Abstract
p-Cyanophenylalanine is an extremely useful fluorescence probe of protein structure which can be recombinantly and chemically incorporated into proteins. The probe has been used to study protein folding, protein-membrane interactions, protein-peptide interactions and amyloid formation, however the factors that control its fluorescence are not fully understood. Hydrogen bonding to the cyano group is known to play a major role in modulating the fluorescence quantum yield, but the role of potential side-chain quenchers has not yet been elucidated. A systematic study on the effects of different side-chains on p-cyanophenylalanine fluorescence is reported. Tyr is found to have the largest effect followed by deprotonated His, Met, Cys, protonated His, Asn, Arg, and protonated Lys. Deprotonated amino groups are much more effective fluorescence quenchers than protonated amino groups. Free neutral imidazole and hydroxide ion are also effective quenchers of p-cyanophenylalanine fluorescence with Stern-Volmer constants of 39.8 M−1 and 22.1 M−1, respectively. The quenching of p-cyanophenylalanine fluorescence by specific side-chains is exploited to develop specific, high sensitivity, fluorescence probes of helix formation. The approach is demonstrated with Ala based peptides that contain a p-cyanophenylalanine-His or a p-cyanophenylalanine-Tyr pair located at positions i and i+4. The p-cyanophenylalanine-His pair is most useful when the His side-chain is deprotonated and is, thus, complimentary to Trp-His pair which is most sensitive when the His side-chain is protonated.
Keywords: Fluorescence Quenching, p-Cyanophenylalanine, Resonance Energy Transfer, Protein Folding, Protein Dynamics, Unnatural Amino Acid
Fluorescence spectroscopy is widely used to study proteins and peptides, with the vast majority of studies making use of the naturally occurring fluorophores, Tyr and Trp (1, 2). However, there is considerable interest in the incorporation of novel fluorophores, because they can offer site specific probes and, in many cases, they can be chosen to allow selective excitation and detection (3–7). Recently p-cyanophenylalanine (FCN) fluorescence has been developed as a robust probe of protein folding, protein-peptide interactions, protein-membrane interactions, ligand binding and amyloid formation (Figure 1) (8–15).
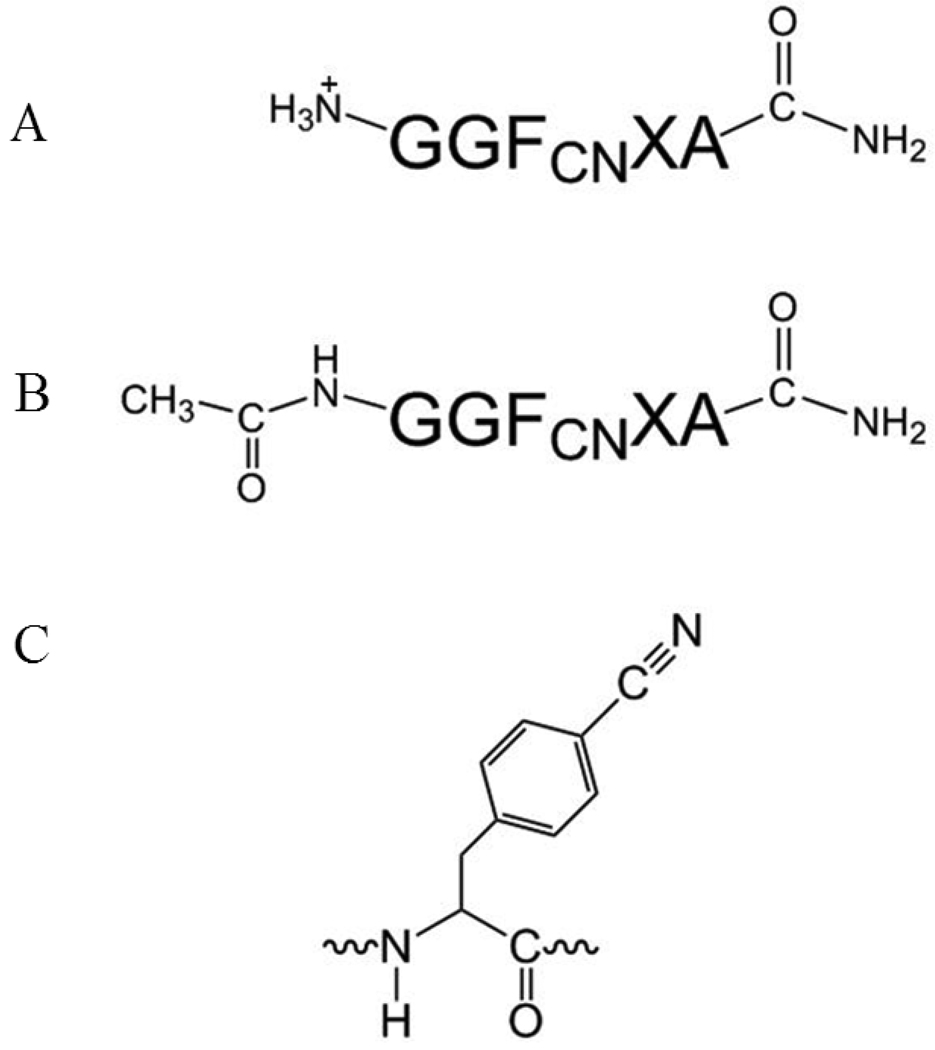
Figure 1.
General sequence of the peptides studied. (A) Several variants have an amidated C-terminus and a free N-terminus. (B) An additional three peptides GGFCNAA, GGFCNHA and GGFCNKA were synthesized with an acetylated N-terminus. (C) The structure of the p-cyanophenylalanine side-chain.
The utility of p-cyanophenylalanine is due to three features. First, it can be incorporated into proteins recombinantly, using the so called 21st pair methodology, or by solid-phase peptide synthesis (11, 16). Secondly, it represents a relatively conservative substitution for the aromatic amino acids since it is much more similar in shape and size than many other fluorophores and the polarity of the cyano group is between that of a methylene and an amide group. This intermediate polarity allows FCN to be accepted in both a hydrophobic and hydrophilic environments in a protein. Third, the photophysical properties of FCN nicely complement existing fluorophores. It can be selectively excited in the presence of Trp and Tyr and it forms a useful resonance energy transfer (RET) probe with both Tyr and Trp (7, 14, 17, 18). However, the factors that control its quantum yield are not completely understood. It is known that the fluorescence is high when the cyano group is hydrogen bonded and fluorescence can be quenched by RET to Tyr or Trp, but unfortunately the effect of other amino acid side-chains is not known (14, 19). A detailed understanding of the factors that control FCN fluorescence is required to fully exploit this very promising probe and to avoid misinterpretation. A striking example is provided by our recent application of FCN to study the folding of NTL9, a small globular protein. The fluorescence of FCN in NTL9 was very low in the folded state suggesting that the cyano group was sequestered from solvent, however IR measurements showed that it was exposed, and further investigation revealed that the FCN fluorescence was quenched by interactions with a Tyr side-chain in the native state and showed that the cyano group was, in fact, exposed to solvent (14).
