Abstract
Background
There are several studies reporting association of alpha-4 nicotinic acetylcholine receptors (encoded by CHRNA4) with nicotine dependence (ND). A meta-analysis of genomewide linkage studies for ND implicated a single chromosomal region, which includes CHRNA4, as genomewide significant.
Methods
After establishing that common variants are unlikely to completely account for this linkage, we investigated the distribution of CHRNA4 rare variants by sequencing the coding exons and flanking intronic regions of CHRNA4 in 209 European American (EA) ND cases and 183 EA controls. Because most of the rare variants that we detected (and all nonsynonymous changes) were in exon 5, we sequenced exon 5 in an additional 1000 ND cases and 1000 non-ND comparison subjects, both of which included equal numbers of EAs and African Americans (AAs).
Results
Comparison subjects had a higher frequency of rare nonsynonymous variants in the exon 5 region (encoding the large intercellular loop of the α4 subunit) (Fisher’s exact test p=0.009; association test p=0.009, OR=0.43; weighted-sum method p=0.014), indicating a protective effect against ND. Considering data from the two stages combined and only nonsynonymous variants predicted to alter protein function, the association was stronger (Fisher’s exact test p=0.005; association test p=0.008, OR=0.29). SPECT imaging results were consistent with functionality.
Conclusions
CHRNA4 functional rare variants may reduce ND risk. This is the first demonstration that rare functional variants at a candidate locus protect against substance dependence, suggesting a novel mechanism of substance dependence heritability that is potentially of general importance.
Keywords: Nicotine dependence, rare variants, nonsynonymous, CHRNA4, imaging genetics, deep sequencing
Introduction
It is estimated that 18.1% of total US deaths were caused by tobacco in 2000, which makes tobacco smoking the leading cause of mortality (1). Despite increasing awareness of the health risks caused by smoking and efforts at tobacco-use prevention and control, the prevalence of smoking has declined slowly.
The difficulty people experience in quitting smoking is an expression of nicotine addiction. The reinforcing actions of nicotine are mediated by binding to nicotinic acetylcholine receptors (nAChRs) in the central nervous system. nAChRs are ligand-gated ion channels that respond to the neurotransmitter acetylcholine (ACh), as well as to nicotine. Chronic exposure of nAChRs to nicotine stabilizes a desensitized, closed state, decreasing receptor function, and increases the assembly of high-affinity receptors (2). Assembled, functional nAChRs are composed of combinations of five subunits that are encoded by 17 genes. In the brain, the majority of nAChRs are composed of α4β2 hetero-oligomers that have high affinity for nicotine (3).
Studies in rodent models and human subjects have demonstrated that α4β2 nAChRs play an important role in nicotine dependence (ND). First, activity of α4β2 nAChRs contributes to nicotine self-administration. Pretreatment with a nicotinic receptor antagonist dihydro-β-erythroidine, which blocks the α4β2 subtype, decreases nicotine self-administration in rats (4). Consistent with this finding, β2 knock-out mice, which lack high-affinity nAChRs, do not maintain self-administration of nicotine (5). Second, α4β2 receptors activate mesolimbic dopamine systems that contribute to the rewarding effects of nicotine. For example, activation of α4β2 receptors in the ventral tegmental area increases extracellular dopamine levels in the nucleus accumbens (6,7), and neither β2 nor α4 knock-out mice exhibit nicotine-elicited increases in dopamine levels (5,8). Third, experiments with gain-of-function α4* nAChR knock-in mice demonstrate that α4* nAChR activation is sufficient for nicotine-induced reward, tolerance, and sensitization (9). Finally, in humans, a partial agonist of α4β2 nAChRs, varenicline, is efficacious for smoking cessation (10).
In recent years, genome-wide association studies (GWAS) identified several regions containing genes associated with smoking-related behaviors, especially a cluster of three nAChR subunit genes, CHRNA5, CHRNA3 and CHRNB4 (11-13). Despite the important functions of α4* nAChR in ND, no GWAS yielded a significant signal near CHRNA4, though several candidate gene studies reported significant associations of CHRNA4 and smoking behaviors (14-20).
Genomewide linkage studies (GWLS) identified numerous chromosomal regions linked to ND. A recent meta-analysis of GWLS of the maximum number of cigarettes smoked in a 24-hour period yielded only one genome-wide significant linkage signal at 20q13.12-q13.32 in European-ancestry samples; this peak overlies CHRNA4 (21).
Rare variants may be important reservoirs of risk for complex genetic traits, including neuropsychiatric traits (22,23). However, the role of rare variants is largely unexplored for ND. Because of the inconsistency of the evidence common polymorphisms in CHRNA4 as contributors to ND risk and the functional importance of the α4 subunit of the nAChR for nicotine actions, we hypothesized that rare variants in CHRNA4 modulate risk for ND.
In this study, we investigated rare sequence variants of CHRNA4 in nearly 1200 ND cases and 1200 comparison subjects. The effect of some of the rare variants identified on availability of α4β2 nAChRs was assessed by means of iodide 123-labeled 5-iodo-A-85380 ([123I]5-IA) single-photon emission computed tomography (SPECT) imaging. [123I]5-IA binds with relative selectivity to α4β2 nAChRs (24) and thus can be used to measure the availability of α4β2 nAChRs (25). Here, we report that nonsynonymous rare variants in the exon 5 region encoding the cytoplasmic loop are protective for ND, with the strongest evidence for the African American (AA) population. We also found that at least a subset of these rare variants in CHRNA4 alters availability of high affinity nAChRs in human brain.
Methods and Materials
Subjects
Subjects were recruited for linkage and association studies of the genetics of cocaine, opioid, and alcohol dependence (26,27). The participants were recruited at 5 sites: the University of Connecticut Health Center (538 ND cases and 467 comparison subjects), Yale University School of Medicine (432 ND cases and 428 comparison subjects), the Medical University of South Carolina (54 ND cases and 255 comparison subjects), the University of Pennsylvania School of Medicine (133 ND cases and 16 comparison subjects), and McLean Hospital of Harvard Medical School (52 ND cases and 17 comparison subjects). Written informed consent was obtained from all participants. The institutional review board at each of the participating sites approved the study protocol. All subjects were interviewed by trained interviewers using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) (28,29), which yields diagnoses for a variety of psychiatric and substance use disorders. The SSADDA diagnosis of ND is lifetime and based on DSM-IV. The reliability of the SSADDA for the lifetime diagnosis of ND was excellent, with interrater and test-retest reliability [κ] of 0.77 and 0.97, respectively (28). Subjects affected with major psychiatric illnesses, such as schizophrenia, schizoaffective disorder, or mental retardation, were excluded.
We divided the subjects into 2 groups: the “discovery” sample and the “test” sample. The discovery group contained 209 ND cases and 183 controls, all of whom were self-reported European Americans (EAs), whose population assignment was confirmed by the analysis of ancestry-informative markers (AIMs) using STRUCTURE software (30-32). The 209 ND cases were affected siblings recruited from 114 families. Selecting cases with affected family members was expected to increase power, because rare risk variants could co-segregate with disease status, and thus the probability of cases carrying rare risk variants would increase. The majority of the cases (87%) had comorbid cocaine, opioid, or alcohol dependence. The controls were all unrelated and unaffected individuals. The mean age of the cases (53.1% male) was 37.2 (SD = 9.8) years, and of the controls (53.0% male) was 37.6 (SD = 15.0) years.
