M, Kurimasa A, Hiratsuka M, Katoh M, Oshimura M. SIRT2 downregulation confers resistance to microtubule inhibitors by prolonging chronic mitotic arrest. Cell Cycle. 2009;8:1279–1291. doi: 10.4161/cc.8.8.8245. [DOI] [PubMed] [Google Scholar]
- Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Peloponese JM, Jr, Li Y, Ward JM, Benezra R, Jeang KT. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–6. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- Jacobs KM, Pennington JD, Bisht KS, Aykin-Burns N, Kim HS, Mishra M, Sun L, Nguyen P, Ahn BH, Leclerc J, et al. SIRT3 interacts with the daf-16 homolog FOXO3a in the Mitochondria, as well as increases FOXO3a Dependent Gene expression. Int J Biol Sci. 2008;4:291–299. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Kim YJ, Kim DW, Baek KH, Kang BY, Yeo CY, Lee KY. Sirt2 interacts with 14-3-3 beta/gamma and down-regulates the activity of p53. Biochem Biophys Res Commun. 2008;368:690–695. doi: 10.1016/j.bbrc.2008.01.114. [DOI] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Lee DJ, Oh SP, Park HD, Nam HH, Kim JM, Lim DS. Mouse emi1 has an essential function in mitotic progression during early embryogenesis. Mol Cell Biol. 2006;26:5373–81. doi: 10.1128/MCB.00043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol. 2009;185:983–994. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Shin YH, Hou L, Huang X, Wei Z, Klann E, Zhang P. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat Cell Biol. 2008;10:1083–1089. doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhang P. The function of APC/CCdh1 in cell cycle and beyond. Cell Div. 2009;4:2. doi: 10.1186/1747-1028-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kazgan N. Mammalian sirtuins and energy metabolism. Int J Biol Sci. 2011;7:575–587. doi: 10.7150/ijbs.7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZZ, Jeng YM, Hu FC, Pan HW, Tsao HW, Lai PL, Lee PH, Cheng AL, Hsu HC. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B Overexpression in HCC. BMC Cancer. 2010;10:461. doi: 10.1186/1471-2407-10-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel C, Broxmeyer HE. Sirtuin 1, stem cells, aging, and stem cell aging. Curr Opin Hematol. 2008;15:326–331. doi: 10.1097/MOH.0b013e3283043819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Bieman M, Th’ng J, Lemieux M. The absence of SIR2alpha protein has no effect on global gene silencing in mouse embryonic stem cells. Mol Cancer Res. 2003;1:402–409. [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–9. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Moskalev AA, Plyusnina EN, Shaposhnikov MV. Radiation hormesis and radioadaptive response in Drosophila melanogaster flies with different genetic backgrounds: the role of cellular stress-resistance mechanisms. Biogerontology. 2011;12:253–263. doi: 10.1007/s10522-011-9320-0. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Nadler Y, Camp RL, Schwartz C, Rimm DL, Kluger HM, Kluger Y. Expression of Aurora A (but not Aurora B) is predictive of survival in breast cancer. Clin Cancer Res. 2008;14:4455–4462. doi: 10.1158/1078-0432.CCR-07-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS ONE. 2007;2:e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouaissi M, Sielezneff I, Silvestre R, Sastre B, Bernard JP, Lafontaine JS, Payan MJ, Dahan L, Pirro N, Seitz JF, et al. High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann Surg Oncol. 2008;15:2318–2328. doi: 10.1245/s10434-008-9940-z. [DOI] [PubMed] [Google Scholar]
- Pan HW, Chou HY, Liu SH, Peng SY, Liu CL, Hsu HC. Role of L2DTL, cell cycle-regulated nuclear and centrosome protein, in aggressive hepatocellular carcinoma. Cell Cycle. 2006;5:2676–87. doi: 10.4161/cc.5.22.3500. [DOI] [PubMed] [Google Scholar]
- Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, Fuchter MJ, Hsiao CD, Lam EW. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. 2010;9:844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Pines J. The APC/C: a smorgasbord for proteolysis. Mol Cell. 2009;34:135–136. doi: 10.1016/j.molcel.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Ruggieri A, Barbati C, Malorni W. Cellular and molecular mechanisms involved in hepatocellular carcinoma gender disparity. Int J Cancer. 2010;127:499–504. doi: 10.1002/ijc.25298. [DOI] [PubMed] [Google Scholar]
- Saeki T, Ouchi M, Ouchi T. Physiological and oncogenic Aurora-A pathway. Int J Biol Sci. 2009;5:758–762. doi: 10.7150/ijbs.5.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- Schmit TL, Zhong W, Nihal M, Ahmad N. Polo-like kinase 1 (Plk1) in non-melanoma skin cancers. Cell Cycle. 2009;8:2697–2702. doi: 10.4161/cc.8.17.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Sen S, Mancini MA, Brinkley BR. Dynamic association of a tumor amplified kinase, Aurora-A, with the centrosome and mitotic spindle. Cell Motil Cytoskeleton. 2003;55:134–146. doi: 10.1002/cm.10120. [DOI] [PubMed] [Google Scholar]
- Taga M, Hirooka E, Ouchi T. Essential roles of mTOR/Akt pathway in Aurora-A cell transformation. Int J Biol Sci. 2009;5:444–450. doi: 10.7150/ijbs.5.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E, Hashimoto Y, Ito T, Okumura T, Kan T, Watanabe G, Imamura M, Inazawa J, Shimada Y. The clinical significance of Aurora-A/STK15/BTAK expression in human esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1827–1834. doi: 10.1158/1078-0432.CCR-04-1627. [DOI] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008a;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, Lee MH, Xiao C, Vassilopoulos A, Chen W, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008b;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T, Deng CX. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–7158. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- Wirth KG, Ricci R, Giménez-Abián JF, Taghybeeglu S, Kudo NR, Jochum W, Vasseur-Cognet M, Nasmyth K. Loss of the anaphase-promoting complex in quiescent cells causes unscheduled hepatocyte proliferation. Genes Dev. 2004;18:88–98. doi: 10.1101/gad.285404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Au Q, Zhang M, Barber JR, Ng SC, Zhang B. Identification of a small molecule SIRT2 inhibitor with selective tumor cytotoxicity. Biochem Biophys Res Commun. 2009;386:729–733. doi: 10.1016/j.bbrc.2009.06.113. [DOI] [PubMed] [Google Scholar]
- Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.