Here we systematically examine the ability of other amino acids and the termini of polypeptides to modulate FCN fluorescence by examining the fluorescence of FCN in a set of peptides of general sequence GGFCNXA where X represents Ala, Cys, His, Lys, Met, Asn, Arg, and Tyr. Based on these results we demonstrate how such interactions can be used as a highly sensitive probe of helix formation. We also show that free imidazole and hydroxide ion are effective quenchers of FCN fluorescence.
Materials and Methods
Peptide Synthesis and Purification
A set of peptides with a general sequence GGFCNXA (X = A, C, H, K, M, N, R, or Y) were synthesized using standard Fmoc solid-phase methods on an Applied Biosystems 433A peptide synthesizer. PAL-PEG-PS resin was used which leads to an amidated C-terminus. Unless noted, the peptides have a free N-terminus. Additional GGFCNXA peptides (X = A, H or K) were synthesized with an acetylated N-terminus and an amidated C-terminus. Two 21 residue-long Ala based helical peptides were prepared with the sequence D-PA-A-K-A-A-A-K-A-A-X-A-A-A-FCN-A-A-A-K-A where X is His or Tyr. These peptides were amidated at their C termini and acetylated at their N termini. The peptides were cleaved from the resin using 91% (v/v) trifluoroacetic acid (TFA), 3% (v/v), anisole, 3% (v/v) thioanisole, 3% (v/v) 1,2-ethanedithiol, precipitated using cold ethyl ether, and scavengers were removed under vacuum. The crude peptides were purified with reverse-phase HPLC using a Vydac C18 semi-preparative column. An A–B gradient system was used, with buffer A composed of a 0.1% (v/v) solution of TFA and buffer B composed of 90% (v/v) acetonitrile, 9.9% (v/v) water and 0.1% (v/v) TFA. The gradient was 0–90% B in 90 min. The identities of the purified peptides were confirmed by MALDI-TOF mass spectroscopy. The expected and observed molecular weights for the GGFCNXA peptides are; GGFCNAA 446.5 Da and 446.8 Da; GGFCNYA 538.6 Da and 538.6 Da; GGFCNMA 506.6 Da and 507.8 Da; GGFCNCA 478.5 Da and 478.5 Da; GGFCNHA 512.5 Da and 512.8 Da; GGFCNKA 503.6 Da and 504.0 Da; GGFCNNA 489.5 Da and 489.8 Da; and GGFCNRA 531.5 Da and 531.3 Da, respectively. The expected and observed molecular weights for the Ac-GGFCNXA peptides are; Ac-GGFCNAA 488.5 Da and 488.1 Da; Ac-GGFCNHA 554.5 Da and 554.2 Da, and Ac-GGFCNKA 545.6 Da and 545.3 Da, respectively. The observed masses of the Ala based peptides were 1961.0 Da for the H-FCN peptide and 1987.3 Da for the Y-FCN peptide. The expected masses were 1961.1 Da for the H-FCN peptide and 1987.2 Da for the Y-FCN peptide.
Fluorescence Spectroscopy
Fluorescence emission spectra of the peptides were collected using an Applied Photon Technologies fluorimeter. The peptide concentration was determined by measuring the absorbance of the sample at 280 nm. The extinction coefficient for FCN is 850 M−1cm−1 and for Tyr is 1490 M−1cm−1 (9). Fluorescence was excited at 240 nm and the emission signal was recorded from 250–350 nm. For the pH dependent fluorescence experiments, the excitation wavelength for the FCN group was 240 nm, and the emission signal was followed at 291 nm. The peptide concentration for both the fluorescence emission spectra and pH dependent fluorescence experiments was 20 uM.
Emission spectra of the Ala based peptides were collected in buffer and 8 M urea over the range of 250 to 350 nm with excitation at 240 nm. The peptide concentration was 25 uM. The Tyr-FCN Ala rich peptide was prepared at pH 5.5 in 10 mM sodium acetate. Fluorescence emission spectra of the His-FCN peptide were taken as a function of pH. The His-FCN Ala rich peptide was examined in 10 mM sodium acetate at pH 5.5 and 10 mM Tris at pH 8.3.
Circular Dichroism
Circular dichroism spectra of the 21 residue Ala based peptides were collected using a Chirascan Applied Photophysics CD spectrophotometer. The peptides were in 10mM sodium acetate at pH 5.5 or 10 mM tris at pH 8.3 for studies of the folded form. The unfolded peptides were in 8 M urea and either 10 mM sodium acetate at pH 5.5 or 10 mM Tris at pH 8.3. The urea concentration was determined using refractive index measurements. Wavelength scans were recorded at 25 °C from 190–260 nm for the folded peptide and from 210–260 nm for the urea unfolded peptide.
Results and Discussion
A set of peptides with the general sequence GGFCN XA (X= A, C, H, K, M, N, R, or Y) were synthesized to probe the ability of different side-chains to modulate FCN fluorescence. These side-chains include all of the potential quenching groups in proteins with the exception of Trp which has already been analyzed. Ser was not examined because it is known that hydrogen bonding to the cyano group leads to high fluorescence. The potential effect of protonated and deprotonated carboxylic acid were examined by a Stern-Volmer analysis of the quenching by acetic acid and sodium acetate (supporting information). All of the peptides have amidated C-terminus. Two GGFCNAA peptides were prepared, one with a free N-terminus and the other with an acetylated N-terminus (Figure 1). The structure of FCN is shown in Figure 1. The GGFCNAA peptide sequence is the parent (reference) peptide, and was chosen as such since the Ala side-chain will not quench the fluorescence.