The test group contained 1000 unrelated ND cases and 1000 unrelated comparison subjects, including controls and subjects without ND, but with another substance dependence. Five hundred cases and 500 comparison subjects were EAs, and an additional 500 cases and 500 comparison subjects were AAs. As with the discovery sample, the race of the subjects was confirmed by the analysis of AIMs data. For ND cases, 97.8% were comorbid with cocaine, opioid, or alcohol dependence (49.4% had opioid dependence, 54.3% had alcohol dependence and 85.1% had cocaine dependence). Of the comparison subjects, 27.9% had another substance dependence diagnosis (mostly alcohol dependence). The mean age of the cases (56.7% male) was 40.0 (SD = 9.8) years, and of the comparison subjects (42.1% male) it was 38.8 (SD = 13.1) years.
A total of 139 subjects (92 smokers and 47 non-smokers) participated in [123I]5-IA SPECT scans. Subjects had no history of psychiatric, neurological, or medical disease and no history of drug or alcohol dependence except for ND. Smokers were required to abstain from smoking for 7-9 days prior to their SPECT scans and were instructed that they could not use any forms of nicotine replacement therapy or medication throughout the study. Abstinence from smoking was confirmed by urine cotinine and breath carbon monoxide levels monitored daily for the first 8 days of smoking cessation and a minimum of twice weekly thereafter. Subjects were administered equivalent doses of [123I]5-IA as a bolus to constant infusion at a ratio of 7.0 for 8 hours, and were imaged at equilibrium, between 6 and 8 hours. Detailed methods for the imaging study were described previously (25). Written informed consent was obtained from all participants.
Sequencing and Genotyping
The CHRNA4 gene is more than 17 kb long and contains 6 exons spanning a total of 5,486 bp. The mRNA is 1884 bp, encoding a protein of 627 amino acids. The largest exon among the 6 coding exons is exon 5, which is 1375 bp. As with other nAChR subunits, the polypeptide chain of α4 has a large extracellular N-terminal domain, which is glycosylated. It contains 4 transmembrane domains; a large cytoplasmic loop is between the third and fourth transmembrane domains. A short C-terminal domain is extracellular. According to Uniprot, there are 270 amino acids in the cytoplasmic loop, and exon 5 codes the first 256 of these amino acids.
We designed primers for polymerase chain reaction (PCR) (see Supplement) based on the Ensembl sequences (ENSG00000101204). After PCR amplification, products were purified using Exonuclease I and shrimp alkaline phosphatase. Sequence analysis was carried out on Applied Biosystems 3730 capillary instruments at the Yale Keck core facility. In the discovery stage, PCR was performed for all of the coding exons and flanking intronic regions of CHRNA4. Based on the results from the discovery samples, only exon 5 and its flanking intronic regions were sequenced in the test stage and the imaging study. All rare variants detected were confirmed by independent PCR and sequencing from the other direction of the amplicons. Some PCR conditions were more difficult to optimize than others based on the quality of the DNA template, so the number of individuals for whom we could get Sanger sequencing data differed for each amplicon. The success rate for the sequencing was 80-99.6%.
Forty-one AIMs were genotyped to analyze the race of each subject. These 41 markers included 36 short tandem repeats markers and 5 SNPs, all of which are highly ancestry-informative. The detailed information has been described previously (33,34).
Data analysis
STRUCTURE software (30-32) set at 500,000 burn-in iterations and 500,000 repeats, was used to analyze the AIMs data. Subjects with ancestry proportion scores ≥ 0.50 were classified as AA, and those with ancestry proportion score <0.50 were classified as EA. The results were highly concordant with self-reported race.
DNA sequences were analyzed by SeqScape v2.6 (ABI). Automated sequence analysis was performed as follows to validate the results from SeqScape: Sequence chromatograms were loaded into CodonCode Aligner (CodonCode Corperation) to determine homozygous and heterozygous calls using the built-in phred program. Phred-derived sequences were then converted into fasta, and uploaded into the Galaxy (35) tool for sequence alignment against the CHRNA4 genomic sequence.
To increase statistical power, we used a collapsing method, in which the total number of rare nonsynonymous variants in cases and comparison subjects was compared using Fisher’s exact test (FET). The total number of chromosome sequenced was based on the assumption that each individual has two chromosomes (and two copies) of this gene. To account for the variable sample size sequenced for each rare variant, the sample size was adjusted using harmonic mean: N = n /(∑1/ Ni ) , where Ni is the sample size of the i th variant and n is the number of variants. The number of each rare variant was adjusted as ∑ pi * N , where pi is the frequency of the i th variant. In addition to the collapsing method, we also used a weighted-sum method (36) (WSM) to test for the association of rare variants and ND.
PolyPhen (37) was used to predict the effects of nonsynonymous variants on protein structure and function. PolyPhen gives three predictions: benign, possibly damaging, and probably damaging. We considered rare nonsynonymous variants with a prediction of possibly damaging or probably damaging to be functional.
For the imaging study, images were reconstructed and analyzed as previously described including a nonuniform attenuation correction (25). We chose brain regions that were known to express α4β2 nAChRs, including frontal, parietal, anterior cingulate, temporal and occipital cortices, thalamus, striatum (an average of caudate and putamen) and cerebellum. The outcome measure VT/fP (regional activity divided by free plasma parent between 6 and 8 hours) was used (38). For each rare variant carrier, the availability of α4β2 nAChRs was compared to a group of 4 to 6 age- and smoking status-matched non-carriers.
Results
Sequencing the coding exons of CHRNA4 in the discovery sample set
We identified 31 unique single-nucleotide variants in the coding exons and flanking intronic regions of CHRNA4 in the discovery cohort. Of these variants, 21 were exonic, including 8 types of SNPs (minor allele frequencies (MAF) > 5%) and 13 types of rare variants (MAF ≤ 1%, Figure 1A). None of the variants were found to have MAF between 1% and 5%. Six of the 13 rare variants identified were nonsynonymous substitutions, and all were missense mutations. Two individuals from the control group seemed to be homozygous for rare nonsynonymous variants. However, due to the rarity of these variants, the probability of being homozygous is low; it is possible that these two controls had a deletion (e.g., an overlapping CNV) in one chromosome over that region. The following statistical analyses considered these “homozygous” sites as monoallelic, to be conservative. In total, we observed 11 rare nonsynonymous variants (Table 1), which did not differ in frequency between cases and controls (FET p=0.56, WSM p=0.30).
