Abstract
Bacteriophages occupy a unique position in biology, representing an absolute majority of all organisms in the biosphere. Because their genomes are relatively small, elucidating the genetic diversity of the phage population, deciphering their origins, and identifying the evolutionary mechanisms that shape the population would seem readily feasible. And yet the pace of phage genome characterization has slowed over the past three years, reflecting in part a need to transition from sequencing known and well-characterized bacteriophages to the isolation and comparative analysis of new isolates. The current state of bacteriophage genomics shows that the genetic diversity of the population is very high, that phages have been actively evolving for billions of years with active engagement of horizontal genetic exchange, and that their genomes are consequently pervasively mosaic in their architectures. But we have barely scratched the surface and the next years of phage genome exploration promise to be especially revealing.
Introduction
Bacteriophages offer a special perspective on the diversity, origins, and evolution of viruses, not only in their tremendous abundance – there are more than 1031 phage particles in the biosphere – but in their distant origins, likely more than three billion years ago [1,2]. Moreover, viral ecologists calculate that there are about 1023 phage infections per second on a global scale, indicating that the population is not only large and old but also highly dynamic [3].
Unfortunately, there is no bacteriophage fossil record as such and the best promise for understanding phage origins and evolution is the comparative analysis of phages present in the environment today. There are essentially three types of populations available for study. First, individual phages can be isolated in the laboratory that infect a particular bacterial host used for their propagation. Numerous different hosts have been employed and it seems reasonable that phages exist for the vast majority of bacterial species that can be propagated in the lab. The number of individual phages isolated is substantial [>5,000; [4]], but the complete genome sequences of only about 750 of these have been determined. About 70% of these sequenced phages correspond to only 12 different bacterial hosts, and this modest collection likely represents just a small portion of the overall diversity. Furthermore, there may be phages that cannot be recovered using standard laboratory approaches and are missed using this approach. A notable advantage of this genome-by-genome approach, however, is that individual phages with sequenced genomes become available for further genetic, biochemical and structural dissection.
A second general approach is viral metagenomics, in which the viral population is harvested en masse from an environmental source, concentrated, and large numbers of DNA segments sequenced at random [5,6]. With high throughput sequencing approaches becoming generally and relatively cheaply available, it has become possible to sample substantial numbers of metagenomic samples and to compare viral populations as a functions of geography and time, providing considerable insights into the dynamics of phage populations [6,7]. While there are a number of advantages of these metagenomic approaches – including the vast amounts of sequence data that are obtained– the disadvantage is that the output is pure data, and no biological materials for further experimentation are recovered [8].
A third approach is mining prophage and phage-related sequences embedded in sequenced bacterial genomes. Sequencing of complete bacterial genomes has become relatively simple and there are over 1,600 completely sequenced bacterial chromosomes. A substantial portion of these contain at least one prophage and prophages can occupy up to 20% of the chromosomal content [9–11]; this prevalence of prophages is not unexpected given the finding that a large proportion of isolated bacteriophages are temperate. However, accurately identifying prophages and phage-related objects remains a challenge – especially for those bacteria for which few if any viruses have been characterized – and it is not easy to predict from genome information alone which of these can generate infectious particles through prophage induction. The issue is further complicated by the presence of genome sequences that are phage-derived or phage-associated but play biological roles separate from viral lytic growth. These include Gene Transfer Agents [12], pathogenicity islands [13], encapsuins [14] and large bacteriocins [15], among others.
Diversity of the bacteriophage population
At the time of writing the total number of unique sequenced bacteriophage genomes is about 750. These encompass many types of virion morphologies and nucleic acid compositions, but the large majority are double stranded DNA (dsDNA) tailed phages (Caudovirales), reflecting predictions from virion morphology surveys [4]. For example there are fewer than 50 each of completely sequenced RNA phages and ssDNA phages, and of the more than 500 sequenced dsDNA tailed phages, ~55% are morphologically members of the Siphoviridae with long flexible non-contractile tails; the remainder are Myoviridae with contractile tails and Podoviridae with short stubby tails (~25% and ~20% respectively). Overall, phage genomes represent only about 15% of all viruses with known unique sequences and are thus vastly under-represented in the genome databases.