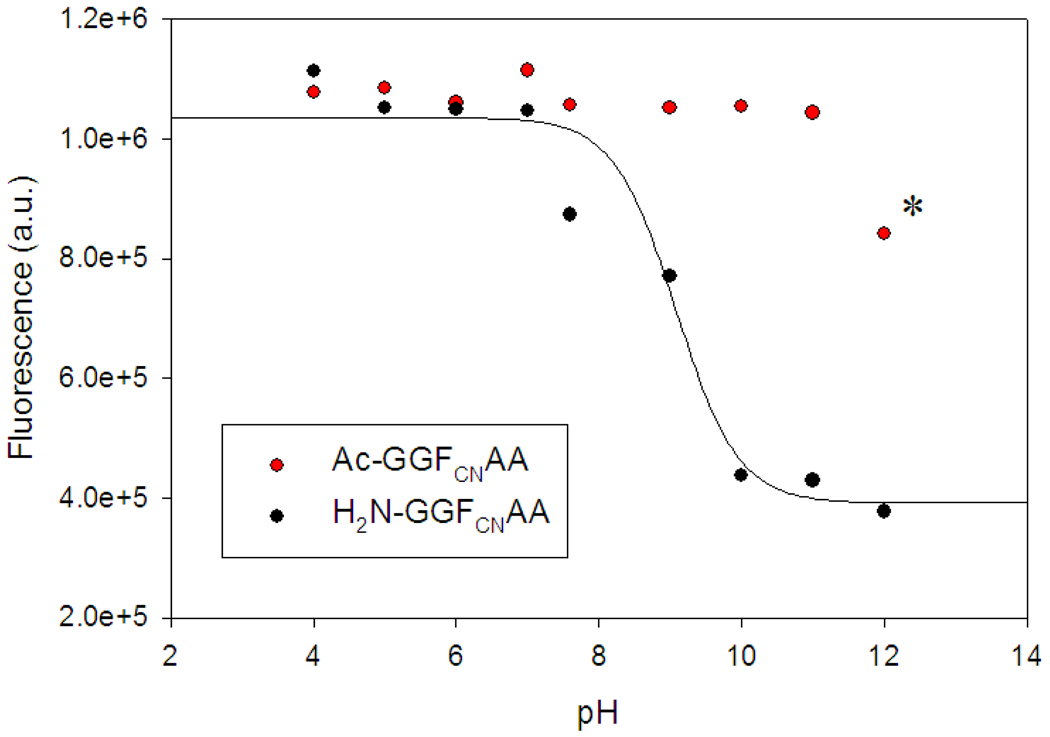
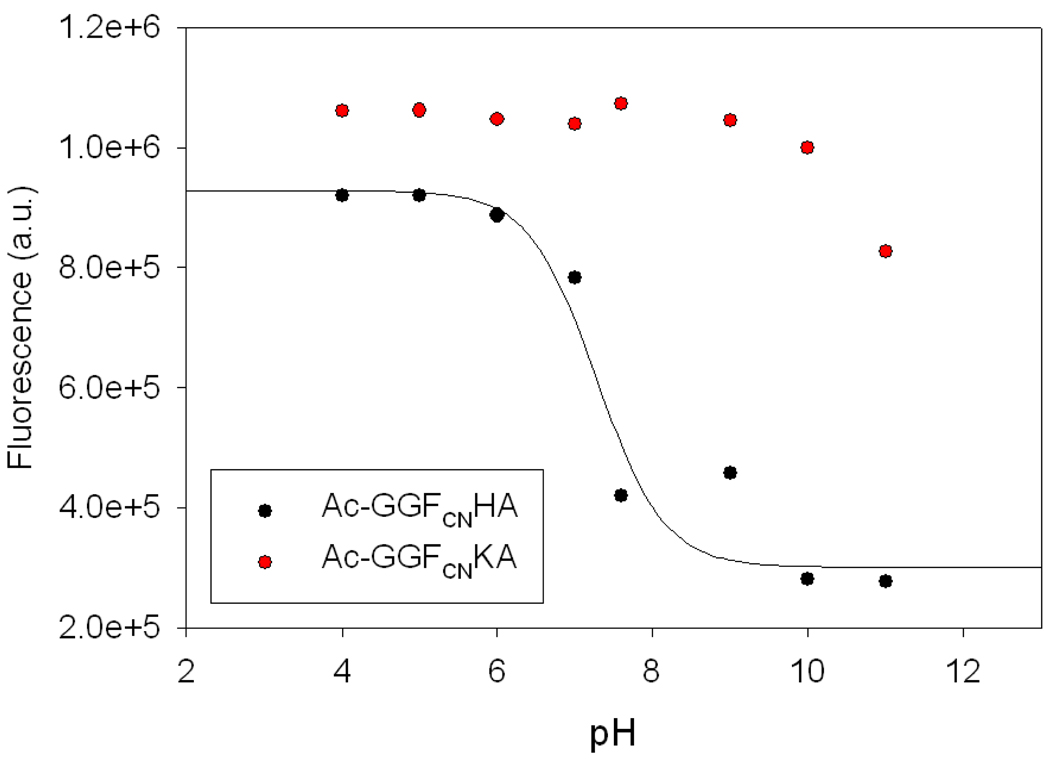
In order to determine the potential effects of the N-terminus we examined the fluorescence of the GGFCNAA variant with a free N-terminus as a function of pH (Figure 2). The fluorescence intensity titrates with a pKa of 9.0, consistent with that of the amino group, decreasing as the amino group is deprotonated. This observation is consistent with reports on benzonitrile fluorescence quenching with diethylamine (20). As a control, a parent peptide with an acetylated N-terminus was also examined and showed no change in fluorescence up to pH 12. The decrease at pH 12 is reproducible and is due to quenching by hydroxide ion (Supporting information).
Figure 2.
Comparison of the pH dependence of the fluorescence emission intensity for GGFCNAA peptide with a free N-terminus (black) and with an acetylated N-terminus (Ac-GGFCNAA, red). The solid black line is a fit of the data to the Henderson-Hasselbalch equation with an apparent pKa of 9.0. The decrease in intensity for the Ac-GGFCNAA peptide above pH 11 is due to quenching by hydroxide ion (labeled with a star). Fluorescence was excited at 240 nm and the emission was collected at 291 nm. The peptide concentration was 20 uM. The experiments were conducted in water at the indicated pH values.
