Abstract
There is increasing evidence that epigenetic information can be inherited across generations in mammals, despite extensive reprogramming both in the gametes and in the early developing embryo. One corollary to this is that disrupting the establishment of epigenetic state in the gametes of a parent, as a result of heterozygosity for mutations in genes involved in reprogramming, could affect the phenotype of offspring that do not inherit the mutant allele. Here we show that such effects do occur following paternal inheritance in the mouse. We detected changes to transcription and chromosome ploidy in adult animals. Paternal effects of this type have not been reported previously in mammals and suggest that the untransmitted genotype of male parents can influence the phenotype of their offspring.
Epigenetic modifications are generally considered to be cleared on passage through the germline, and the idea that epigenetic state can be inherited during meiosis in humans remains controversial1. The strongest evidence for transgenerational epigenetic inheritance in mammals comes from studies of a handful of transgenes and endogenous genes, such as agouti viable yellow (Avy) in mice. At these loci, there seems to be incomplete erasure of epigenetic state between generations2. There is a paucity of evidence for similar effects in humans, and this may be associated with the difficulties of working with outbred populations. If even a small number of genes were subject to transgenerational epigenetic inheritance in humans, it could represent a major source of phenotypic variation and a major shift in our understanding of inheritance of disease.
One consequence of an inherent failure to clear epigenetic marks between generations would be that disruption of the epigenetic state in germ cells could affect the epigenetic state in the next generation. To test this hypothesis, we studied wild-type offspring from male mice heterozygous for mutations in genes involved in epigenetic processes. In other words, we asked whether such genes show paternal effects. Paternal effect genes are those in which a mutation in the male parent affects wild-type offspring. A few cases have been reported in Drosophila melanogaster in which these genes cause embryonic lethality3. Maternal effect genes (that is, genes whose disruption in the female parent affects wild-type offspring) have been reported in most eukaryotic model organisms, including mammals, and they encode proteins required in the egg for the development of the embryo4.
The epigenetic states of alleles that show transgenerational epigenetic inheritance, such as Avy, are sensitive to changes in dosage of chromatin proteins and enzymes involved in epigenetic reprogramming. For example, the tendency to establish a silent state can be influenced by the presence of two X chromosomes, presumably because X inactivation acts as a sink for heterochromatin proteins5. Studies in D. melanogaster have also shown that epigenetic modifiers function in a dynamic equilibrium, as silencing events are sensitive to fluctuations in their cellular concentration6. In D. melanogaster, the heterochromatic Y chromosome acts as a sink for proteins associated with gene silencing7. This dynamic equilibrium is likely to be particularly important in the zygote, in which dramatic reprogramming of the genome occurs.
The variegated expression of transgenes is a well-established model of epigenetic gene silencing in many organisms8. Transgenes and alleles such as Avy are termed metastable epialleles because the establishment of epigenotype is to some degree stochastic, resulting in variegated expression within a tissue9. We have carried out a dominant screen to identify genes involved in reprogramming of the epigenome, using N-ethyl-N-nitrosourea (ENU) mutagenesis on mice carrying a variegating green fluorescent protein (GFP) transgene5. The transgene carries an erythroid-specific promoter to direct expression to a cell type that can be analyzed at the single-cell level. Similar screens have been carried out in D. melanogaster using flies that display variegated expression of the w locus to identify modifiers of position effect variegation10. The mouse screen has produced strains in which we can test our hypothesis that epigenetic modifiers will show paternal effects. Knockouts of a number of known modifiers of epigenetic state are already available, but in most cases it is not known if heterozygosity results in a detectable phenotype. Because the mouse screen is dominant, it will specifically identify such genes. The experiments described below investigate the effects of mutations in such genes in the male parent on a maternally inherited reporter allele in the offspring (that is, effects in trans). This approach avoids the possibility of subtle confounding genetic changes at the reporter locus in the gametes of the mutant sire. Here we report paternal effects of two genes, identified in the mutagenesis screen, on expression of a maternally inherited Avy allele.
The epigenetic state of the genome has a role not only in transcriptional regulation but also in genome stability11–13. Genome-wide hypomethylation of DNA is associated with the loss or gain of whole chromosomes or parts of chromosomes, so we searched for such events in wild-type offspring of sires haploinsufficient for epigenetic modifiers. We detected paternal effects on sex chromosome ploidy, including loss of the maternal X chromosome. Together, these results suggest that in mammals, the untransmitted genotype of the male parent can influence the phenotype of his offspring through epigenetic information carried in his gametes.
RESULTS
Momme D4 carries a mutation in Smarca5, encoding Snf2h
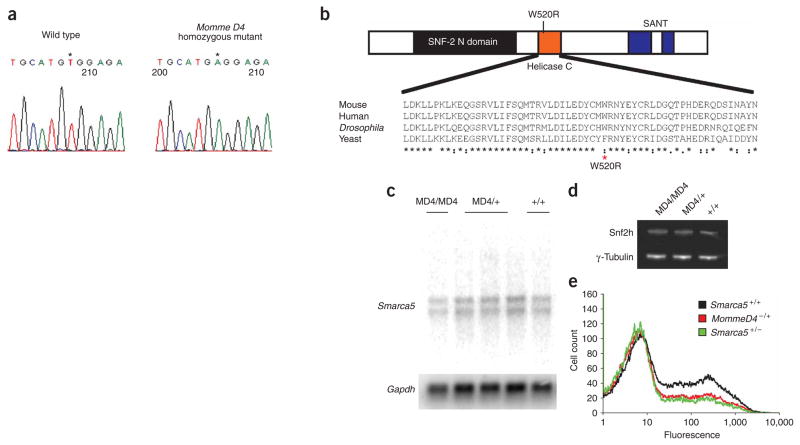
One of the mutant lines produced from the dominant screen, Momme (modifier of murine metastable epiallele) D4, is an enhancer of variegation. In the presence of the Momme D4 mutation, the percentage of red blood cells expressing GFP decreases5. The Momme D4 mutation is semidominant with respect to transgene variegation and is homozygous lethal5. The linked interval is between the markers D8Sar2 (79064467 bp in Ensembl build 34) and D8Mit77 (81460261 bp in Ensembl build 34)5. Experiments in additional mice, now totaling 174 mutants and 102 wild-type animals, confirmed this location (data not shown). This 2.4-Mb interval contains ten genes (Supplementary Table 1 online), including Smarca5, also known as Snf2h. This gene belongs to the imitation switch (ISWI) family of chromatin remodelers. We designed primers for Smarca5 covering all exons, including exon-intron junctions. We obtained PCR products from two wild-type mice and two homozygous mutant embryos, generated from a C57/FVB Momme D4 F1 intercross and identified by linkage. We found a single T-to-A mutation (at position 1921 of the transcript) in exon 12 of both mutant embryos, which we subsequently verified with DNA from heterozygotes. This results in a nonconservative amino acid change, tryptophan (hydrophobic) to arginine (basic), in the helicase domain of the protein (residue 520) (Fig. 1a,b). Based on a homology model that we termed ‘mut-snf2h’ (generated using Swiss-Model and the coordinates of the Rad54 protein (PDB code 1Z3I)), the mutated residue lies in a highly conserved α-helix on the surface of the protein (Supplementary Fig. 1 online). The mutation is unlikely to affect the overall fold of the protein but may affect local structure and/or the protein’s ability to participate in biomolecular interactions.