Phage genome size varies enormously, ranging from the ~3,300 nucleotide ssRNA viruses of Escherichia coli [16] to the almost 500 kbp genome of Bacillus megaterium phage G (our unpublished data). The smallest of the dsDNA tailed phages genomes are ~11.5 kbp [e.g. Mycoplasma phage P1 [17]], ~21kbp [e.g. Lactococcus phage c2, [18]], and ~ 30 kbp [e.g. Pasteurella phage F108 [19]] for the Podoviridae, Siphoviridae, and Myoviridae respectively, but there are broad size ranges among these. In general, these genomes are packaged at similar densities into their capsids and the size of the capsid varies as a function of genome size. Because virion infectivity is influenced by the amount of DNA packaged within any given capsid – either too little or too much leads to loss of virion stability – there are evolutionary pressures to either gain or lose DNA to accommodate packaging and virion stability. This selection for genome size plays an important role in bacteriophage evolution, providing a mechanism for DNA gain and loss that is independent of gene function. Newly acquired DNA thus provides a reservoir of genetic information for potential future use, rather than being selected for immediate utility. This represents a notable departure from bacterial genome evolution, where flexibility in the cell membrane and cell wall does not impose any obvious constraint on genome size.
Nucleotide sequence comparison of bacteriophage genomes reveals them to be enormously diverse [1]. For example, unless any given phage genome has a known close relative that infects the same host, or there is a closely-related prophage, it is unusual to find extensive nucleotide sequence similarity to other database entries. This suggests that host preferences represent a significant barrier to genetic exchange, and although phages clearly have the capacity to switch hosts – perhaps more easily the more closely related the hosts are – by a variety of mechanisms, it is unclear at what rate this occurs in natural populations. Phage infecting a common host can also exhibit substantial diversity suggesting that there are additional barriers to genetic exchange [20–22]. However, some caution is warranted in interpreting this, because the ‘natural’ or ‘preferred’ host – the one the virus was associated with in its most recent ecological and evolutionary past – may not be the same as the host used for its isolation. It is notable that the GC% content of the large collection of mycobacteriophages varies between 55% and 70%, which is consistent with this consideration [20].
Mosaicism: The hallmark feature of bacteriophage genomes
Perhaps the most striking feature emerging from phage genome comparative analyses is that they are pervasively mosaic – with different segments having distinct evolutionary histories [23]. A simple general explanation is that horizontal genetic exchange plays a dominant role in shaping these genome architectures. Mosaicism is of course not confined to phage genomes as bacteria also acquire DNA by horizontal genetic transfer; but the extent appears to be much greater in bacteriophage evolution.
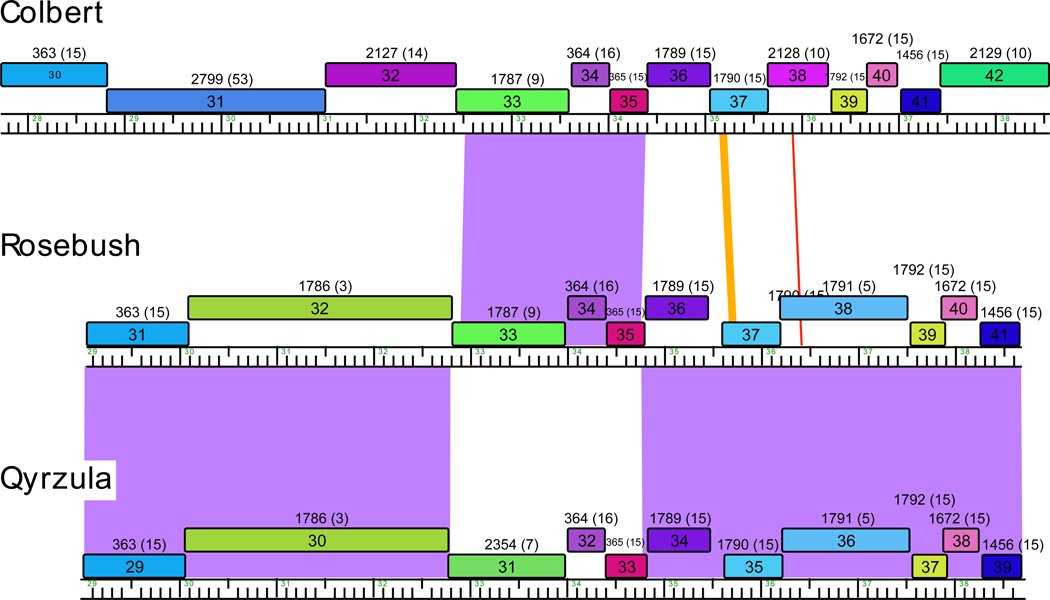
Exchange events occurring in relatively recent evolutionary time can occasionally be seen through whole genome nucleotide comparisons (Fig. 1). For example, mycobacteriophages Rosebush and Qyrzula are closely related and share extensive nucleotide sequence similarity. Phage Colbert is a more distant relative sharing similar overall genome organization and many common genes but without a high level of nucleotide sequence similarity. However, a ~1.8 kbp segment of Colbert is closely related to Rosebush (94% nucleotide identity) and was presumably acquired relatively recently from a Rosebush-like phage. Interestingly, the same region in Rosebush is from that in Qyrzula (Fig. 1).