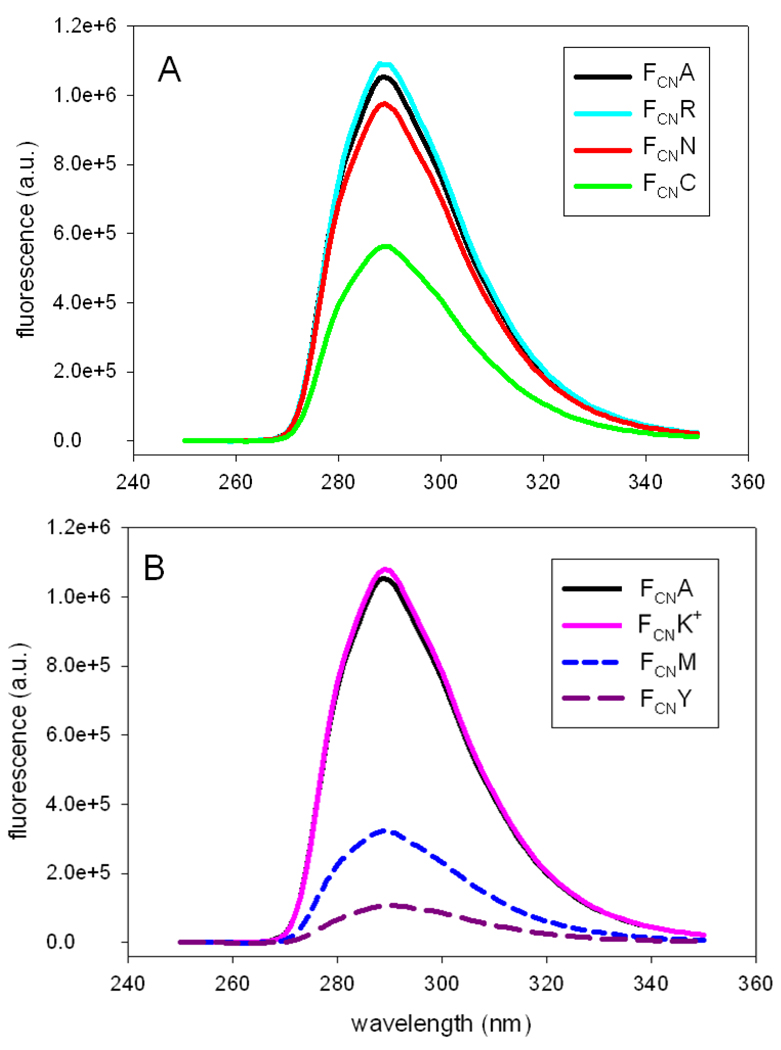
The ability of individual side-chains to modulate FCN fluorescence was examined in the context of the GGFCNAA peptides at pH 5. The peptides have a free N-terminus, but the pH dependent studies of the parent peptide with the acetylated and unblocked N-terminus demonstrate that a protonated amino group has no effect, thus alleviating any potential concerns about the interpretation of the spectra, since quenching is dominated by the side-chain. Figure 3 displays fluorescence emission spectra of various peptides. Each panel of the figure includes the spectrum of the parent peptide, GGFCNAA for comparison. At pH 5.0 the rank order of effectiveness in reducing the FCN fluorescence is Y > M > C > H+ > N > R = K+, where H+ denotes a positively charged His side-chain and K+ a positively charged Lys side-chain. Arg and positively charged Lys had no effect on FCN fluorescence.
Figure 3.
Fluorescence emission spectra of GGFCNXA peptides with a free N-terminus at pH 5. X being (A) Ala, Arg, Asn, and Cys; (B) Ala, Met, Lys, and Tyr. Fluorescence was excited at 240 nm. The experiments were conducted in water.
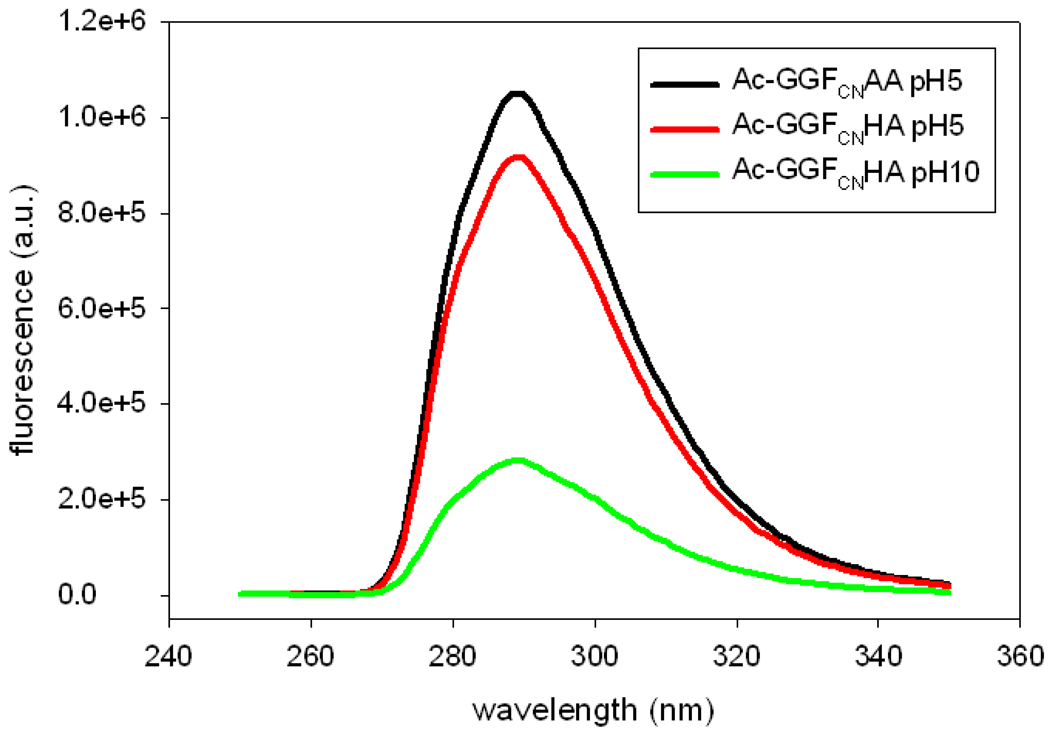
The pH dependent data collected for the GGFCNAA peptide suggests that it is worthwhile to examine the pH dependence of the quenching by His and Lys. In order to examine the effect of the protonation state of the His and Lys side-chains, we recorded fluorescence emission spectra of N-terminally acetylated variants of the GGFCNAA, GGFCNHA, and GGFCNKA peptides. Acetylation blocks the N-terminus and the resulting amide linkage is not an effective quencher. This allows the effect of the side-chain to be probed without complications from quenching by a deprotonated N-terminal amino group at high pH. The data indicates that a deprotonated His side-chain is a much more effective quencher than a protonated one (Figure 4). Figure 5 displays the fluorescence emission of His and Lys peptides with an acetylated N-terminus as a function of pH. In the His peptide, the FCN fluorescence decreases as the His side-chain is deprotonated at higher pH and the intensity vs. pH profile is well fit to an apparent pKa of 7.3. It is interesting to note that protonated His is a much more effective quencher of Trp fluorescence than neutral His, but the situation is reversed here (21, 22). Likewise, for the Lys peptide, the FCN fluorescence intensity does not change until the pH approaches the pKa of the Lys side-chain and then decreases. It was not possible to accurately monitor FCN fluorescence of the Lys peptide over the entire pH range, because quenching by hydroxide is significant at pH 12 and above. However, our observation that a deprotonated N-terminus is an effective quencher indicates that a deprotonated Lys side-chain will be a better quencher than a protonated Lys side-chain. Of course, the effect is likely to be of little practical significance in most studies of proteins given that the intrinsic pKa of Lys is above 10. We examined the ability of protonated and deprotonated carboxylic acids to quench FCN fluorescence by conducting Stern-Volmer plots of the quenching by acetic acid and sodium acetate. Both are inefficient quenchers with Stern-Volmer constants of 0.31 M−1 and 0.25 M−1 (supporting information).