Figure 1.
Momme D4 is caused by a mutation in Smarca5, which encodes the ISWI chromatin remodeler Snf2h. (a) A single–base pair mutation (a T-to-A transversion) occurs in exon 12 of Smarca5 in homozygous Momme D4 embryos (MD4/MD4). (b) Schematic of the Snf2h protein structure. The point mutation in Momme D4 causes a nonconservative amino acid substitution of a large aromatic residue (tryptophan) with a basic residue (arginine). This residue is highly conserved in ISWI members across species. (c) RNA blot analysis performed on poly(A)+ mRNA does not show any difference in Smarca5 mRNA levels or transcript size between homozygous (MD4/MD4), heterozygous (MD4/+) and wild-type (+/+) individuals. Membranes were stripped and hybridized with mouse Gapdh. (d) Whole-cell lysate from 17.5-d.p.c. homozygous (MD4/MD4), heterozygous (MD4/+) and wild-type (+/+) Momme D4 embryos, probed with anti-Snf2h (top row). Membranes were also probed with anti-γ-tubulin (bottom row). (e) FACS data of offspring from a cross between Momme D4 heterozygotes and a Smarca5+/− knockout allele. The GFP transgene expression profile of erythrocytes from 3-week-old mice is shown. The x axis represents the erythrocyte fluorescence on a logarithmic scale, and the y axis is the number of cells detected at each fluorescence level.
To determine the consequences of this mutation on transcript and protein stability, we isolated RNA and protein from 17.5-d post coitum (d.p.c.) embryos from a Momme D4 heterozygous intercross. Parallel studies showed that homozygous mutant embryos were present throughout gestation. We developed a PCR genotyping assay based on the fact that the point mutation disrupts an NspI site (see Methods). RNA blot analysis demonstrated that the size and amount of Smarca5 mRNA transcripts were similar in wild-type, heterozygous and homozygous embryos (Fig. 1c). Protein blot analysis of lysates from wild-type, heterozygous and homozygous embryos with an antibody to Snf2h (anti-Snf2h) uncovered a protein of approximately 120 kDa, consistent with the expected mass, and the amount seemed unchanged across the different genotypes (Fig. 1d). A knockout allele of Smarca5 has been reported, and null embryos (Smarca5−/−) die at or around implantation14. We used genetic complementation to confirm that the W520R alteration was responsible for the Momme D4 phenotype. We mated female Momme D4 heterozygotes (Momme D4+/−) to male Smarca5 knockout heterozygotes (Smarca5+/−) and genotyped offspring at weaning. In a total of 51 offspring, we did not observe any compound heterozygotes. The ratios of the other genotypes, wild-type (n = 16), Momme D4+/− (n = 15) and Smarca5+/−(n = 20), were as expected. Furthermore, the Smarca5 knockout allele had a similar effect on transgene expression as that observed with the Momme D4 mutation, decreasing the percentage of expressing cells (Fig. 1e). We conclude that the Momme D4 allele is the result of a mutation in Smarca5 that we termed Smarca5MommeD4.
We performed embryonic dissections at 14.5 and 17.5 d.p.c. after a heterozygous intercross (n = 14 litters). We found homozygous Smarca5MommeD4 embryos in the expected proportions at both stages (Supplementary Table 2 online). At 17.5 d.p.c., 11 of 12 homozygous embryos showed abnormalities, including craniofacial defects and growth retardation (data not shown), suggesting that the homozygotes are unlikely to survive beyond birth. Given the survival of homozygous Smarca5MommeD4 embryos into late gestation, we believe that the T-to-A mutation has produced a hypomorphic allele.
Heterozygotes for the Smarca5MommeD4 mutation were viable and fertile but did show a subtle phenotype. At weaning, we weighed all offspring produced from a heterozygous Smarca5MommeD4 sire and wild-type female, and on average, both male and female Smarca5MommeD4 individuals were 10% smaller than wild-type littermates (Supplementary Table 2).
Smarca5MommeD4 shows a paternal effect
We recovered the Smarca5MommeD4 mutation from the screen based on its ability to modify variegation of a multicopy transgene array in red blood cells. We were keen to discover the effect of all mutations recovered from this screen on the expression of agouti viable yellow (Avy), a single-copy gene that, like the transgene, is known to be sensitive to epigenetic state. Transcription at Avy initiates in the long terminal repeat (LTR) of an intracisternal A-particle (IAP) retrotransposon15, and the protein product, agouti, influences coat color: the more agouti produced, the more yellow the coat. The coat color in isogenic Avy littermates varies from yellow through mottled to agouti and correlates with the level of DNA methylation at the LTR promoter16 (Fig. 2a). Furthermore, penetrance at the locus (that is, the proportion of yellow offspring) is sensitive to strain background17–20. The Avy allele is maintained on a C57BL/6J background, and because C57 mice are null (a/a) for agouti (hence the black coat), the single Avy allele in Avy/a individuals is responsible for the yellowness of the coat.
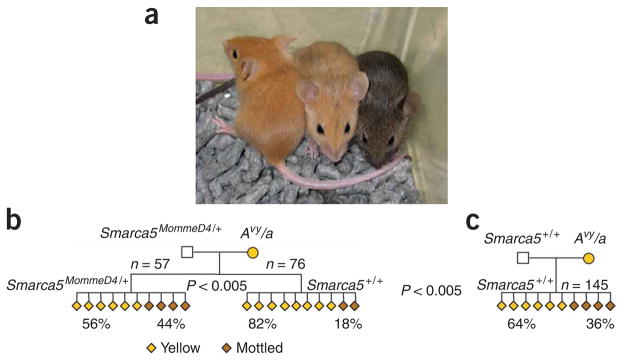
Figure 2.