Figure 1. Recent recombination events giving rise to phage genome mosaicism.
Genetic exchange events giving rise to genome mosaicism are usually only observed at the nucleotide sequence level when the events have occurred relatively recently in evolutionary time. Mycobacteriophages Colbert, Rosebush and Qyrzula share similar overall genome architectures and many genes, but only Rosebush and Qyrzula have extensive nucleotide sequence similarity. A segment of Colbert containing genes 33–35 appears to have been acquired recently from a Rosebush-like phage, and the conserved sequences share 94% nucleotide identity. The extent of nucleotide similarity is displayed by coloring between the genomes, color-coded by spectrum with violet being the most similar and red the least. Predicted genes are shown as boxes, with gene numbers in the boxes and the sequence phamilies [24] above, with the number of phamily members shown in brackets; phamilies correspond to groups of related genes [24]. Genes are colored according to their phamily membership. Note that the apparent sites of recombination are located close to gene boundaries. The functions of most of these genes are not known but are predicted to be involved in tail assembly. Maps were generated using the program Phamerator (S. Cresawn, manuscript submitted).
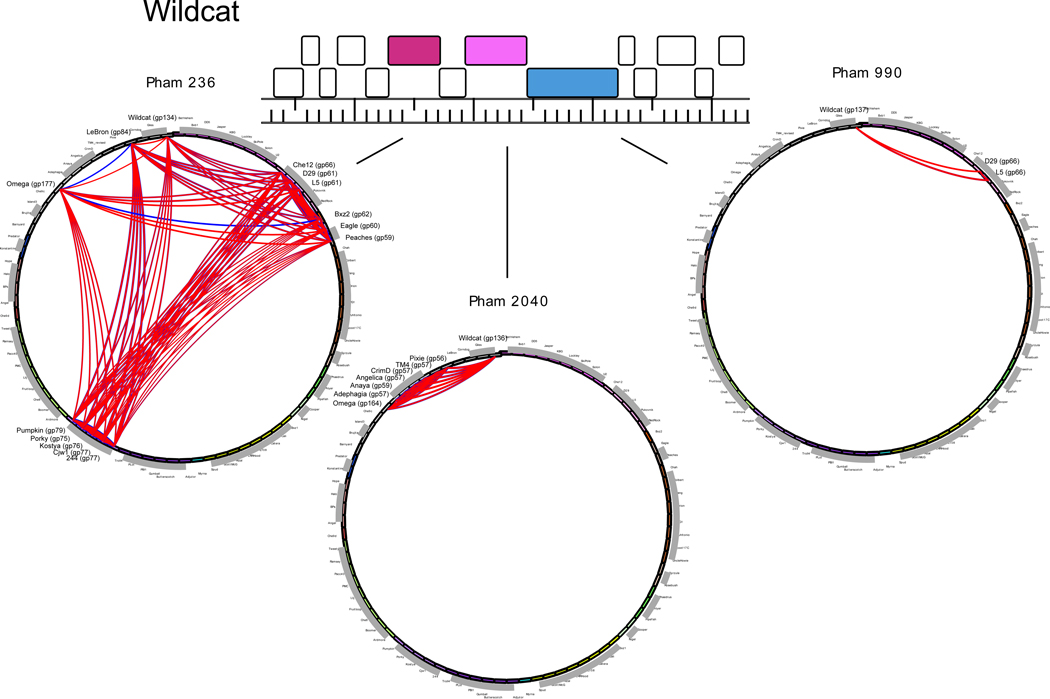
Genome mosaicism can also be observed by comparing genes at the amino acid sequence level, identifying genes of common ancestry that diverged sufficiently long ago that nucleotide sequence similarity is no longer recognizable (Fig. 2). With a sufficiently large collection of diverse genome sequences, homologues can be identified that are present in otherwise far-distantly related phages and establish their phylogenetic relationships [24]. In many cases, the modules that are being exchanged correspond to single genes (Fig. 2). While these relationships are clearly evident among the larger collections of phages of a common host, it seems likely that this is a general property of bacteriophage genomes. This pervasive mosaicism and the multiple phylogenies of different genomic segments renders it difficult to impose whole genome phylogenetic relationships and reticulate approaches are likely to be more informative [25,26].