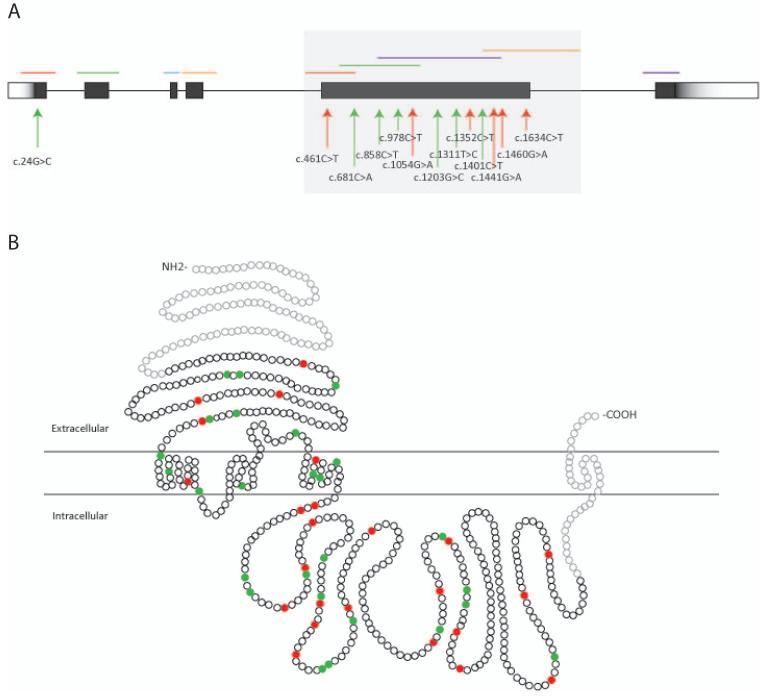
Figure 1.
Locations of CHRNA4 rare variants identified in the discovery and the test samples.
(A) Schematic genomic structure of CHRNA4 spanning 17 Kb and 6 coding exons (dark boxes). Red arrows point to the identified rare nonsynonymous variants in the discovery samples; green arrows point to the identified rare synonymous variants in the discovery samples. The bars on the top show the PCR products. The dark grey region covering exon 5 was sequenced in the test samples.
(B) Schematic depiction of the α4 nAChR subunit. Exon 5 was sequenced in the test samples, which encodes 114 amino acids of the extracellular domain, all of the first three transmembrane domains, and 256 amino acids of the cytoplasmic loop. Amino acids encoded by the gene regions not sequenced in the test stage are depicted in light grey. Red dots represent the amino acids affected by rare nonsynonymous variants identified in the test samples; green dots are those affected by rare synonymous variants identified in the test samples.
Table 1.
Rare variants observed in the discovery stage
| Exon | Chr20 Location bp GRCh 37 |
Base Change |
Reported in dbSNP |
AA Change |
Substitution Type |
Protein Domain | cases | controls | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr # | Total Chr # |
MAF | Chr # | Total Chr # |
MAF | |||||||
| 1 | 61992494 | G>C | yes | A8A | synonymous | Signal peptide | 0 | 1 | 362 | 0.00276 | ||
| 5 | 61982302 | C>T | no | P154L | missense | Extracellular | 1 | 418 | 0.00239 | 1 | 366 | 0.00273 |
| 5 | 61982082 | C>A | yes | A227A | synonymous | Extracellular | 0 | 1 | 352 | 0.00284 | ||
| 5 | 61981905 | C>T | no | T286T | synonymous | TMD 2 | 3 | 416 | 0.00721 | 0 | ||
| 5 | 61981785 | C>T | yes | F326F | synonymous | TMD 3 | 0 | 2 | 352 | 0.00568 | ||
| 5 | 61981709 | G>A | no | V352M | missense | Cytoplasmic | 0 | 1 | 328 | 0.00305 | ||
| 5 | 61981560 | G>C | yes | L401L | synonymous | Cytoplasmic | 1 | 406 | 0.00246 | 0 | ||
| 5 | 61981452 | T>C | no | A437A | synonymous | Cytoplasmic | 0 | 1 | 328 | 0.00305 | ||
| 5 | 61981411 | C>T | yes | P451L | missense | Cytoplasmic | 1 | 406 | 0.00246 | 3 | 328 | 0.00915 |
| 5 | 61981362 | C>T | yes | S467S | synonymous | Cytoplasmic | 4 | 406 | 0.00985 | 4 | 328 | 0.01220 |
| 5 | 61981322 | G>A | yes | G481S | missense | Cytoplasmic | 0 | 1 | 328 | 0.00305 | ||
| 5 | 61981303 | G>A | no | R487Q | missense | Cytoplasmic | 1 | 406 | 0.00246 | 0 | ||
| 5 | 61981129 | C>T | no | T545M | missense | Cytoplasmic | 2 | 400 | 0.005 | 0 | ||
| Sum: | 13 | 15 | ||||||||||
Rare nonsynonymous variants are shown in bold. Chr # is the number of chromosomes sequenced for the region.
Sequencing the exon 5 of CHRNA4 in the test sample set
Because 12 of the 13 types of rare variants observed were located in exon 5 of CHRNA4, including all of the rare nonsynonymous variants, we sequenced this exon in an additional 1000 ND cases and 1000 comparison subjects. Per Uniprot, exon 5 encodes 114 amino acids of the extracellular domain, all of the first three transmembrane domains, and 256 amino acids of the cytoplasmic loop.
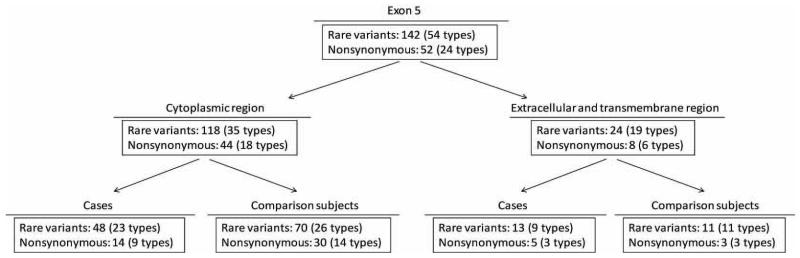
In the test stage, we identified 142 rare variants in exon 5, of 54 different types with MAF < 1% (Figure 1B, Table 2). Seven of the 12 types of rare variants identified in the discovery stage in exon 5 were observed again in the test stage. Of the 54 unique rare variants, 35 were observed only once, and eight were observed more than five times. Most of the rare variants (64.8% of the 54 types of rare variants, and 83.1% of the total variants) were located in the region encoding the cytoplasmic loop of the α4 subunit. Twenty-four of the 54 unique rare variants were nonsynonymous variants, which were all missense, with 18 of them located in the region encoding the cytoplasmic loop. Of these cytoplasmic missense rare variants, 30 were found in comparison subjects, and 14 were found in cases (Figure 2). Comparison subjects had a significantly higher frequency of rare nonsynonymous variants in the cytoplasmic loop region than the cases (FET p=0.009; association test p=0.009, OR=0.43, 95%CI=0.23-0.81; WSM p=0.014). The frequency of rare nonsynonymous variants located in the exon 5 region encoding the extracellular and transmembrane domains did not differ between cases and comparison subjects (FET p=0.73, WSM p=0.80). In addition, the frequency of rare synonymous variants in exon 5 did not differ between cases and comparison subjects (FET p=0.39, WSM p=0.40).