Coat-color phenotypes after paternal transmission of Momme D4. (a) Isogenic mice carrying the agouti viable yellow allele show different coat colors (yellow, mottled and agouti) depending on the epigenetic state of the LTR promoter. (b) Pedigree produced from a Smarca5MommeD4/+ sire and yellow Avy/a dam. Offspring heterozygous for the Momme D4 mutation demonstrated a shift in penetrance at Avy toward mottled when compared with wild-type littermates (P < 0.005) (a subset of this data is shown in ref. 5). (c) The coat color phenotypes of wild-type offspring from a wild-type sire were compared with the wild-type offspring from a Smarca5MommeD4/+ sire. Both sets of offspring were genetically identical (Smarca5+/+; Avy/A); the only difference was that one group originated from gametes produced in a Smarca5MommeD4/+ sire. Wild-type offspring from Smarca5MommeD4/+ sires showed a significant increase in the proportion of animals with yellow coats when compared with the wild-type offspring from wild-type sires.
We set up crosses between FVB/NJ males heterozygous for the Smarca5MommeD4 allele with yellow C57BL/6J females heterozygous for the Avy allele. As reported previously, in offspring that inherit the Smarca5MommeD4 allele, we observed a shift in penetrance at Avy in a manner consistent with its role as an enhancer of variegation at the transgene locus. In other words, Smarca5MommeD4/+ offspring are less likely to be yellow than their wild-type littermates (ref. 5 and Fig. 2b). In this pedigree, the offspring are isogenic C57/FVB F1 hybrids. FVB/NJ mice carry a wild-type agouti locus, A, that is not a metastable epiallele and is not sensitive to epigenetic state. Avy is dominant over A20. We compared the wild-type animals (Smarca5+/+) with wild-type animals from a control cross (that is, one in which both parents were wild-type for Smarca5) and uncovered a paternal effect not previously reported (Fig. 2b,c). Namely, the range of coat colors of wild-type (Smarca5+/+) offspring produced from a Smarca5MommeD4/+ sire was not the same as the range seen in wild-type offspring from a wild-type (Smarca5+/+) sire, despite the fact that in all cases the offspring were genetically identical. They differed only with respect to the untransmitted genotype of the sire.
The wild-type animals from heterozygous sires were more likely to be yellow than the wild-type individuals from wild-type sires. This indicates that the role of paternally derived Snf 2h protein (or a Snf 2h-regulated product) in the early preimplantation embryo may differ from that of Snf 2h seen later in development. Snf 2h is known to be part of multiprotein complexes associated with both transcriptional activation and repression21. This paternal effect must be a direct or indirect consequence of mutant Snf 2h protein in wild-type male gametes.
Snf2h protein is present in spermatocytes and late spermatids
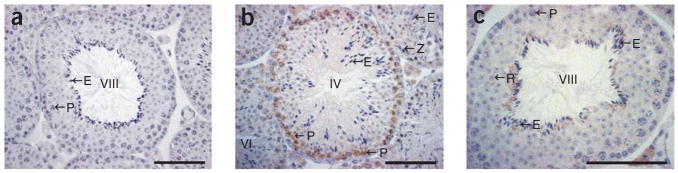
Snf2h is much more highly expressed in testes than in any other tissue22. Immunohistochemistry on testes of wild-type male mice showed that Snf2h protein was present in prophase I spermatocytes to stage VII pachytene spermatocytes (that is, before segregation of homologous chromosomes into haploid gametes) (Fig. 3). Modifications to the epigenome made at this stage would affect chromosomes entering both wild-type and mutant gametes, and those differences may be retained. We also found Snf2h in late elongated spermatids (Fig. 3). The progeny of each maturing spermatogonium remain connected to one another by cytoplasmic bridges, providing a second opportunity for sharing of molecules between mutant and wild-type gametes.
Figure 3.
Localization of Snf2h during spermatogenesis. (a) Preabsorbed control to indicate the specificity of immunohistochemical staining. (b,c) Snf2h protein was present in prophase I spermatocytes to stage VII pachytene spermatocytes and in terminally differentiating elongating spermatids. All other germ cell types and Sertoli cells were unstained. However, we observed Snf2h staining in the cytoplasm of Leydig cells within the interstitial space. Z = zygotene spermatocyte, P = pachytene spermatocyte, r = round spermatid, E = elongated spermatid. Scale bars = 100 μm.
The paternal effect reported here occurs as a result of events in the Smarca5MommeD4/+ testes, but the reporter allele, Avy, is inherited with the maternal set of chromosomes. This implies a trans effect some time after fertilization. Unlike the oocyte, sperm have relatively little cytoplasmic contribution to the fertilized zygote, but they do contribute RNA, and the only similar phenomenon previously reported in mammals is thought to be mediated by RNA molecules in the mature sperm23. Mice heterozygous for a lacZ insertion in Kit can transfer a silent epigenetic state to offspring with a wild-type Kit allele via Kit transcripts23. Recently, the presence of small (~30 nt) germline-specific RNAs have been reported in mouse testes24–27, some of which are homologous to the IAP LTR found at Avy (ref. 27). We prepared RNA from the testes of 4-week-old male mice from the Smarca5MommeD4 colony; RNA blot analysis did not uncover any abnormal small IAP transcripts (data not shown) or large IAP transcripts (Supplementary Fig. 2).
Although this does not preclude an RNA-based mechanism, we favor the idea that the paternal effect is mediated through a mechanism involving dosage of chromatin proteins. The paternal set of chromosomes from sperm produced from a Smarca5MommeD4/+ male, both wild-type and mutant, may carry abnormal populations of chromatin proteins (or epigenetic marks) into the zygote. Snf2h has been reported to have a critical role, along with Tif1α (also known as Trim28), in modulation of embryonic transcription in the mouse zygote28. Thus, reduced amounts of functional chromatin proteins could alter (either directly or indirectly) the epigenetic reprogramming that takes place at Avy after fertilization.