Figure 4.
Comparison of the effects of a protonated and deprotonated His side-chain on FCN fluorescence measured for the GGFCNXA peptides with an acetylated (blocked) N-terminus. Black, X=Ala at pH 5; red X=His at pH 5; blue, X=His at pH 10. All spectra were recorded in water at the indicated pH. Sample concentration was 20 uM and the excitation was at 240 nm.
Figure 5.
Fluorescence emission intensity of the Ac-GGFCNHA (black) and Ac-GGFCNKA (red) N-teminal acetylated peptides as a function of pH. The FCN fluorescence is quenched as the pH approaches the pKa of His, indicating that deprotonated His is a better quencher of FCN fluorescence than protonated His. A decrease in fluorescence intensity of the Ac-GGFCNKA peptide is observed as the pH approaches the pKa of Lys. Data is not reported above pH 11 because quenching by OH− is significant. The solid curve is a fit of the His data to the Henderson Hasselbalch equation and yields an apparent pKa of 7.3.
Combining the peptide data with the pH dependent studies shows that the rank order of quenching is Y > H° > M > deprotonated N-terminus > C > H+ > N > R = K+ where H° denotes a neutral His side-chain. As noted, the quenching by a neutral Lys side-chain could not be measured, but it is expected to be comparable to that of the deprotonated N-terminus.
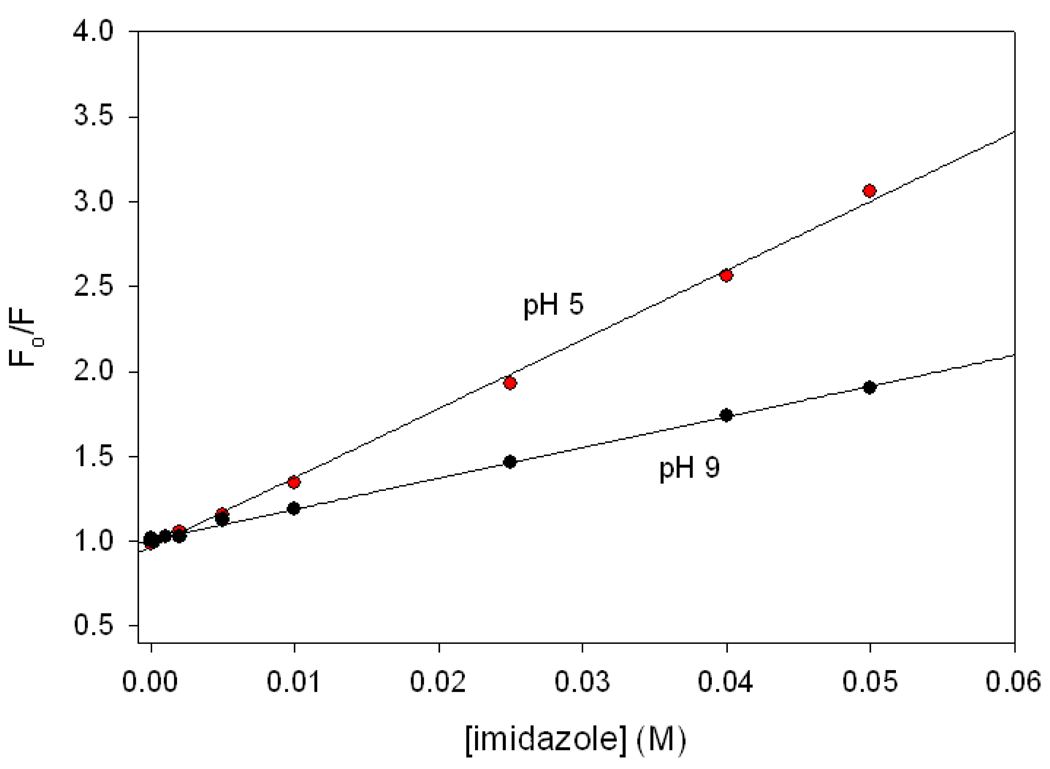
The results with the His peptide indicate that free imidazole should be a quencher of FCN fluorescence, and suggest that the neutral form should be more efficient than the protonated form. This was confirmed by Stern-Volmer analysis of quenching data for the acetylated GGFCNAA peptide (Figure 6). A Stern-Volmer plot of the ratio of the FCN fluorescence in the absence and presence of the quencher, F0/F vs. quencher concentration is linear. The slope of the curve, the Stern-Volmer constant (KSV), is a measure of the efficiency of a quencher. KSV for imidazole at pH 5 is 18.3 M−1 and at pH 9, it is 39.8 M−1, confirming that neutral imidazole is a significantly better quencher of FCN fluorescence than protonated imidazole. For comparison the Ksv value for hydroxide ion is 22.1 M−1 (supporting information). Chloride ion is the only other reported quencher of FCN fluorescence and its KSV value is 9.3 M−1 (9, 23).
Figure 6.
Stern-Volmer plots of the quenching of FCN fluorescence of the acetylated GGFCNAA (Ac-GGFCNAA) peptide by imidazole at pH 5 (black) and pH 9 (red). The Stern-Volmer constants are 18.3 M−1 at pH 5 and 39.8 M−1 at pH 9.