Table 2.
Rare variants observed in the test stage
| Exon | Chr20 Location bp GRCh 37 |
Base Change |
Reported in dbSNP |
AA Change |
Substitution Type |
Protein Domain | cases | controls | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr # | Total Chr # |
Race | Chr # | Total Chr # |
Race | |||||||
| 5 | 61982321 | C>T | no | R148W | missense | Extracellular | 2 | 1992 | all EA | 0 | ||
| 5 | 61982301 | G>A | no | P154P | synonymous | Extracellular | 3 | 1992 | all AA | 1 | 1952 | AA |
| 5 | 61982259 | C>T | no | F168F | synonymous | Extracellular | 1 | 1992 | AA | 0 | ||
| 5 | 61982253 | C>T | no | F170F | synonymous | Extracellular | 1 | 1992 | AA | 0 | ||
| 5 | 61982178 | C>G | no | H195Q | missense | Extracellular | 2 | 1992 | all AA | 0 | ||
| 5 | 61982146 | G>C | no | S206T | missense | Extracellular | 0 | 1 | 1940 | AA | ||
| 5 | 61982070 | G>A | no | P231P | synonymous | Extracellular | 1 | 1990 | AA | 0 | ||
| 5 | 61982058 | T>C | no | Y235Y | synonymous | Extracellular | 0 | 1 | 1940 | AA | ||
| 5 | 61982056 | C>T | no | A236V | missense | Extracellular | 0 | 1 | 1940 | EA | ||
| 5 | 61982034 | G>A | no | P243P | synonymous | TMD 1 | 0 | 1 | 1940 | EA | ||
| 5 | 61982010 | C>T | no | I251I | synonymous | TMD 1 | 0 | 1 | 1940 | AA | ||
| 5 | 61981988 | T>C | no | C259R | missense | TMD 1 | 1 | 1990 | EA | 0 | ||
| 5 | 61981964 | C>T | no | L267L | synonymous | TMD 1 | 0 | 1 | 1940 | EA | ||
| 5 | 61981905 | C>T | no | T286T | synonymous | TMD 2 | 0 | 1 | 1940 | EA | ||
| 5 | 61981848 | C>T | no | L305L | synonymous | TMD 3 | 1 | 1990 | AA | 0 | ||
| 5 | 61981815 | C>T | no | F316F | synonymous | TMD 3 | 0 | 1 | 1940 | EA | ||
| 5 | 61981810 | C>T | no | T318I | missense | TMD 3 | 0 | 1 | 1940 | EA | ||
| 5 | 61981800 | C>T | no | I321I | synonymous | TMD 3 | 1 | 1990 | EA | 0 | ||
| 5 | 61981785 | C>T | yes | F326F | synonymous | TMD 3 | 0 | 1 | 1940 | EA | ||
| 5 | 61981762 | C>T | no | S334L | missense | Cytoplasmic | 0 | 1 | 1608 | AA | ||
| 5 | 61981761 | G>A | no | S334S | synonymous | Cytoplasmic | 1 | 1746 | EA | 1 | 1608 | AA |
| 5 | 61981757 | C>T | yes | R336C | missense | Cytoplasmic | 1 | 1746 | EA | 0 | ||
| 5 | 61981716 | G>A | yes | L349L | synonymous | Cytoplasmic | 2 | 1746 | all AA | 6 | 1608 | all AA |
| 5 | 61981710 | C>T | yes | I351I | synonymous | Cytoplasmic | 2 | 1746 | all AA | 3 | 1608 | all AA |
| 5 | 61981697 | C>T | no | L356F | missense | Cytoplasmic | 0 | 1 | 1608 | AA | ||
| 5 | 61981677 | C>T | yes | S362S | synonymous | Cytoplasmic | 0 | 1 | 1608 | AA | ||
| 5 | 61981676 | G>A | no | V363M | missense | Cytoplasmic | 0 | 1 | 1608 | AA | ||
| 5 | 61981654 | G>A | no | R370Q | missense | Cytoplasmic | 1 | 1746 | AA | 0 | ||
| 5 | 61981620 | C>T | yes | A381A | synonymous | Cytoplasmic | 3 | 1746 | all AA | 7 | 1608 | all AA |
| 5 | 61981605 | C>G | yes | P386P | synonymous | Cytoplasmic | 4 | 1746 | EA, 3AA | 5 | 1608 | all AA |
| 5 | 61981603 | A>G | yes | E387G | missense | Cytoplasmic | 4 | 1746 | EA, 3AA | 5 | 1608 | all AA |
| 5 | 61981594 | G>A | no | G390E | missense | Cytoplasmic | 1 | 1746 | AA | 1 | 1608 | AA |
| 5 | 61981579 | C>T | no | T395M | missense | Cytoplasmic | 1 | 1746 | AA | 0 | ||
| 5 | 61981563 | C>T | no | S400S | synonymous | Cytoplasmic | 1 | 1746 | AA | 0 | ||
| 5 | 61981560 | G>C | yes | L401L | synonymous | Cytoplasmic | 1 | 1746 | EA | 2 | 1608 | all EA |
| 5 | 61981539 | C>T | no | F408F | synonymous | Cytoplasmic | 1 | 1746 | AA | 0 | ||
| 5 | 61981535 | G>A | no | V410I | missense | Cytoplasmic | 1 | 1746 | EA | 0 | ||
| 5 | 61981498 | G>A | no | C422Y | missense | Cytoplasmic | 0 | 1 | 1608 | AA | ||
| 5 | 61981497 | C>T | no | C422C | synonymous | Cytoplasmic | 0 | 1 | 1608 | EA | ||
| 5 | 61981411 | C>T | yes | P451L | missense | Cytoplasmic | 1 | 1746 | EA | 5 | 1608 | all EA |
| 5 | 61981410 | G>A | yes | P451P | synonymous | Cytoplasmic | 3 | 1746 | all AA | 7 | 1608 | all AA |
| 5 | 61981404 | C>T | no | H453H | synonymous | Cytoplasmic | 1 | 1746 | AA | 0 | ||
| 5 | 61981390 | C>T | no | P458L | missense | Cytoplasmic | 2 | 1746 | all AA | 3 | 1608 | all AA |
| 5 | 61981362 | C>T | yes | S467S | synonymous | Cytoplasmic | 7 | 1746 | 4EA,3AA | 3 | 1608 | 2EA, AA |
| 5 | 61981361 | G>A | no | V468I | missense | Cytoplasmic | 0 | 1 | 1608 | EA | ||
| 5 | 61981338 | C>T | no | G475G | synonymous | Cytoplasmic | 1 | 1746 | EA | 0 | ||
| 5 | 61981332 | G>A | yes | A477A | synonymous | Cytoplasmic | 1 | 1746 | AA | 0 | ||
| 5 | 61981322 | G>A | yes | G481S | missense | Cytoplasmic | 2 | 1746 | EA,AA | 3 | 1608 | all AA |
| 5 | 61981303 | G>A | no | R487Q | missense | Cytoplasmic | 0 | 4 | 1608 | EA, 3AA | ||
| 5 | 61981138 | C>T | no | P542L | missense | Cytoplasmic | 0 | 1 | 1724 | AA | ||
| 5 | 61981128 | G>A | no | T545T | synonymous | Cytoplasmic | 0 | 1 | 1724 | EA | ||
| 5 | 61981102 | C>T | yes | P554L | missense | Cytoplasmic | 0 | 1 | 1724 | EA | ||
| 5 | 61981101 | G>A | no | P554P | synonymous | Cytoplasmic | 6 | 1602 | EA, 5AA | 3 | 1724 | all AA |
| 5 | 61981052 | G>A | no | V571I | missense | Cytoplasmic | 0 | 2 | 1724 | all AA | ||
| Sum: | 61 | 81 | ||||||||||
Rare nonsynonymous variants are shown in bold.