Momme D2 carries a mutation in DNA methyltransferase 1
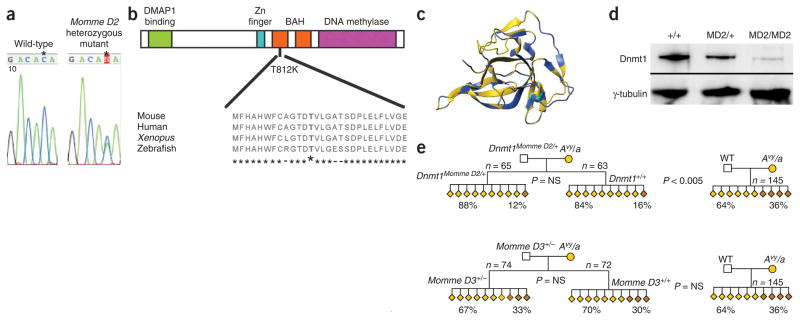
Momme D2 is a suppressor of variegation. In the presence of the Momme D2 mutation, the percentage of red blood cells expressing GFP increases5. The Momme D2 mutation is semidominant with respect to transgene variegation and is homozygous lethal5. Linkage analysis has been reported5, and experiments on 298 additional mice (168 mutants and 130 wild-types) confirmed this location. The linked interval is between the markers D9Mit62 (19242043 bp in Ensembl build 36) and D9Sar1 (21697069 bp in Ensembl build 36). This 2.5-Mb interval contains 77 genes, including Dnmt1, encoding DNA methyltransferase 1. Dnmt1 is the maintenance DNA methyltransferase in mice29. Sequencing the exons and exon-intron junctions of Dnmt1 in heterozygous Momme D2 mice uncovered a C-to-A transversion in exon 25 (Fig. 4a). This results in a nonconservative amino acid change (threonine to lysine) in a highly conserved region of the protein (Fig. 4b). Homology mapping (generated using Swiss-Model and the coordinates of the proximal bromo-adjacent homology (BAH) domain of the BAH1 protein (PDB code 1W4S)) shows that this residue lies in the center of the BAH domain of Dnmt1 and is likely to disrupt the structure of the protein (Fig. 4c). Protein analysis with anti-Dnmt1 on nuclear extracts from 9.5-d.p.c. embryos showed greatly reduced levels in the homozygous mutants (Fig. 4d), suggesting that the mutation has reduced the stability of the protein.
Figure 4.
Momme D2 is caused by a mutation in Dnmt1. (a) A single–base pair mutation (C-to-A transversion) occurs in exon 25 of Dnmt1 in heterozygous Momme D2 mice. (b) A schematic of the Dnmt1 protein structure. The point mutation in Momme D2 causes a nonconservative amino acid substitution in which a small polar residue (threonine) is replaced with a larger, basic residue (lysine). This residue is highly conserved in Dnmt1 orthologs. (c) Homology mapping, generated using Swiss-Model and the coordinates of the proximal bromo-adjacent homology (BAH) domain of the BAH1 protein (PDB code 1W4S) shows the relevant amino acid in the center of the BAH domain. (d) Nuclear lysate from individual 9.5-d.p.c. wild-type embryos (+/+), heterozygous embryos (MD2/+) and four pooled homozygous (MD2/MD2) embryos, probed with anti-Dnmt1 (top row). As a loading control, blots were probed with anti-γ-tubulin (bottom row). (e) Coat color phenotypes following paternal transmission of Momme D2. Pedigree produced from a Dnmt1MommeD2/+ sire and a yellow Avy dam. Wild-type offspring from a Dnmt1MommeD2/+ sire were more likely to be yellow than wild-type offspring from a wild-type sire. Both sets of offspring were genetically identical (Dnmt1+/+; Avy/A), except that one group originated from gametes produced in a Dnmt1MommeD2/+ sire. (f) Coat-color phenotypes following paternal transmission of Momme D3. Pedigree produced from a Momme D3+/− sire and a yellow Avy dam showed no paternal effects.
A knockout allele of Dnmt1 (Dnmt1n) that severely reduces expression of the protein has been reported; homozygous mutant embryos die around 9.5 d.p.c.30. We used genetic complementation to confirm that the T812K alteration was responsible for the Momme D2 phenotype. We mated female Momme D2 heterozygotes (Momme D2+/−) to male Dnmt1n heterozygotes (Dnmt1n/+) and genotyped offspring after birth. From a total of 39 offspring, we did not observe any compound heterozygotes. The proportions of the other genotypes (wild-type (n = 17), Momme D2+/− (n = 11) and Dnmt1n/+ (n = 11)) were as expected. We conclude that the Momme D2 allele is the result of a mutation in Dnmt1 that we termed Dnmt1MommeD2.
After a Dnmt1MommeD2 heterozygous intercross, all homozygous mutant embryos were severely developmentally delayed at 9.5 d.p.c. (n = 39 out of 119 embryos). DNA from 8.5-d.p.c. Dnmt1MommeD2 homozygous embryos showed marked hypomethylation of repeat sequences, including minor satellite and IAP retrotransposons (Supplementary Fig. 3 online). These findings are similar to those reported for Dnmt1n homozygous embryos30.
Dnmt1MommeD2 shows a paternal effect on expression of Avy
The Dnmt1MommeD2 allele showed a similar paternal effect as was seen with Smarca5MommeD4. Wild-type offspring from a Dnmt1MommeD2/+ sire were more likely to be yellow than wild-type offspring from a control cross in which both parents were wild-type for Dnmt1 (Fig. 4e). Dnmt1 protein is expressed at high levels in early prophase spermatocytes, providing an opportunity for haploinsufficiency to modify both the wild-type and the mutant gametes31. The effect of paternal haploinsufficiency of Dnmt1 is in the expected direction. The chromosomes may enter the zygote in a more active transcriptional state, with fewer ‘silencing’ factors, and may behave as sinks for such factors in the chromatin proteome of the zygote, sequestering them from sensitive alleles such as Avy. A study looking at the effects of reduced levels of Dnmt1 on expression of a related allele, Aiapy, did not detect paternal effects18. The latter study was carried out in a C57BL/6J background, and the Aiapy allele was transmitted paternally, not maternally. For reasons that are unclear, parent-of-origin effects and transgenerational epigenetic inheritance are more exaggerated in F1 hybrids19.
In this pedigree, we did not observe any difference in the range of coat colors in heterozygous mutants compared with their wild-type littermates (Fig. 4e). This may be a consequence of the fact that between fertilization and implantation, Dnmt1 is supplied mainly by the egg. Consistent with this idea, we and others have reported a maternal effect5,18.
Not all mutant lines produced in the dominant screen show paternal effects of this type. Momme D3 is a suppressor of variegation, is semidominant with respect to transgene variegation and is homozygous lethal5. The 6-Mb linked interval, between D11Mit320 (70565924 bp in Ensembl build 34) and D11Mit116 (76549381 bp in Ensembl build 34), contains over 144 genes, and the underlying gene has not yet been identified. Following paternal inheritance of Momme D3 and maternal inheritance of the Avy allele, we did not detect a paternal effect (Fig. 4f).
Dnmt3l shows paternal effects on sex chromosome ploidy
Dnmt3l is involved in the establishment of DNA methylation in germ cells, and expression of Dnmt3l is restricted to diploid prospermatogonia, which undergo a number of mitoses before meiosis. Complete loss of Dnmt3l causes demethylation of dispersed repeats and paternally imprinted loci and increased expression of retrotransposons in the premeiotic genome, followed by meiotic failure32,33. Males haploinsufficient for Dnmt3l are phenotypically normal and fertile, and as such, they provide an opportunity to search for paternal effects. Although abnormal meiosis has not been reported in Dnmt3l haploinsufficient testes, subtle changes to the methylation or chromatin state of the genome in premeiotic prospermatogonia could occur and be retained during and after meiosis. Hypomethylation in mice is associated with chromosome aneuploidy12,34. Although most aneuploidies are embryonic lethal and would not be detectable in adult offspring, abnormal numbers of sex chromosomes are compatible with life, and so we have focused our attention on the X chromosome.