FCN Based Site Specific Probes of Helix Formation
The short range nature of the quenching suggests that suitable X-FCN pairs could be used to probe local structure formation. We tested this hypothesis by examining de novo designed helical peptides which contained either a Tyr-FCN pair or a His-FCN pair located one helical turn apart. Two 21 residue Ala based peptides were designed to test the ability of FCN fluorescence quenching to probe helix formation. The designed peptides each contain an Asp residue at position one since it is an excellent capping residue and a Pro at position two since prolines are favorable at this position in a helix (24). Several Lys residues were included to ensure solubility and the termini were capped to avoid unfavorable interactions with the helix dipole. The method will be easiest to interpret quantitatively in terms of helix formation when the FCN group is exposed to solvent in both states since the fluorescence quantum yield is influenced by hydrogen bonding to the cyano group as well as by quenching from other protein side-chains. If the FCN group were buried in one conformation, but not in the other, then the observed intensity change would be a convolution of the effects due to changes in side-chain quencher interactions and the effects of changes in solvation (18). However, note that both quenching and side-chain burial reduce FCN fluorescence. Thus, the pair can still be used as a probe of folding although deciphering the effects due to helix formation from those due to side-chain quenching could be difficult.
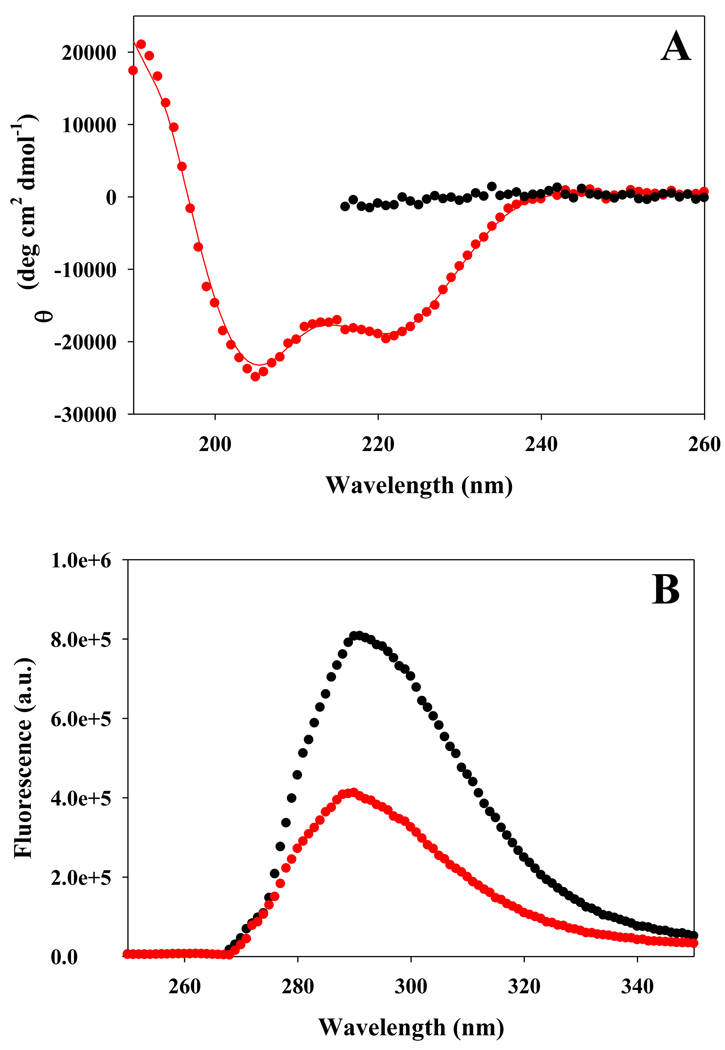
We first examined the use of a Tyr-FCN pair. In the present case, Tyr was incorporated at residue-12 and FCN at residue-16. The distance between the Tyr and FCN residues, when folded into a helical conformation is well within the Förster distance of 12 Ǻ (see supporting information for the Förster distance calculation) and the cyano group is exposed to solvent. The CD spectrum indicates that the Tyr-FCN peptide is partially helical in buffer. The spectrum shows the characteristic helical trend in ellipticity at 208 nm and 222 nm. The mean residue ellipticity is −19,300 deg cm2 dmol−1 at 222 nm, which corresponds to a helical content of 55%, as estimated using the method of Baldwin (Figure 7a) (25). CD reveals, as expected, that the peptide is unstructured in 8 M urea. The change in the conformation of the peptide is reflected by significant changes in its fluorescence properties. The fluorescence intensity is high in the urea induced unfolded state, but is significantly reduced in the partially folded state in buffer, owing to helix formation which brings the Tyr-FCN pair into closer proximity (Figure 7b). The ratio of the fluorescence of the partially folded form to that of the unfolded form is 0.50. The data demonstrates that a solvent exposed Tyr-FCN pair can be used as a local fluorescence probe of helix formation.
Figure 7.
(A) CD spectra of the Tyr-FCN peptide indicates that it is partially helical in buffer (red), but is unfolded in 8 M urea (black). Spectra were recorded at 25°C at pH 5.5. (B) Fluorescence emission spectra confirm that the Tyr-FCN pair provides a probe of helix formation. Fluorescence emission spectra of the Tyr-FCN peptide in the partially helical state (red) and in the urea unfolded state (black) are shown.