Figure 2.
Rare variants found in the test samples, showing the locations of the rare variants and the distributions among cases and controls.
Combining results from the two stages
When results from the discovery and the test stages were combined, we observed a total of 53 (20 types) rare nonsynonymous variants in the exon 5 region that encodes the cytoplasmic loop (Table 3). Of these rare nonsynonymous variants, 35 were in comparison subjects and 18 were in ND cases. Traces of Sanger sequencing results can be found in the Supplement.
Table 3.
Rare missense variants found in exon 5 region coding the cytoplasmic loop (data combined from discovery and test stages)
| Exon | Chr20 Location bp GRCh 37 |
Base Change |
Reported in dbSNP |
AA Change |
Substitution Type |
Protein Domain |
cases | controls | Polyphen Predict |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr # |
Total Chr # |
Race | Chr # |
Total Chr # |
Race | ||||||||
| 5 | 61981762 | C>T | no | S334L | missense | Cytoplasmic | 0 | 1 | 1936 | AA | damaging | ||
| 5 | 61981757 | C>T | yes | R336C | missense | Cytoplasmic | 1 | 2152 | EA | 0 | damaging | ||
| 5 | 61981709 | G>A | no | V352M | missense | Cytoplasmic | 0 | 1 | 1936 | EA | benign | ||
| 5 | 61981697 | C>T | no | L356F | missense | Cytoplasmic | 0 | 1 | 1936 | AA | damaging | ||
| 5 | 61981676 | G>A | no | V363M | missense | Cytoplasmic | 0 | 1 | 1936 | AA | benign | ||
| 5 | 61981654 | G>A | no | R370Q | missense | Cytoplasmic | 1 | 2152 | AA | 0 | benign | ||
| 5 | 61981603 | A>G | yes | E387G | missense | Cytoplasmic | 4 | 2152 | EA, 3AA | 5 | 1936 | all AA | benign |
| 5 | 61981594 | G>A | no | G390E | missense | Cytoplasmic | 1 | 2152 | AA | 1 | 1936 | AA | benign |
| 5 | 61981579 | C>T | no | T395M | missense | Cytoplasmic | 1 | 2152 | AA | 0 | benign | ||
| 5 | 61981535 | G>A | no | V410I | missense | Cytoplasmic | 1 | 2152 | EA | 0 | benign | ||
| 5 | 61981498 | G>A | no | C422Y | missense | Cytoplasmic | 0 | 1 | 1936 | AA | damaging | ||
| 5 | 61981411 | C>T | yes | P451L | missense | Cytoplasmic | 2 | 2152 | all EA | 8 | 1936 | all EA | damaging |
| 5 | 61981390 | C>T | no | P458L | missense | Cytoplasmic | 2 | 2152 | all AA | 3 | 1936 | all AA | damaging |
| 5 | 61981361 | G>A | no | V468I | missense | Cytoplasmic | 0 | 1 | 1936 | EA | benign | ||
| 5 | 61981322 | G>A | yes | G481S | missense | Cytoplasmic | 2 | 2152 | EA,AA | 4 | 1936 | EA, 3AA | benign |
| 5 | 61981303 | G>A | no | R487Q | missense | Cytoplasmic | 1 | 2152 | EA | 4 | 1936 | EA, 3AA | damaging |
| 5 | 61981138 | C>T | no | P542L | missense | Cytoplasmic | 0 | 1 | 2032 | AA | damaging | ||
| 5 | 61981129 | C>T | no | T545M | missense | Cytoplasmic | 2 | 2002 | EA | 0 | benign | ||
| 5 | 61981102 | C>T | yes | P554L | missense | Cytoplasmic | 0 | 1 | 2032 | EA | benign | ||
| 5 | 61981052 | G>A | no | V571I | missense | Cytoplasmic | 0 | 2 | 2032 | all AA | benign | ||
| Sum: | 18 | 35 | |||||||||||
As expected, AAs carried more rare variants than EAs (because they represent an older population), and most variants were population-specific. In both populations, comparison subjects had a higher frequency of rare nonsynonymous variants in the exon 5 cytoplasmic loop region than cases. The difference was significant in AAs (FET p=0.01, WSM p=0.008), but not in EAs (FET p=0.29, WSM p=0.35). However, a missense mutation p.Pro451Leu, which was exclusively observed in EAs, was found in 8 comparison subjects, but only in 2 ND cases. The difference in frequency of this rare variant is marginally significant (FET p=0.05), potentially indicating a protective effect against ND.
Functional analysis
PolyPhen (37) was used to predict the effects of rare nonsynonymous variants on protein function. Among the 20 types of rare missense variants observed in the exon 5 cytoplasmic loop region, PolyPhen predicted 8 to be possibly or probably damaging (Table 3). Comparison subjects had a total of 19 alleles of these variants and cases had 6, a significant difference (FET p=0.005; association test p=0.008, OR=0.29, 95%CI=0.11-0.72; WSM p=0.005), suggesting that functional rare variants in this region had protective effects against ND.
In the discovery stage, 209 ND cases from 114 families were included, most of whom were affected siblings. We only observed one pair of affected siblings both carrying a missense rare variant; this variant was not predicted to be damaging to α4 subunit function by PolyPhen. For protective variants, this design choice should not affect power.
Imaging study
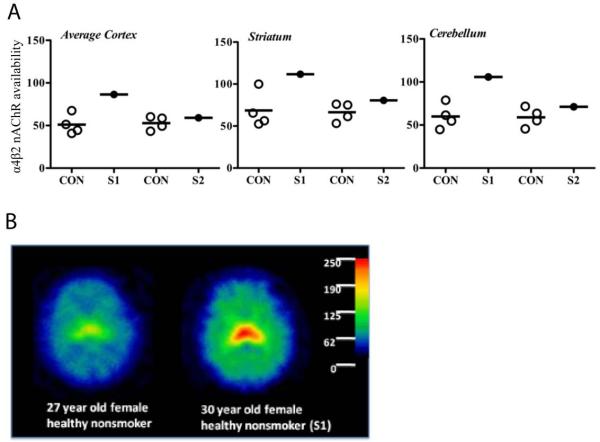
Among the 139 subjects (92 smokers and 47 nonsmokers) who participated in the [123I]5-IA SPECT imaging study, one nonsmoker and two smokers carried different nonsynonymous rare variants in the exon 5 cytoplasmic loop region. The nonsmoker, who was a 30-year-old EA female, carried a p.Arg336Cys rare variant. Compared to a group of four age-matched control nonsmokers, the regional α4β2 nAChR availability in the brain of this rare variant carrier was 69.3%, 63.0% and 76.7% higher in the cortical areas, striatum, and cerebellum, respectively (Figure 3A, subject 1 (S1), and Figure 3B). The α4β2 nAChR availabilities among the two smokers who carried nonsynonymous rare variants (p.Pro452Ser and p.Arg487Gln) and their age-matched control smokers were similar.