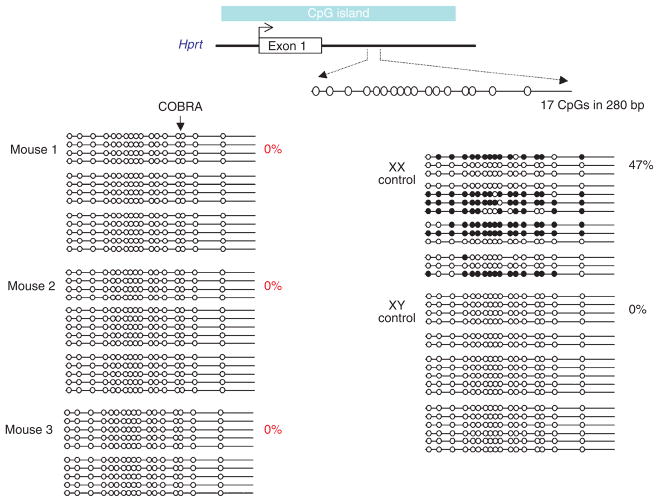
We backcrossed a Dnmt3l (also known as Dnmt3L) knockout line33 for ten generations onto C57BL/6J and maintained the colony using Dnmt3l+/− males crossed to wild-type females. We screened DNA from 148 adult female offspring (both Dnmt3l+/+ and Dnmt3l+/−) for abnormal methylation at Hprt on the X chromosome using combined bisulfite restriction analysis (COBRA) and identified three animals with an unmethylated profile (data not shown). We confirmed the lack of methylation by bisulfite sequencing (Fig. 5). We also tested the DNA at another X-linked locus (Mtm1) and, again, found virtually no methylation (1 in 168 CpG sites) (Supplementary Fig. 4 online). As expected, samples from normal 40,XX females contain some methylated clones at both regions (Fig. 5 and Supplementary Fig. 4). All three animals were negative for Sry and wild-type for Dnmt3l. Two of the three were sterile, and the fertility status of the third was unknown. These results suggest that these three animals carried only one X chromosome, as having two active X chromosomes leads to embryonic lethality35. Genome-wide copy number analysis on one of the samples using a comparative genomic hybridization (CGH) array verified a 39,XO genotype equivalent to Turner’s syndrome in humans (Fig. 6). A COBRA screen carried out on 214 adult females from a wild-type C57B/6J colony did not produce any such individuals.
Figure 5.
Hypomethylation of the X-linked Hprt CpG island in adult females from Dnmt3l+/− sires. Bisulfite sequence analysis of a 280-bp region located within intron 1 of the mouse Hprt locus, encompassing 17 CpG dinucleotides. Methylated (filled circles) and unmethylated (open circles) CpG dinucleotides are shown for a number of independently sequenced templates (horizontal lines). Each group of horizontal lines represents an independent bisulfite conversion and PCR reaction. The percentage of methylation in each mouse is shown (calculated from the number of methylated CpGs divided by the total CpGs sequenced, multiplied by 100). Clones were included in the samples only if they could be distinguished from others in the sample by non-CpG methylation. Any clones with >5% non-CpG methylation (an indication of incomplete bisulfite conversion) were excluded from the data set.
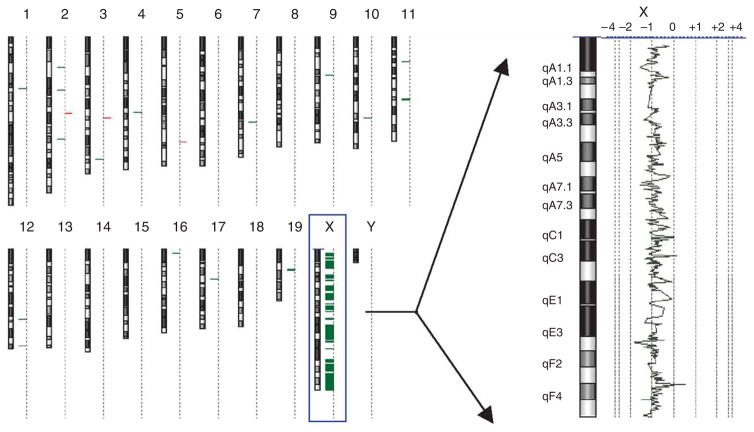
Figure 6.
A single sex chromosome in an adult female from a Dnmt3l+/− sire. Array comparative genomic hybridization (CGH) analysis of mouse 2. The genome view (left) and expanded X chromosome view (right) were created using Agilent CGH analytics software. The genome view shows an ideogram of each mouse chromosome, with a dotted vertical line beside each ideogram. Horizontal bars that extend to the right and left of this dotted line indicate gain and loss calls, respectively, by the software. The chromosome view shows a moving average plot with a 0.5-Mb window. The moving average is shifted from 0 to −1 along the entire X chromosome, indicating a single X chromosome.
In the cross described above, it was not possible to determine whether it was the maternal or the paternal X that was missing. To answer this question, we analyzed 10.5-d.p.c. embryos from FVB/NJ females mated to Dnmt3l+/− sires using X-linked microsatellite markers. Of 123 female embryos (as determined by absence of Sry), three (all phenotypically normal) showed loss of heterozygosity (LOH) at two independent X-linked markers (Table 1). All three were wild-type for Dnmt3l. In one case, the maternal X was absent, and in two cases, the paternal X or Y was absent (Table 1). Genome-wide analysis carried out on all three samples confirmed a 39,XO genotype (Fig. 7 and data not shown). We did not detect any loss of heterozygosity (LOH) of the X chromosome in 148 female embryos from wild-type C57B/6J sires mated to FVB/NJ females.
Table 1.
Stochastic sex chromosome LOH in F1 hybrid embryos
| XmY | XmXp | Xm | Xp | Total | |
|---|---|---|---|---|---|
| Dnmt3l+/− | 59 | 61 | 0 | 0 | 120 |
| Dnmt3l+/+ | 57 | 59 | 2 | 1 | 119 |
| Total | 116 | 120 | 2 | 1 | 239 |
Genomic DNA from F1 hybrid offspring of C57BL/6 Dnmt3l+/− sires crossed with FVB/N dams was prepared at 10.5 d.p.c. The presence or absence of the Y chromosome was determined by Sry PCR, and the Dnmt3l genotype was determined by PCR (see Methods). Two polymorphic microsatellite markers (DXMit117 and DXMit1) were used to determine parental origin of the X chromosomes. The maternal (Xm) and paternal (Xp) X chromosomes are indicated.