Figure 2. Illustrations of phage genome mosaicism.
A small segment of the mycobacteriophage Wildcat genome is shown encompassing genes 129–142. Genes are drawn and annotated as in Figure 1; those that have no homologues among the collection of mycobacteriophage genomes are shown as white boxes. The phylogenetic relationships of the three genes (134, 136 and 137) are represented as phamily circles, in which all genomes within the Phamerator database (S. Cresawn, manuscript submitted) are positioned around the circumference of the circle, and arcs are drawn between those phages sharing members of the phamily, with the thickness of the line reflecting the strength of sequence similarity. Each of the genes in Phams 236, 2040 and 990 clearly has a distinct evolutionary history.
The junctions between phage genome segments with distinct phylogenies often correspond to gene boundaries (Fig. 1). Several types of recombination events are postulated to contribute to this. First, there are examples of short conserved sequences at gene boundaries that could serve to target homologous recombination to these positions [27,28]. While these likely contribute to mosaicism, it is far from clear that this is a general mechanism, because most phage gene boundaries are not associated with conserved sequence segments sufficiently long to be recognized by the homologous recombination machinery [29]. Additional plausible mechanisms are by transposition, site-specific recombination, and homing endonucleases and while all these contribute they are likely also minor components. A likely major contributor is illegitimate recombination, or recombination between short conserved sequences (a few bases), coupled with functional selection of genes [30]. Furthermore, phage-encoded homologous recombinases have properties that are well-suited to facilitating this process [31].
Differential gene mobility
Phage genomes are mosaic, but not all genes in a given genome participate in mosaicism to the same degree. This is seen most strikingly in the head gene regions of most phages, where there is little or no evidence of horizontal swapping of genes within this group of genes, even though other parts of the genome may be flamboyantly mosaic. Similar groups of genes that “travel together through evolution” may include the tail genes, or lysis genes, among others. We understand the evolutionary coherence of these groups of genes in terms of the biological functions of the proteins they encode. The head genes, for example, code for proteins that interact intimately with each other in building the head structure, and these genes must co-evolve with each other to maintain those interactions. As a result, if recombination generates a hybrid by joining parts of head gene sets from two different phages, the recombinant would be non-functional and so lost from the population, even though each of the genes was fully functional in its original context. The tendency for genes whose proteins function together to stay together reaches an extreme in the group of phages related to coliphage T4. Here a majority of the so-called core genes—the genes that are shared by all members of the group—fail to engage in horizontal exchange relative to the other genes in the group [32,33]. The genes that “travel together” include head genes, tail genes, DNA replication genes, and nucleotide metabolism genes. As with the head genes discussed above, this more extensive case of genetic linkage can be understood in terms of the interactions of the encoded proteins: T4’s DNA replication proteins form a complex, the nucleotide metabolism genes probably form a complex that feeds precursors into the DNA replication machine. The observation that the corresponding genes travel together with the head and tail genes is less easily explained but may reflect well-documented interactions among recombination proteins and DNA replication initiation and between recombination proteins and DNA packaging [34].
The other genes of a phage genome—the “non-core” genes—provide a striking contrast to the core genes discussed above [33]. These are (by definition) not found in all members of a group of related phages, and often only one example of a particular gene of this sort is present in known phage genomes. Non-core genes are found in all tailed phage genomes where there is enough information to define the core genes; they are often in small clusters of genes, with the clusters interspersed among the clusters of core genes. In most cases the functions of the non-core genes are unknown, and in some well-studied phages like coliphage λ, they have been deleted without adverse effect on phage growth under laboratory conditions. Their average size is substantially less than that of core genes, often by nearly a factor of two, and it has been suggested that they may correspond to individual functional protein domains [35,36]. The possibility that some of these small genes provide no selective benefit to the phage that carries them cannot be ruled out, but their generally orderly arrangement on the genome, with good translation start sequences argues against their being disorganized “junk” DNA. An intriguing possibility, whether they are providing a selective benefit or not, is that these genes could serve as a “gene nursery”, where novel genetic functions could be built by recombination and mutation among genetic sequences that have no essential role in phage survival. In a few cases, enzymatic or even biological functions have been found for non-core genes, and some of these appear to provide small, non-essential benefits to phage growth or to be essential only in certain hosts or environments [37]. This leads to the view that the non-core genes may optimize the phage to occupy a certain ecological niche, and that the changing repertoire of these genes gives the phage population access to new niches.