Figure 3.
SPECT scans using 123-labeled 5-iodo-A-85380 ([123I]5-IA) for two nonsmoking rare variant carriers (S1 with a nonsynonymous rare variant and S2 with two synonymous variants) compared to their age-matched nonsmoking controls.
(A) α4β2 nAChR availability (VT/fP) in cortical regions (average of parietal, frontal, anterior cingulate, temporal and occipital cortices), striatum (average of caudate and putamen) and cerebellum in S1 and S2 compared to age-matched controls (CON).
(B) Parametric images (α4β2 nAChR availability, VT/fP) of S1, a 30-year-old female healthy nonsmoker and an age- and sex-matched control). The color bar shows corresponding VT/fP values.
Another three subjects (one nonsmoker and two smokers) were found to carry synonymous rare variants in the exon 5 cytoplasmic loop region. Similar to what was observed for the nonsmoker with the nonsynonymous rare variant, the nonsmoker who carried two synonymous rare variants (p.Pro451Pro and p.Pro554Pro) also had higher α4β2 nAChR availability in the cortical areas (11.9%), striatum (21.4%), and cerebellum (20.8%), than a group of four age-matched control nonsmokers (Figure 3A, subject 2 (S2)); while no difference was observed between the two smokers who carried the synonymous rare variants p.Phe326Phe and p.Leu520Leu and their age-matched control smokers.
Discussion
In this study, we investigated the distribution of rare variants in CHRNA4 in more than 2000 ND cases and comparison subjects. Compared to cases, controls had a significantly higher frequency of rare nonsynonymous variants located in the exon 5 region encoding the cytoplasmic loop of the α4 nAChR (FET p=0.009; association test p=0.009, OR=0.43, 95%CI=0.23-0.81; WSM p=0.014). Functional in-silico analysis (PolyPhen) further demonstrated that the comparison subjects carried significantly more potentially damaging rare nonsynonymous variants than the cases (FET p=0.005; association test p=0.008, OR= 0.29, 95%CI=0.11-0.72; WSM p=0.005), suggesting that some rare functional variants exert a protective effect against ND. In addition, although we observed that comparison subjects had a higher frequency of rare nonsynonymous variants in the exon 5 cytoplasmic loop region in both EAs and AAs, the difference was significant only in AAs.
The basic structure of different nAChR subunits is quite similar, but the large cytoplasmic loop is highly divergent, which accounts in part for the functional differences and differences in the distribution (39), assembly (40), and other characteristics of nAChR subunits. This cytoplasmic loop is also important for interaction with intercellular proteins, which could increase the stability of α4β2 nAChRs (41), as well as post-translational modification of the α4 subunit (42). Our imaging study showed that for nonsmokers, specific rare variants in the CHRNA5 gene region encoding the cytoplasmic loop could increase the availability of α4β2 nAChRs in multiple brain regions. In addition, a nonsynonymous rare variant seems to have a bigger effect than the synonymous rare variants (Figure 3). One possible explanation for the difference in nAChR availability observed in the subject with the nonsynonymous rare variant is that increased availability of α4β2 nAChRs compensates for damaged protein function. Alternatively, an increase in stability of the mutated α4 protein could increase baseline nAChR function, providing protection against further increases by the nicotine in tobacco. The results from this imaging study are preliminary, and these hypotheses remain to be tested. For smokers carrying nonsynonymous rare variants, Chronic exposure to nicotine up-regulates α4β2 nAChRs (43), and this may mask the effect of rare variants. It is hard to generalize based on these findings however, because it is plausible that each specific variant could have a unique effect on function. These findings should therefore be viewed as illustrative of possible effects rather than predictive of all.
Animal experiments have demonstrated that activation of α4β2 nAChRs is involved in the development of ND (5,8,9). Therefore, current understanding is consistent with the notion that nonsynonymous rare variants s in the cytoplasmic loop of the α4 subunit may have a protective effect against ND. Evaluation of the role of these rare variants in the α4 subunit in nAChR function by making point mutations in the functional consequence using in vitro or in vivo models will help identify the structure-activity changes resulting from these changes.
Recently, Wessel et al. resequenced 10 nAChR subunit genes and found a significant association between rare variants in CHRNA4 and FTND score (44). However, due to the small sample size in that study (N=430), they observed a total of only 21 rare variants, including 5 nonsynonymous rare variants in CHRNA4. Because they were unable to perform more detailed analyses on nonsynonymous variants, the results of their study were not conclusive. In contrast, we used a two stage case-control design. When results from the two stages were combined, a total of 169 rare variants of 59 types were observed in exon 5. Among them, we found a total of 63 nonsynonymous rare variants of 27 types. This sample size proved to be adequate to provide significant evidence that functional nonsynonymous rare variants located in the cytoplasmic loop of the α4 subunit play a protective role in ND, especially in the AA population.
Among the 20 types of missense rare variants we detected in the exon 5 cytoplasmic loop region, 12 were observed only once (MAF=0.025%), with the most frequent missense rare variants having an MAF of 0.25%. Although the joint analysis of all rare variants provided sufficient power to detect differences between ND cases and controls, this approach assumes the same direction of effect (protective, deleterious or neutral) for all of the variants on ND risk, otherwise it could adversely affect power (45). To address this concern, we conducted a bioinformatic analysis to predict the effect of each nonsynonymous rare variant on protein function. This analysis suggested that eight of the 20 types of rare variants were predicted to be damaging to α4 subunit function. Restricting the comparisons to the aggregate effect of these eight variants confirmed the results of the analysis of all 20 nonsynonymous rare variants.
Even though the proportion of nonsynonymous variants in the exon 5 cytoplasmic region did not differ significantly between EA ND cases and comparison subjects, we identified a missense rare variant, p.Pro451Leu, that was significantly more frequent in controls and may have a protective effect with respect to ND risk in that population. p.Pro451Leu is predicted by PolyPhen to be damaging, and was exclusively observed in EA subjects. In the discovery sample, this variant was found in 3 controls and 1 case; in the test sample, it was observed in 5 comparison subjects and 1 case. The difference is marginally significant (FET p=0.05).