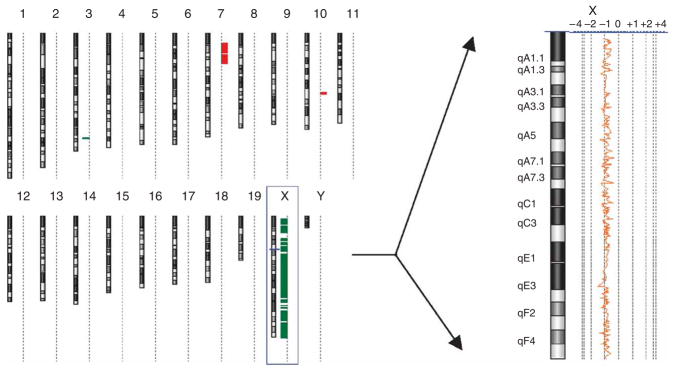
Figure 7.
A single sex chromosome in an F1 hybrid embryo from a Dnmt3l+/− sire. Array CGH analysis of a mouse with paternal X chromosome only (see Table 1). The genome view (left) and expanded X chromosome view (right) were created using Agilent CGH analytics software. The genome view shows an ideogram of each mouse chromosome, with a dotted vertical line beside each ideogram. Horizontal bars that extend to the right and left of this dotted line indicate gain and loss calls, respectively, by the software. The chromosome view shows a moving average plot with a 0.5-Mb window. In this mouse, the moving average is shifted from 0 to −1 along the entire X chromosome, indicating a single X chromosome.
The spontaneous occurrence of XO in the mouse is reported to be extremely low36, and our data are consistent with this: we did not find any in 214 females from the control cross (<0.5%). We show that spontaneous sex chromosome aneuploidy occurs more frequently in offspring from Dnmt3l haploinsufficient sires: it occurs in 2% of females in the inbred background and 2.4% in the F1 hybrid background. In at least one case, we know that the event must have occurred post-fertilization, as it is associated with loss of the maternal X. All offspring that showed abnormal sex chromosome complements were wild-type for Dnmt3l; hence, these are paternal effects. This study was designed to detect nonmosaic chromosome loss in somatic cells of the individual, (that is, either germline or zygotic loss). Additional mosaic loss of X resulting from post-zygotic events is possible.
These studies raise the possibility that decreased DNA methylation at repeats and imprinted loci would be detectable in the sperm of males haploinsufficient for Dnmt1 or Dnmt3l. Mice null for Dnmt3l show such effects32. We have analyzed the methylation state of these regions in DNA purified from sperm of Dnmt1MommeD2/+ and Dnmt3l+/− males and did not detect any changes (Supplementary Fig. 5 online). The paternal effects reported here at Avy are subtle; this allele is known to be particularly sensitive to epigenetic state5, and the paternal effects on sex chromosome aneuploidy are infrequent. Hence, we were not surprised at our failure to detect global methylation changes elsewhere. Furthermore, we and others have not detected changes in the methylation state of dispersed repeats in somatic tissues of mice haploinsufficient for these proteins (Supplementary Fig. 3 and ref. 34).
DISCUSSION
Here we have used mouse mutants produced in a dominant screen for modifiers of epigenetic reprogramming to test the hypothesis that such genes will show paternal effects. It is commonly believed that if a genetic mutation is not passed on to the offspring, the offspring will be unaffected. In fact, there are a handful of reports that maternal deficiency of genes encoding proteins required in early embryogenesis can result in lethality for genetically wild-type offspring4,37–42. These are called maternal effect genes. Equivalent paternal effect genes have been reported previously in D. melanogaster but have not been reported in mammals, to our knowledge. The four paternal effect genes in D. melanogaster are sneaky, K81, paternal loss (pal) and horka; with the exception of pal, all are associated with male sterility3. Notably, the K81 protein is believed to have a role in the repackaging of sperm chromatin. PAL, a small basic protein that localizes to the chromosomes, is detected in the nuclei of elongating spermatids. A proportion of progeny of pal mutant males show paternal chromosome loss. The horka mutation, like pal, induces mitotic loss of paternal chromosomes in the progeny43.
Here we report that Smarca5 (which encodes chromatin remodeler Snf2h) and Dnmt1 (which encodes a DNA methyltransferase), genes that were identified initially because of their role in epigenetic reprogramming, are paternal effect genes in the mouse. At these loci, the untransmitted mutant genotype of the sire influences the phenotype of his offspring through epigenetic changes initially established in his gametes that go on to act on gene expression at sensitive alleles in his offspring. This finding is consistent with the notion that epigenetic information is inherited across generations in mammals.
The haploid genome is compacted within the sperm head by the replacement of histones with protamines, and protamines undergo posttranslational modifications, the function of which is unknown. Moreover, some histones and other chromatin proteins, such as heterochromatin protein 1 (HP1), remain44,45. Furthermore, despite the marked demethylation of the paternal genome in the zygote, there is evidence that some regions are exempt46. The paternal effects on transcriptional activity of a maternally inherited Avy allele are effects in trans and are consistent with the notion that the epigenome is in a dynamic equilibrium: a change in the amount of modifier proteins can shift the balance between euchromatin and heterochromatin at a reporter locus6,7. Recent studies in cell lines using fluorescence recovery after photobleaching (FRAP) have shown that HP1 is highly dynamic within the nucleus47. The reprogramming of the paternal set of chromosomes in the zygote is extensive and dependent on the proteome of the zygote. Although we cannot rule out the possibility that RNA is involved, the simplest hypothesis is that disruption to the amount or functionality of proteins involved in establishing and maintaining epigenetic state have modified the epigenome of wild-type gametes, bringing a compromised set of chromosomes into the zygote.
We have also found that Dnmt3l shows paternal effects on sex chromosome aneuploidy in adult and embryonic offspring. In one case, this loss must have occurred after fertilization, suggesting that the gamete had a full set of chromosomes. In two cases, it involves loss of the paternal X, which may have occurred in the gamete of the haploinsufficient testis or later (zygotic) as a consequence of an epigenetically compromised set of chromosomes. The fact that disruption to the epigenome affects genome stability is well recognized and reminiscent of the paternal effect gene phenotypes seen in D. melanogaster.
In this study, we have specifically focused on adult phenotypes. Nevertheless, it is likely that paternal effects of this type will increase the risk of early embryonic death, as reported in D. melanogaster and as such may turn out to be associated with reduced fertility. Such effects have not been systematically studied in humans. The notion that untransmitted alleles can have such effects is rarely considered, but if true, it would provide another opportunity to improve estimates of disease risk in extended families and explain sporadic disease in some cases.