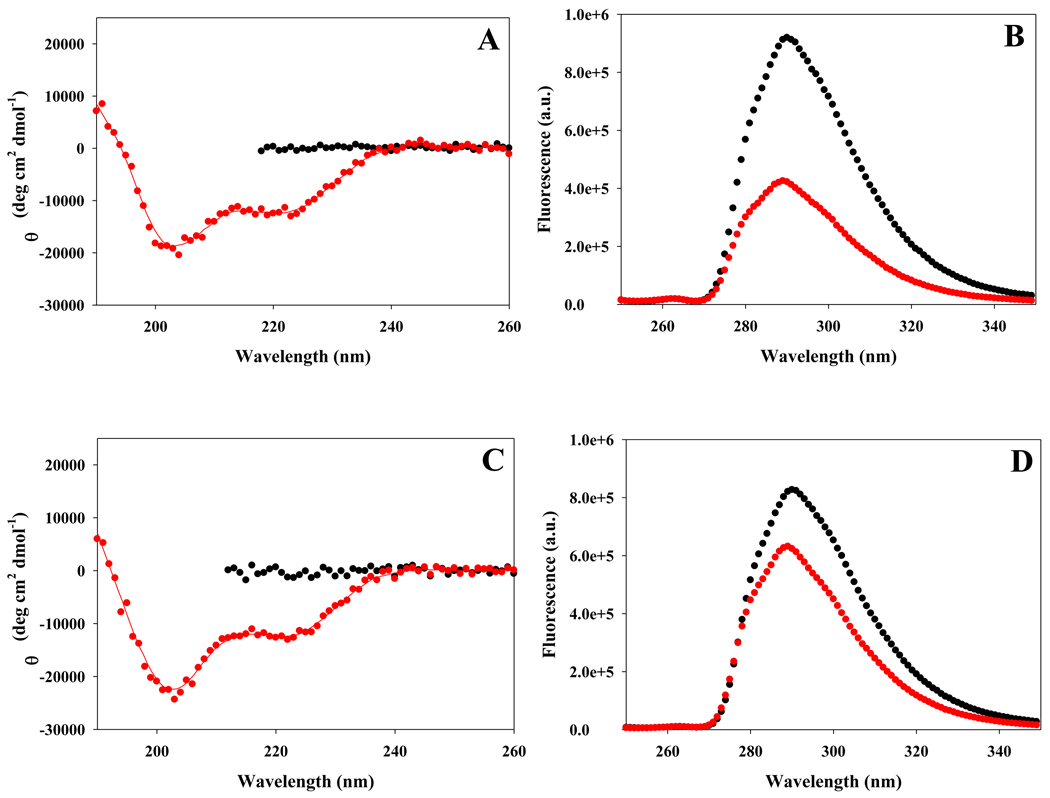
The data collected on the GGFCNHA peptide suggests that a solvent exposed His-FCN pair could also be exploited to provide a fluorescence based probe of helix formation. Our analysis of the small peptides indicates that the method should be more sensitive at pH values where the His sidechain is neutral. The quenching of Trp by protonated His has been used to probe protein conformation changes, but is limited to pHs where His is protonated (26). Thus, a His-FCN pair should be complimentary since it will be most effective when the His side-chain is neutral. We examined the fluorescence properties of the His-FCN containing Ala based peptide at pH 5.5, where the His side-chain is predominantly protonated, and at pH 8.3, where it is largely deprotonated. The peptide is partially helical in buffer at both pH 5.5 and pH 8.3 as judged by CD (Figure-8). The helical content is less than observed for the Tyr containing peptide, but it is still significant. The shape of the spectra suggests that the His containing peptide is slightly more helical at the higher pH, although the helical content is on the order of 37 to 42% for both pHs. The addition of 8 M molar urea disrupts the helical structure (Figure-8). The fluorescence is lower in the partially folded state than in the unfolded state at both pHs, however the change is much larger at pH 8.3, where the ratio of the fluorescence of the partially folded form to that of the unfolded form is 0.46 (Figure 8b). At pH 5.5 the ratio is 0.74 (Figure-8d). The data confirms that solvent exposed FCN-His pairs can be effectively exploited to probe helical structure and demonstrates that the method is most sensitive when the His sidechain is neutral.
Figure 8.
(A) CD spectra of the His-FCN peptide at pH 8.3 indicates that it is partially helical in buffer (red), but is unfolded in 8 M urea (black). The red curve in the CD spectra is included as an aid to visualize. (B) Fluorescence emission spectra confirm that the His-FCN pair provides a sensitive probe of helix formation when the His side-chain is neutral. Fluorescence emission spectra of the His-FCN peptide in the partially helical state (red) and in the urea-unfolded state (black) are shown at pH 8.3. (C) CD spectra of the His-FCN peptide at pH 5.5 indicates that it is partially helical in buffer (red), but is unfolded in 8 M urea (black). (D) Fluorescence emission spectra confirm that the His-FCN pair is a less sensitive probe of helix formation when the His side-chain is charged. Fluorescence emission spectra of the His-FCN peptide in the partially helical state (red) and in the urea unfolded state (black) are shown at pH 5.5.
Conclusions
The data presented here provides a comprehensive catalog of the effect of protein side-chains on FCN fluorescence. The short range of the quenching by His and Tyr can be exploited to design fluorescence-based sensors of specific elements of secondary structure. The strategy was demonstrated here using α-helical peptides, but can obviously be applied to globular proteins as well. In this case the FCN-Tyr/His pair should be located in solvent exposed sites so that the change in fluorescence intensity upon unfolding is dominated by changes in the quencher flurophore distance and not by changes in the solvation of the cyano group, which may arise if the probe were incorporated into a site that is buried in the native state but exposed in the unfolded state. The change in fluorescence should be even larger than observed for the Ala-rich peptides, since the helical structure will be fully formed in a folded globular protein, but is only partially formed in the peptides. The approach is clearly not limited to helices and could be used to follow β-hairpin or β-sheet formation. The quenching studies of the pentapeptides indicate that FCN-Met pair could also be exploited to follow local structure formation.
Supplementary Material
Table 1.
Effect of various side-chains on p-cyanophenylalanine fluorescence. Measurements were performed in water at pH 5 if not otherwise indicated. Acetylated peptides were used to obtain the data for both of the His peptides.
| Side-chain | Intensity ratio relative to control: GGFCNXA / GGFCNAA |
|---|---|
| Tyr | 0.10 |
| His (pH:10) | 0.27 |
| Met | 0.31 |
| Cys | 0.54 |
| His+ (pH 5) | 0.85 |
| Asn | 0.93 |
| Lys+ (pH 5) | 1.00 |
| Arg | 1.00 |
Acknowledgements
We thank Mr. Vadim Patsalo for helpful discussions and data analysis.
Grant Sponsor: NIH grant GM078114 and NSF grant MCB0919860 to D.P.R. and NSF grant CBET 1080909 to I.C.