A characteristic of our sample is a high level of comorbidity with other kinds of substance dependence and ND, a consequence of our recruitment methods and also of our detailed phenotypic assessment protocol. Notwithstanding this fact, we note that we have replicated ND findings first reported by other groups (for example, regarding the CHRNA3/A5/B4 gene cluster (46)), and our findings have been replicated by others (for example, ANKK1/TTC12 ((47-49)). This holds not only for association findings, but for linkage findings as well. Thus, existing evidence supports our hypothesis that ND in this sample is similar to ND in other samples. Of the 33 subjects from the comparison group who carried missense rare variants in the exon 5 cytoplasmic loop region (two of whom had double missense rare variants at different loci), two were alcohol dependent, and one was cocaine dependent. All of the other individuals were healthy, with no psychiatric disorders or substance dependence. All ND cases who carried missense rare variants in the cytoplasmic loop region were comorbid with alcohol, cocaine or opioid dependence.
In conclusion, we observed that ND cases had significantly fewer rare nonsynonymous variants in the cytoplasmic loop of the α4 nAChR subunit than comparison subjects, especially among AAs. We hypothesize that these rare variants have protective effects against ND. Further genetic and functional studies are needed to test this hypothesis.
Supplementary Material
Acknowledgements
Ann Marie Lacobelle provided technical assistance. We thank the individuals and families participating in this work and the interviewers at all the participating sites for collecting the data. This study is supported by NIH grants R01 DA12690, R01 DA12849, R01 AA11330, R01 AA017535, K01 DA020651, R01 DA015577,2P50-AA012870, K05 AA017435, R01 DA030976and the VA VISN1 MIRECC, VA Alcohol Research Center, and the Clinical Neuroscience Division of the VA National Center for PTSD.
Footnotes
Financial Disclosures Dr. Kranzler has received consulting fees from Alkermes, GlaxoSmithKline, and Gilead, and research support from Merck. Dr. Anton reports for the last two years, being a consultant for Eli Lilly, GlaxoSmithKline, and Alkermes. Drs. Kranzler and Anton also report associations with Eli Lilly, Merck, Janssen, Schering Plough, Lundbeck, Alkermes, GlaxoSmithKline, Abbott, and Johnson & Johnson, as these companies provide support to the ACNP Alcohol Clinical Trials Initiative (ACTIVE) and they receive support from ACTIVE. Dr. Picciotto has received an honorarium from Targacept. Dr. Krystal reports serving as a scientific consultant and/or on the scientific advisory board to the following companies: Aisling Capital, AstraZeneca Pharmaceuticals, Brintnall & Nicolini, Easton Associates, Gilead Sciences, GlaxoSmithKline, Janssen Pharmaceuticals, Lundbeck Research USA, Medivation, Merz Pharmaceuticals, MK Medical Communications, F. Hoffmann-La Roche, SK Holdings Co., Takeda Industries, Teva Pharmaceutical Industries, Abbott Laboratories, Bristol-Myers Squibb, Eisai, Eli Lilly and Co, Lohocla Research Corporation, Naurex and Pfizer Pharmaceuticals. Dr Krystal holds less than US$150 in exercisable warrant options with Tetragenex Pharmaceuticals, being the principal investigator of a multicenter study in which Janssen Research Foundation has provided drug and some financial support to the Department of Veterans Affairs, and a cosponsor for 2 patents under review for glutamatergic agents targeting the treatment of depression, including 1 that involves the antidepressant effects of ketamine. The other authors report no biomedical financial interests or potential conflicts of interest.
Accession Numbers The dbSNP submitted SNP (ss) numbers of CHRNA4 reported in this paper are: 270137503, 270137504, 270137505, 270137506, 270137507, 270137508, 270137509, 270137510, 270137511, 270137512, 270137513, 270137514, 270137515, 270137516, 270137517, 270137518, 270137519, 270137520, 270137521, 270137522, 270137523, 270137524, 270137525, 270137526, 270137527, 270137528, 270137529, 270137530, 270137531, 270137532, 270137533, 270137534, 270137535, 270137536, 270137537, 270137538, 270137539, 270137540, 270137541, 270137542, 270137543, 270137544, 270137545.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. Jama. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 2.Govind AP, Vezina P, Green WN. Nicotine-induced upregulation of nicotinic receptors: underlying mechanisms and relevance to nicotine addiction. Biochemical pharmacology. 2009;78:756–765. doi: 10.1016/j.bcp.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flores CM, Rogers SW, Pabreza LA, Wolfe BB, Kellar KJ. A subtype of nicotinic cholinergic receptor in rat brain is composed of alpha 4 and beta 2 subunits and is up-regulated by chronic nicotine treatment. Molecular pharmacology. 1992;41:31–37. [PubMed] [Google Scholar]
- 4.Corrigall WA, Coen KM, Adamson KL. Self-administered nicotine activates the mesolimbic dopamine system through the ventral tegmental area. Brain research. 1994;653:278–284. doi: 10.1016/0006-8993(94)90401-4. [DOI] [PubMed] [Google Scholar]
- 5.Picciotto MR, Zoli M, Rimondini R, Lena C, Marubio LM, Pich EM, et al. Acetylcholine receptors containing the beta2 subunit are involved in the reinforcing properties of nicotine. Nature. 1998;391:173–177. doi: 10.1038/34413. [DOI] [PubMed] [Google Scholar]
- 6.Laviolette SR, van der Kooy D. Blockade of mesolimbic dopamine transmission dramatically increases sensitivity to the rewarding effects of nicotine in the ventral tegmental area. Molecular psychiatry. 2003;8:50–59. 59. doi: 10.1038/sj.mp.4001197. [DOI] [PubMed] [Google Scholar]
- 7.Nisell M, Nomikos GG, Svensson TH. Systemic nicotine-induced dopamine release in the rat nucleus accumbens is regulated by nicotinic receptors in the ventral tegmental area. Synapse (New York, NY. 1994;16:36–44. doi: 10.1002/syn.890160105. [DOI] [PubMed] [Google Scholar]
- 8.Marubio LM, Gardier AM, Durier S, David D, Klink R, Arroyo-Jimenez MM, et al. Effects of nicotine in the dopaminergic system of mice lacking the alpha4 subunit of neuronal nicotinic acetylcholine receptors. The European journal of neuroscience. 2003;17:1329–1337. doi: 10.1046/j.1460-9568.2003.02564.x. [DOI] [PubMed] [Google Scholar]
- 9.Tapper AR, McKinney SL, Nashmi R, Schwarz J, Deshpande P, Labarca C, et al. Nicotine activation of alpha4* receptors: sufficient for reward, tolerance, and sensitization. Science (New York, NY. 2004;306:1029–1032. doi: 10.1126/science.1099420. [DOI] [PubMed] [Google Scholar]
- 10.Jorenby DE, Hays JT, Rigotti NA, Azoulay S, Watsky EJ, Williams KE, et al. Efficacy of varenicline, an alpha4beta2 nicotinic acetylcholine receptor partial agonist, vs placebo or sustained-release bupropion for smoking cessation: a randomized controlled trial. Jama. 2006;296:56–63. doi: 10.1001/jama.296.1.56. [DOI] [PubMed] [Google Scholar]