METHODS
Mouse strains and genotyping
Wild-type inbred C57BL/6J and FVB/NJ mice were purchased from ARC Perth. All F1 hybrid offspring were produced by natural matings, and detection of a vaginal plug was called 0.5 d.p.c. Procedures were approved by the Animal Ethics Committee of the University of Sydney and the Animal Ethics Committee of the Queensland Institute of Medical Research. The ENU screen was carried out in the FVB/NJ inbred transgenic line, as described previously5. The resultant Momme D mutant lines were maintained in this background, unless stated otherwise. The Smarca5 knockout line was obtained and maintained on a C57BL/6J background and genotyped for the presence of the modified locus, as described previously14. The Dnmt3l knockout line was produced on a mixed 129/Sv, C57BL/6J background and was backcrossed for ten generations onto C57BL/6J. The Dnmt3l knockout allele was detected by PCR using primers specific for the neo cassette, as described at the Jackson Laboratory website (see URL below). The Avy allele was obtained and maintained on a C57BL/6J background. All agouti-colored mice were genotyped by PCR to assess whether they were Avy/A or A/a, as reported19. The presence of the Sry gene was detected by PCR, as described48. DXMit117 and DXMit1 PCRs were performed as described previously5 (Supplementary Table 3 online) with an annealing temperature of 55 °C.
Bisulfite analyses
For Hprt, we performed bisulfite conversion of DNA as previously described19, except that bisulfite treatment was for 40 h. Nested PCR was used to amplify a section of the mouse Hprt 5′ CpG island (N. Brockdorff, personal communication). The first-round primers were Hprt4-L and Hprt4-R, and the second-round primers were Hprt4-Lint and Hprt4-Rint (Supplementary Table 3). The expected PCR products were 389 bp (first round) and 280 bp (second round). Cycling parameters for both included an initial denaturation at 94 °C (5 min) followed by 30 rounds of denaturation at 94 °C (1 min), annealing for 1 min and extension at 72 °C for 1 min. A final extension step was carried out at 72 °C for 5 min. The annealing temperature was 50 °C for the first round and 57 °C for the second round. For Mtm1, we used nested PCR to amplify a section of the mouse Mtm1 CpG island for bisulfite sequencing (N. Brockdorff, personal communication). The first-round primers were Mtm1SEQ-L and Mtm1SEQ-R. Two different sets of second-round primers were used. They are Mtm1SEQLshort and Mtm1SEQRshort or Mtm1L and Mtm1R (Supplementary Table 3). The expected PCR product was 851 bp for the first round and either 430 bp (using primers Mtm1SEQL and Mtm1SEQRshort) or 409 bp (using primers Mtm1L and Mtm1R) for the second round. Cycling parameters for both included an initial denaturation at 94 °C for 5 min followed by 30 rounds of denaturation at 94 °C for 1 min, annealing for 1 min and extension at 72 °C for 1 min. A final extension step was carried out at 72 °C for 5 min. The annealing temperature was 68 °C for the first round and either 60 °C (using primers Mtm1SEQL and Mtm1SEQR-short) or 57 °C (using primers Mtm1L and Mtm1LR) for the second round. We carried out sperm COBRA analysis as described in ref. 49, with the exception of the DNA bisulfite conversion, which was carried out as described in ref. 50.
Genotyping Smarca5MommeD4 and Dnmt1MommeD2
Once the point mutation in Momme D4 had been identified, we carried out genotyping by NspI digestion of a PCR product from primers MD4L and MD4R (Supplementary Table 3). Once the point mutation had been identified in Momme D2, we carried out genotyping by HpyCH4III digestion of a PCR product from primers MD2L and MD2R (Supplementary Table 3).
Crosses between Momme D mutants and mice carrying the Avy allele
We mated FVB/NJ male mice heterozygous for the Momme D4 or Momme D2 mutation and homozygous for the GFP transgene with C57BL/6J mice heterozygous for the Avy allele. The coat color phenotype was classified at weaning by a trained observer. This observer had no knowledge of the genotype (with respect to the Momme mutation). We determined the percentage of yellow, mottled and agouti coat color on each mouse (front and back) and placed the mouse into one of five categories: yellow (>95% yellow Y), yellowmottled (between 75–95% yellow Y), mottled (<75% yellow and >25% yellow M), pseudoagoutimottled (between 75–95% agouti M) or pseudoagouti (>95% agouti ΨA).
These were then pooled into three categories (Y, M or ΨA). Subsequently, we assessed GFP expression by flow cytometry. FVB/NJ mice carry the A locus, and C57BL/6J carry the a locus. Yellow and mottled mice carried the Avy allele. We genotyped all agouti-colored mice by PCR to determine whether they were Avy/A or A/a19.
Protein analysis
We prepared nuclear extracts from 9.5-d.p.c. Dnmt1MommeD2 embryos genotyped from DNA prepared from the yolk sac and 17.5-d.p.c. Snf2hMommeD4 embryos genotyped from a tissue sample after heterozygous intercrosses. Approximately 5 μg were separated by SDS-PAGE on a 4%–12% gradient gel (Invitrogen) and were analyzed with a polyclonal antibody to Dnmt1 (sc-20701, Santa Cruz Biotechnology) or Snf2h (ab3749, Abcam).
DNA blot analysis of DNA methylation
After a heterozygous intercross, DNA from Momme D2 wild-type, heterozygous and homozygous mutant embryos was digested with MspI and HpaII, separated on an agarose gel and hybridized with a minor satellite and IAP probe (a 1.6-kb PstI-EcoRI fragment of a IΔ1 IAP (accession number DQ436346)).
RNA blot analysis
Poly(A)+ RNA was purified from 17.5-d.p.c. whole embryos after a Smarca5MommeD4 heterozygous intercross or from 4-week old testes. RNA was separated, transferred and hybridized with a fragment amplified from exon 11 of Smarca5 using primers (Supplementary Table 3) or the IAP probe (a 1.6-kb PstI-EcoRI fragment of an IΔ1 IAP (accession number DQ436346).
Immunohistochemistry of germ cells
Snf2h protein was detected in adult mouse testes fixed with Bouin’s using methods described previously37 using 1 μg ml−1 of anti-Snf2h (Abcam). The specificity of staining was determined by pre-absorbing the antiserum with a 200-fold excess (wt/wt) of the immunizing peptide before the immunohistochemical procedure.
Flow cytometry
GFP fluorescence in red blood cells was analyzed by flow cytometry at weaning. A drop of blood was collected in Osmosol buffer and analyzed on a FACSCalibur (Becton Dickinson) with excitation at 488 and 550 nm. The 488 nm channel predominantly measures GFP fluorescence, and the 550 nm channel measures autofluorescence. The data were analyzed using CELL QUEST software with a GFP-positive gate set to exclude 99.9% of wild-type erythrocytes. Mean fluorescence was calculated using cells within the positive gate. Histograms depict only the GFP fluorescence channel.