Abbreviations
- FCN
p-cyanophenylalanine
- RET
resonance energy transfer
- MALDI-TOF MS
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- CD
circular dichroism
- KSV
Stern-Volmer constant
Footnotes
Supporting information. Fluorescence emission spectrum of FCN, absorbance spectrum of Tyr, Förster distance calculation for the FCN-Tyr pair and Stern-Volmer plots of FCN fluorescence quenching by hydroxide ion, acetic acid and sodium acetate. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Lakowicz JR. Principles of Fluorescence Spectroscopy. LLC New York, NY: Springer Science+Business Media; 2006. [Google Scholar]
- 2.Beechem JM, Brand L. Time-resolved fluorescence of proteins. Annu. Rev. Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- 3.Ross JBA, Szabo AG, Hogue CWV. Enhancement of protein spectra with tryptophan analogs: Fluorescence spectroscopy of protein-protein and proteinnucleic acid interactions. Fluorescence Spectroscopy. 1997;278:151–190. doi: 10.1016/s0076-6879(97)78010-8. [DOI] [PubMed] [Google Scholar]
- 4.Wong CY, Eftink MR. Incorporation of tryptophan analogues into staphylococcal nuclease, its V66W mutant, and Delta 137–149 fragment: Spectroscopic studies. Biochemistry. 1998;37:8938–8946. doi: 10.1021/bi971862o. [DOI] [PubMed] [Google Scholar]
- 5.Twine SM, Szabo AG. Fluorescent amino acid analogs. Method Enzymol. 2003;360:104–127. doi: 10.1016/s0076-6879(03)60108-4. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu. Rev. Biophys.Biomol. Struc. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 7.Rogers JMG, Lippert LG, Gai F. Non-natural amino acid fluorophores for one- and two-step fluorescence resonance energy transfer applications. Anal. Biochem. 2010;399:182–189. doi: 10.1016/j.ab.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucker MJ, Getahun Z, Nanda V, DeGrado WF, Gai F. A new method for determining the local environment and orientation of individual side chains of membrane-binding peptides. J. Am. Chem. Soc. 2004;126:5078–5079. doi: 10.1021/ja032015d. [DOI] [PubMed] [Google Scholar]
- 9.Tucker MJ, Oyola R, Gai F. A novel fluorescent probe for protein binding and folding studies: p-cyano-phenylalanine. Biopolymers. 2006;83:571–576. doi: 10.1002/bip.20587. [DOI] [PubMed] [Google Scholar]
- 10.Tucker MJ, Tang J, Gai F. Probing the kinetics of membrane-mediated helix folding. J. Phys. Chem. B. 2006;110:8105–8109. doi: 10.1021/jp060900n. [DOI] [PubMed] [Google Scholar]
- 11.Aprilakis KN, Taskent H, Raleigh DP. Use of the novel fluorescent amino acid p-cyanophenylalanine offers a direct probe of hydrophobic core formation during the folding of the N-terminal domain of the ribosomal protein L9 and provides evidence for two-state folding. Biochemistry. 2007;46:12308–12313. doi: 10.1021/bi7010674. [DOI] [PubMed] [Google Scholar]
- 12.Marek P, Gupta R, Raleigh DP. The fluorescent amino acid p-cyanophenylalanine provides an intrinsic probe of amyloid formation. Chembiochem. 2008;9:1372–1374. doi: 10.1002/cbic.200800052. [DOI] [PubMed] [Google Scholar]
- 13.Miyake-Stoner SJ, Miller AM, Hammill JT, Peeler JC, Hess KR, Mehl RA, Brewer SH. Probing protein folding using site-specifically encoded unnatural amino acids as FRET donors with tryptophan. Biochemistry. 2009;48:5953–5962. doi: 10.1021/bi900426d. [DOI] [PubMed] [Google Scholar]
- 14.Taskent-Sezgin H, Chung J, Patsalo V, Miyake-Stoner SJ, Miller AM, Brewer SH, Mehl RA, Green DF, Raleigh DP, Carrico I. Interpretation of p-cyanophenylalanine fluorescence in proteins in terms of solvent exposure and contribution of side-chain quenchers: A combined fluorescence, IR and molecular dynamics study. Biochemistry. 2009;48:9040–9046. doi: 10.1021/bi900938z. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Strzalka J, Tronin A, Johansson JS, Blasie JK. Mechanism of interaction between the general anesthetic halothane and a model ion channel protein, II: fluorescence and vibrational spectroscopy using a cyanophenylalanine probe. Biophys. J. 2009;96:4176–4187. doi: 10.1016/j.bpj.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz KC, Supekova L, Ryu YH, Xie JM, Perera R, Schultz PG. A genetically encoded infrared probe. J. Am. Chem. Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 17.Tucker MJ, Oyola R, Gai F. Conformational distribution of a 14-residue peptide in solution: A fluorescence resonance energy transfer study. J. Phys. Chem. B. 2005;109:4788–4795. doi: 10.1021/jp044347q. [DOI] [PubMed] [Google Scholar]
- 18.Glasscock JM, Zhu YJ, Chowdhury P, Tang J, Gai F. Using an amino acid fluorescence resonance energy transfer pair to probe protein unfolding: Application to the villin headpiece subdomain and the LysM domain. Biochemistry. 2008;47:11070–11076. doi: 10.1021/bi8012406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serrano AL, Troxler T, Tucker MJ, Gai F. Photophysics of a fluorescent non-natural amino acid: p-Cyanophenylalanine. Chem. Phys. Lett. 2010;487:303–306. doi: 10.1016/j.cplett.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida M, Kaneko H, Kitamura A, Ito T, Oohashi K, Morikawa N, Sakuragi H, Tokumaru K. Mechanism for light-induced hydrogen isotope exchange in benzonitrile. B. Chem. Soc. Jpn. 1976;49:1697–1700. [Google Scholar]
- 21.Chen Y, Barkley MD. Toward understanding tryptophan fluorescence in proteins. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 22.VanGilst M, Hudson BS. Histidine-tryptophan interactions in T4 lysozyme: 'Anomalous' pH dependence of fluorescence. Biophys. Chem. 1996;63:17–25. doi: 10.1016/s0301-4622(96)02181-3. [DOI] [PubMed] [Google Scholar]
- 23.Marek P, Mukherjee S, Zanni MT, Raleigh DP. Residue specific, real time characterization of lag phase species and fibril growth during amyloid formation: A combined fluorescence and IR study of p-cyanophenylalanine analogs of islet amyloid polypeptide. J. Mol. Biol. 2010 doi: 10.1016/j.jmb.2010.05.041. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aurora R, Rose GD. Helix capping. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo PZ, Baldwin RL. Mechanism of helix induction by trifluoroethanol: A framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 26.Kubelka J, Chiu TK, Davies DR, Eaton WA, Hofrichter J. Sub-microsecond protein folding. J. Mol. Biol. 2006;359:546–553. doi: 10.1016/j.jmb.2006.03.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.