- 11.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nature genetics. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nature genetics. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium TAG. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature genetics. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li MD, Beuten J, Ma JZ, Payne TJ, Lou XY, Garcia V, et al. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Human molecular genetics. 2005;14:1211–1219. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Niu T, Xing H, Xu X, Chen C, Peng S, et al. A common haplotype of the nicotine acetylcholine receptor alpha 4 subunit gene is associated with vulnerability to nicotine addiction in men. American journal of human genetics. 2004;75:112–121. doi: 10.1086/422194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breitling LP, Dahmen N, Mittelstrass K, Rujescu D, Gallinat J, Fehr C, et al. Association of nicotinic acetylcholine receptor subunit alpha 4 polymorphisms with nicotine dependence in 5500 Germans. The pharmacogenomics journal. 2009;9:219–224. doi: 10.1038/tpj.2009.6. [DOI] [PubMed] [Google Scholar]
- 17.Saccone NL, Schwantes-An TH, Wang JC, Grucza RA, Breslau N, Hatsukami D, et al. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes, brain, and behavior. 2010;9:741–750. doi: 10.1111/j.1601-183X.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchison KE, Allen DL, Filbey FM, Jepson C, Lerman C, Benowitz NL, et al. CHRNA4 and tobacco dependence: from gene regulation to treatment outcome. Archives of general psychiatry. 2007;64:1078–1086. doi: 10.1001/archpsyc.64.9.1078. [DOI] [PubMed] [Google Scholar]
- 19.Li MD, Lou XY, Chen G, Ma JZ, Elston RC. Gene-gene interactions among CHRNA4, CHRNB2, BDNF, and NTRK2 in nicotine dependence. Biological psychiatry. 2008;64:951–957. doi: 10.1016/j.biopsych.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han S, Yang BZ, Kranzler HR, Oslin D, Anton RF, Gelernter J. Association of CHRNA4 polymorphisms with smoking behavior in two populations. Am J Med Genet B Neuropsychiatr Genet. 2011 doi: 10.1002/ajmg.b.31177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han S, Gelernter J, Luo X, Yang BZ. Meta-analysis of 15 genome-wide linkage scans of smoking behavior. Biological psychiatry. 2010;67:12–19. doi: 10.1016/j.biopsych.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 23.Knight HM, Pickard BS, Maclean A, Malloy MP, Soares DC, McRae AF, et al. A cytogenetic abnormality and rare coding variants identify ABCA13 as a candidate gene in schizophrenia, bipolar disorder, and depression. American journal of human genetics. 2009;85:833–846. doi: 10.1016/j.ajhg.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhin AG, Gundisch D, Horti AG, Koren AO, Tamagnan G, Kimes AS, et al. 5-Iodo-A-85380, an alpha4beta2 subtype-selective ligand for nicotinic acetylcholine receptors. Molecular pharmacology. 2000;57:642–649. doi: 10.1124/mol.57.3.642. [DOI] [PubMed] [Google Scholar]
- 25.Cosgrove KP, Batis J, Bois F, Maciejewski PK, Esterlis I, Kloczynski T, et al. beta2-Nicotinic acetylcholine receptor availability during acute and prolonged abstinence from tobacco smoking. Archives of general psychiatry. 2009;66:666–676. doi: 10.1001/archgenpsychiatry.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, Poling J, et al. Dense genomewide linkage scan for alcohol dependence in African Americans: significant linkage on chromosome 10. Biological psychiatry. 2009;65:111–115. doi: 10.1016/j.biopsych.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gelernter J, Panhuysen C, Weiss R, Brady K, Poling J, Krauthammer M, et al. Genomewide linkage scan for nicotine dependence: identification of a chromosome 5 risk locus. Biological psychiatry. 2007;61:119–126. doi: 10.1016/j.biopsych.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 28.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug and alcohol dependence. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, et al. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug and alcohol dependence. 2007;91:85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. American journal of human genetics. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang BZ, Zhao H, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genetic epidemiology. 2005;28:302–312. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
- 34.Xie P, Kranzler HR, Poling J, Stein MB, Anton RF, Brady K, et al. Interactive effect of stressful life events and the serotonin transporter 5-HTTLPR genotype on posttraumatic stress disorder diagnosis in 2 independent populations. Archives of general psychiatry. 2009;66:1201–1209. doi: 10.1001/archgenpsychiatry.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome biology. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS genetics. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic acids research. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27:1533–1539. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- 39.Williams BM, Temburni MK, Levey MS, Bertrand S, Bertrand D, Jacob MH. The long internal loop of the alpha 3 subunit targets nAChRs to subdomains within individual synapses on neurons in vivo. Nature neuroscience. 1998;1:557–562. doi: 10.1038/2792. [DOI] [PubMed] [Google Scholar]
- 40.Ren XQ, Cheng SB, Treuil MW, Mukherjee J, Rao J, Braunewell KH, et al. Structural determinants of alpha4beta2 nicotinic acetylcholine receptor trafficking. J Neurosci. 2005;25:6676–6686. doi: 10.1523/JNEUROSCI.1079-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeanclos EM, Lin L, Treuil MW, Rao J, DeCoster MA, Anand R. The chaperone protein 14-3-3eta interacts with the nicotinic acetylcholine receptor alpha 4 subunit. Evidence for a dynamic role in subunit stabilization. The Journal of biological chemistry. 2001;276:28281–28290. doi: 10.1074/jbc.M011549200. [DOI] [PubMed] [Google Scholar]
- 42.Pollock VV, Pastoor TE, Wecker L. Cyclic AMP-dependent protein kinase (PKA) phosphorylates Ser362 and 467 and protein kinase C phosphorylates Ser550 within the M3/M4 cytoplasmic domain of human nicotinic receptor alpha4 subunits. Journal of neurochemistry. 2007;103:456–466. doi: 10.1111/j.1471-4159.2007.04853.x. [DOI] [PubMed] [Google Scholar]
- 43.Picciotto MR, Addy NA, Mineur YS, Brunzell DH. It is not “either/or”: activation and desensitization of nicotinic acetylcholine receptors both contribute to behaviors related to nicotine addiction and mood. Progress in neurobiology. 2008;84:329–342. doi: 10.1016/j.pneurobio.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wessel J, McDonald SM, Hinds DA, Stokowski RP, Javitz HS, Kennemer M, et al. Resequencing of Nicotinic Acetylcholine Receptor Genes and Association of Common and Rare Variants with the Fagerstrom Test for Nicotine Dependence. Neuropsychopharmacology. 2010 doi: 10.1038/npp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. American journal of human genetics. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, et al. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35:1921–1931. doi: 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ducci F, Kaakinen M, Pouta A, Hartikainen AL, Veijola J, Isohanni M, et al. TTC12-ANKK1-DRD2 and CHRNA5-CHRNA3-CHRNB4 Influence Different Pathways Leading to Smoking Behavior from Adolescence to Mid-Adulthood. Biological psychiatry. 2011;69:650–660. doi: 10.1016/j.biopsych.2010.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, et al. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Human molecular genetics. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- 49.Huang W, Payne TJ, Ma JZ, Beuten J, Dupont RT, Inohara N, et al. Significant association of ANKK1 and detection of a functional polymorphism with nicotine dependence in an African-American sample. Neuropsychopharmacology. 2009;34:319–330. doi: 10.1038/npp.2008.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.