Oligonucleotide array CGH
We used mouse genome CGH microarray 44A slides (Agilent Technologies) according to the manufacturer’s instructions. The reference sample in each experiment was DNA isolated from the same tissue of a wild-type female mouse of the same strain background but maintained in separate colonies. Microarrays were scanned by a laser confocal scanner (Agilent Technologies), and Feature Extraction software was used to measure the fluorescence intensities at the target locations (Agilent Technologies).
URLs
Jackson Laboratory: http://www.jax.org
Accession codes
PDB: BAH1, 1W4S; Rad54, 1Z3I. Microarray data can be found at the Gene Expression Omnibus (GEO) under accession code GSE7384.
Supplementary Material
Acknowledgments
This study was supported by National Health and Medical Research Council of Australia grants to E.W., D.D.K., H.S., J.M. and M.O’B. and an Australian Research Council grant to S.C. and E.W. M.B., A.A. and N.Z. were supported by Australian Postgraduate Awards. N.V. was supported by an International Postgraduate Award (University of Sydney). N.Y. and E.W. were supported by fellowships from the Queensland Institute of Medical Research.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
S.C., N.V., A.A., N.Z., N.Y., S.H. and M.B. performed experiments and provided intellectual input. T.S., A.S. and H.S.S. provided backcrossed mice. D.D.K., M.O’B., J.M., H.S.S. and E.W. provided intellectual input. S.C., N.V., A.A., N.Y. and E.W. wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Chong S, Whitelaw E. Epigenetic germline inheritance. Curr Opin Genet Dev. 2004;14:692–696. doi: 10.1016/j.gde.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Rakyan V, Whitelaw E. Transgenerational epigenetic inheritance. Curr Biol. 2003;13:R6. doi: 10.1016/s0960-9822(02)01377-5. [DOI] [PubMed] [Google Scholar]
- 3.Fitch KR, Yasuda GK, Owens KN, Wakimoto BT. Paternal effects in Drosophila: implications for mechanisms of early development. Curr Top Dev Biol. 1998;38:1–34. doi: 10.1016/s0070-2153(08)60243-4. [DOI] [PubMed] [Google Scholar]
- 4.Payer B, et al. Stella is a maternal effect gene required for normal early development in mice. Curr Biol. 2003;13:2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 5.Blewitt ME, et al. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc Natl Acad Sci USA. 2005;102:7629–7634. doi: 10.1073/pnas.0409375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locke J, Kotarski MA, Tartof KD. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics. 1988;120:181–198. doi: 10.1093/genetics/120.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spofford JB. In: The Genetics and Biology of Drosophila. 1c. Ashburner M, editor. Academic; London: 1976. pp. 955–1018. [Google Scholar]
- 8.Fiering S, Whitelaw E, Martin DI. To be or not to be active: the stochastic nature of enhancer action. Bioessays. 2000;22:381–387. doi: 10.1002/(SICI)1521-1878(200004)22:4<381::AID-BIES8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 9.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 10.Reuter G, Spierer P. Position effect variegation and chromatin proteins. Bioessays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- 11.Dodge JE, et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- 12.Gaudet F, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 13.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 14.Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci USA. 2003;100:14097–14102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 16.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 17.Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2006;2:e49. doi: 10.1371/journal.pgen.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudet F, et al. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol Cell Biol. 2004;24:1640–1648. doi: 10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rakyan VK, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolff GL. Influence of maternal phenotype on metabolic differentiation of agouti locus mutants in the mouse. Genetics. 1978;88:529–539. doi: 10.1093/genetics/88.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukiyama T. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat Rev Mol Cell Biol. 2002;3:422–429. doi: 10.1038/nrm828. [DOI] [PubMed] [Google Scholar]
- 22.Lazzaro MA, Picketts DJ. Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J Neurochem. 2001;77:1145–1156. doi: 10.1046/j.1471-4159.2001.00324.x. [DOI] [PubMed] [Google Scholar]
- 23.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 24.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 25.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 26.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres-Padilla ME, Zernicka-Goetz M. Role of TIF1alpha as a modulator of embryonic transcription in the mouse zygote. J Cell Biol. 2006;174:329–338. doi: 10.1083/jcb.200603146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruenbaum Y, Cedar H, Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295:620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 30.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 31.La Salle S, et al. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev Biol. 2004;268:403–415. doi: 10.1016/j.ydbio.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 32.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 33.Webster KE, et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci USA. 2005;102:4068–4073. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 35.Takagi N, Abe K. Detrimental effects of two active X chromosomes on early mouse development. Development. 1990;109:189–201. doi: 10.1242/dev.109.1.189. [DOI] [PubMed] [Google Scholar]
- 36.Cattanach BM. Genetic disorders of sex determination in mice and other mammals. In: Motulsky A, Lenz W, editors. Birth Defects; Proceedings of the Fourth International Conference; Amsterdam: Excerpta Medica; 1974. pp. 129–141. [Google Scholar]
- 37.Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal effect of Hsf1 on reproductive success. Nature. 2000;407:693–694. doi: 10.1038/35037669. [DOI] [PubMed] [Google Scholar]
- 38.Dean J. Oocyte-specific genes regulate follicle formation, fertility and early mouse development. J Reprod Immunol. 2002;53:171–180. doi: 10.1016/s0165-0378(01)00100-0. [DOI] [PubMed] [Google Scholar]
- 39.Gurtu VE, et al. Maternal effect for DNA mismatch repair in the mouse. Genetics. 2002;160:271–277. doi: 10.1093/genetics/160.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howell CY, et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 41.Tong ZB, et al. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000;26:267–268. doi: 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- 42.Wu X, et al. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 2003;33:187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
- 43.Szabad J, Mathe E, Puro J. Horka, a dominant mutation of Drosophila, induces nondisjunction and, through paternal effect, chromosome loss and genetic mosaics. Genetics. 1995;139:1585–1599. doi: 10.1093/genetics/139.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 45.van der Heijden GW, et al. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol. 2006;298:458–469. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 46.Lane N, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 47.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 48.McClive PJ, Sinclair AH. Rapid DNA extraction and PCR-sexing of mouse embryos. Mol Reprod Dev. 2001;60:225–226. doi: 10.1002/mrd.1081. [DOI] [PubMed] [Google Scholar]
- 49.Li JY, Lees-Murdock DJ, Xu GL, Walsh CP. Timing of establishment of paternal methylation imprints in the mouse. Genomics. 2004;84:952–960. doi: 10.1016/j.ygeno.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Arnaud P, et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum Mol Genet. 2003;12:1005–1019. doi: 10.1093/hmg/ddg110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.