The description of core and non-core genes given here implies that the non-core genes are moving in and out of the phage genomes on a much faster time scale than the core genes. We do not suppose that this means recombination occurs more frequently in certain areas of the genome. Rather we suggest that non-homologous recombination occurs rampantly and indiscriminately across the genomes and stringent natural selection for the successful arrangement of the core genes counterselects any gene arrangement that disrupts that, while allowing much more promiscuous reassortments of the non-core genes; a thoroughly Darwinian view of phage evolution.
Drivers of bacteriophage evolution
Bacteriophages genomes arguably also harbor the greatest genetic novelty in the biological world, in that most of their encoded genes (perhaps as much as 80%) are unrelated to known proteins, and are of unknown function. What do all these genes do? Microbial ecology provides a framework for considering this, in that constant infection of bacteria by bacteriophages provides a strong selection for phage resistance, coupled with the necessity to evolve phage variants that overcome resistance. Not surprisingly, there are many different host-mediated protection systems such as restriction-modification [38], CRISPR’s [39], tRNA cleavage [40], and Toxin-Antitoxin systems [41], as well as phage-encoded mechanisms for generating genome diversity at high frequency [42]. Phages not only carry genes that counteract host protection systems such as anti-restriction [43], and RNA repair enzymes [44], but also can provide genes that offer protection from other viruses. It is therefore not surprising that phages sometimes encode their own restriction systems, toxin-antitoxin systems, and immunity systems stolen from other bacteriophages [20]. We predict that many more phage genes participate in these dances for survival, but that answers to these questions will await further dissection of bacteriophages genomes and their biology.
Highlights.
Bacteriophage genomes are enormously diverse
Bacteriophage genomes are pervasively mosaic
Bacteriophage-host dynamics are strong drivers of their evolution
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Hatfull GF. Bacteriophage genomics. Curr Opin Microbiol. 2008;11:447–453. doi: 10.1016/j.mib.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Ackermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 5.Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willner D, Thurber RV, Rohwer F. Metagenomic signatures of 86 microbial and viral metagenomes. Environ Microbiol. 2009;11:1752–1766. doi: 10.1111/j.1462-2920.2009.01901.x. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, Buchanan J, Desnues C, Dinsdale E, Edwards R, et al. Viral and microbial community dynamics in four aquatic environments. Isme J. 2010;4:739–751. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- 8.Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 9.Leplae R, Lima-Mendez G, Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res. 2010;38:D57–D61. doi: 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 11.Canchaya C, Fournous G, Brussow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. 2004;53:9–18. doi: 10.1111/j.1365-2958.2004.04113.x. [DOI] [PubMed] [Google Scholar]
- 12.Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Novick RP, Christie GE, Penades JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol. 2008;15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 15.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol. 2000;38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 16.Friedman SD, Genthner FJ, Gentry J, Sobsey MD, Vinje J. Gene mapping and phylogenetic analysis of the complete genome from 30 single-stranded RNA male-specific coliphages (family Leviviridae) J Virol. 2009;83:11233–11243. doi: 10.1128/JVI.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu AH, Voelker LL, Shen X, Dybvig K. Complete nucleotide sequence of the mycoplasma virus P1 genome. Plasmid. 2001;45:122–126. doi: 10.1006/plas.2000.1501. [DOI] [PubMed] [Google Scholar]
- 18.Lubbers MW, Waterfield NR, Beresford TP, Le Page RW, Jarvis AW. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl Environ Microbiol. 1995;61:4348–4356. doi: 10.1128/aem.61.12.4348-4356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campoy S, Aranda J, Alvarez G, Barbe J, Llagostera M. Isolation and sequencing of a temperate transducing phage for Pasteurella multocida. Appl Environ Microbiol. 2006;72:3154–3160. doi: 10.1128/AEM.72.5.3154-3160.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, Alexander LM, Alfano MB, Alford ST, Amy NE, et al. Expanding the Diversity of Mycobacteriophages: Insights into Genome Architecture and Evolution. PLoS ONE. 2011;6:e16329. doi: 10.1371/journal.pone.0016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A. 2005;102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwan T, Liu J, Dubow M, Gros P, Pelletier J. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J Bacteriol. 2006;188:1184–1187. doi: 10.1128/JB.188.3.1184-1187.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci U S A. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, Gonda RM, Houtz JM, Hryckowian AJ, Kelchner VA, et al. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2006;2:e92. doi: 10.1371/journal.pgen.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawrence JG, Hatfull GF, Hendrix RW. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J Bacteriol. 2002;184:4891–4905. doi: 10.1128/JB.184.17.4891-4905.2002.. Individual phages cannot easily be described as ‘species’, using any of the commonly used definitions of the term.
- 26.Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol Biol Evol. 2008;25:762–777. doi: 10.1093/molbev/msn023. [DOI] [PubMed] [Google Scholar]
- 27.Susskind MM, Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978;42:385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark AJ, Inwood W, Cloutier T, Dhillon TS. Nucleotide sequence of coliphage HK620 and the evolution of lambdoid phages. J Mol Biol. 2001;311:657–679. doi: 10.1006/jmbi.2001.4868. [DOI] [PubMed] [Google Scholar]
- 29.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, et al. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 30.Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. The origins and ongoing evolution of viruses. Trends Microbiol. 2000;8:504–508. doi: 10.1016/s0966-842x(00)01863-1. [DOI] [PubMed] [Google Scholar]
- 31. Martinsohn JT, Radman M, Petit MA. The lambda red proteins promote efficient recombination between diverged sequences: implications for bacteriophage genome mosaicism. PLoS Genet. 2008;4:e1000065. doi: 10.1371/journal.pgen.1000065.. Phage-encoded recombinases can be potent at mediating homeologous recombination and homologous recombination at short DNA sequences, implementing them in the generation of genome mosaicism.
- 32.Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Comeau AM, Bertrand C, Letarov A, Tetart F, Krisch HM. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology. 2007;362:384–396. doi: 10.1016/j.virol.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 34.Mosig G, Eiserling FA. T4 and Related Phages: Structure and Development. In: Calendar R, editor. The Bacteriophages. Oxford University Press; 2006. pp. 225–267. vol 2nd edition. [Google Scholar]
- 35.Stewart CR, Casjens SR, Cresawn SG, Houtz JM, Smith AL, Ford ME, Peebles CL, Hatfull GF, Hendrix RW, Huang WM, et al. The genome of Bacillus subtilis bacteriophage SPO1. J Mol Biol. 2009;388:48–70. doi: 10.1016/j.jmb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko CC, Weber RJ, Patel MC, Germane KL, Edgar RH, et al. Comparative Genomic Analysis of 60 Mycobacteriophage Genomes: Genome Clustering, Gene Acquisition, and Gene Size. J Mol Biol. 2010;397:119–143. doi: 10.1016/j.jmb.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King G, Murray NE. Restriction enzymes in cells, not eppendorfs. Trends Microbiol. 1994;2:465–469. doi: 10.1016/0966-842x(94)90649-1. [DOI] [PubMed] [Google Scholar]
- 39. Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123.. CRISPR systems are widespread throughout bacteria and play important roles in phage infection using interesting mechanistic innovations.
- 40.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A. 2009;106:894–899. doi: 10.1073/pnas.0808832106.. A novel type of Toxin-Antitoxin system is involved in phage resistance through an abortive infection process.
- 42.Medhekar B, Miller JF. Diversity-generating retroelements. Curr Opin Microbiol. 2007;10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMahon SA, Roberts GA, Johnson KA, Cooper LP, Liu H, White JH, Carter LG, Sanghvi B, Oke M, Walkinshaw MD, et al. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 2009;37:4887–4897. doi: 10.1093/nar/gkp478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu H, Yin S, Shuman S. Characterization of polynucleotide kinase/phosphatase enzymes from Mycobacteriophages omega and Cjw1 and vibriophage KVP40. J Biol Chem. 2004;279:26358–26369. doi: 10.1074/jbc.M403200200. [DOI] [PubMed] [Google Scholar]