Abstract
Plasma membrane monoamine transporter (PMAT) is a new polyspecific transporter that interacts with a wide range of structurally diverse organic cations. To map the physicochemical descriptors of cationic compounds that allow interaction with PMAT, we systematically analyzed the interactions between PMAT and three series of structural analogs of known organic cation substrates including phenylalkylamines, n-tetraalkylammonium (n-TAA) compounds, and β-carbolines. Our results showed that phenylalkylamines with a distance between the aromatic ring and the positively charged amine nitrogen atom of ∼6.4 Å confer optimal interactions with PMAT, whereas studies with n-TAA compounds revealed an excellent correlation between IC50 values and hydrophobicity. The five β-carbolines that we tested, which possess a pyridinium-like structure and are structurally related to the neurotoxin 1-methyl-4-phenylpyridinium, inhibited PMAT with high affinity (IC50 values of 39.1–65.5 μM). Cytotoxicity analysis further showed that cells expressing PMAT are 14- to 15-fold more sensitive to harmalan and norharmanium, suggesting that these two β-carbolines are also transportable substrates of PMAT. We then used computer-aided modeling to generate qualitative and quantitative three-dimensional pharmacophore models on the basis of 23 previously reported and currently identified PMAT inhibitors and noninhibitors. These models are characterized by a hydrogen bond donor and two to three hydrophobic features with distances between the hydrogen bond donor and hydrophobic features ranging between 5.20 and 7.02 Å. The consistency between the mapping results and observed PMAT affinity of a set of test compounds indicates that the models performed well in inhibitor prediction and could be useful for future virtual screening of new PMAT inhibitors.
Introduction
Organic cations are structurally diverse endogenous and exogenous compounds that carry a net positive charge at physiological pH. Mammalian cells have evolved complex transport systems to eliminate hydrophilic organic cations from the body, such as the well established organic cation transporters 1 to 3 (OCT1–3, SLC22A1–3), and multidrug and toxin extrusion proteins 1 and 2 (SLC47A1 and 2) (Koepsell and Endou, 2004; Wright and Dantzler, 2004; Otsuka et al., 2005; Fujita et al., 2006). We recently cloned and characterized a novel polyspecific organic cation transporter, designated plasma membrane monoamine transporter (PMAT) (Engel et al., 2004), the fourth member of the equilibrative nucleoside transporter family (SLC29). Unlike other SLC29 family members (i.e., equilibrative nucleoside transporters 1–3), which exclusively transport nucleosides and nucleoside analogs, but similar to the genetically unrelated OCTs, PMAT functions as a polyspecific transporter for a wide range of small hydrophilic organic cations including monoamine neurotransmitters, drugs, and toxins (Engel et al., 2004; Zhou et al., 2007c, 2010). We previously showed that PMAT-mediated organic cation transport is sensitive to membrane potential changes and is also influenced by extracellular pH (Engel et al., 2004; Xia et al., 2007). However, the exact transport mechanism and the role of protons are currently unclear. In humans, PMAT mRNA is most strongly expressed in the brain, but transcripts are also found in other organs such as the kidney, heart, and small intestine (Engel et al., 2004; Zhou et al., 2007c; Xia et al., 2009). Therefore, PMAT may play an important role in brain monoamine clearance and tissue-specific disposition of organic cations (Dahlin et al., 2007; Xia et al., 2007; Zhou et al., 2007a).
Using a collection of structurally diverse organic cations, we previously probed the structural requirements of PMAT substrates and inhibitors (Engel and Wang, 2005). We found that a positive charge (e.g., an amine nitrogen atom) and a hydrophobic moiety are the two basic molecular features for transporter substrate-inhibitor interaction. These observations are consistent with our mutational analysis of PMAT, in which we identified a negatively charged residue (E206) in TM5 that functions as a charge sensor and is critical for cation selectivity (Zhou et al., 2007b). We also recently identified two aromatic residues, Y85 and Y112 in TM 1 and 2, respectively, as important molecular determinants for PMAT substrate interactions (Ho and Wang, 2010).
Whereas many organic cations inhibit PMAT (Engel and Wang, 2005), only a handful of compounds have been confirmed as PMAT substrates, representing three structural categories: 1) primary or secondary alkylamines attached to a planar aromatic ring moiety, such as tyramine, catecholamines, and indolamines (Engel et al., 2004; Engel and Wang, 2005); 2) aliphatic amines without an aromatic feature [e.g., TEA or metformin] (Engel and Wang, 2005; Zhou et al., 2007c); and 3) complex structures including aromatic pyridinium compounds, such as the neurotoxin MPP+ (Engel et al., 2004), which feature positively charged nitrogen atoms within a heterogeneous ring linked with one or more aromatic planar structures.
The goal of this study was to map in detail the molecular descriptors and chemical features of cationic compounds that allow them to interact at the substrate/inhibitor binding site of PMAT. We first systematically analyzed the interactions between PMAT and a series of structural analogs of known organic cation substrates including phenylalkylamines, n-tetraalkylammonium (n-TAA) compounds, and β-carbolines (Fig. 1). Aromatic alkylamine is the core structure shared by many PMAT substrates such as tyramine, catecholamines, and indoleamines (category 1) (Engel et al., 2004; Engel and Wang, 2005). We determined the inhibition potency of a series of phenylalkylamine analogs, which differed only in the distance between the aromatic ring and the positively charged amine nitrogen atom, to identify the optimal distance between the positive charge and the aromatic features for the most favorable interaction with PMAT. n-TAA compounds are aliphatic amines that lack an aromatic moiety (category 2). They are TEA analogs that share a similarity in backbone structure but differ in hydrocarbon chain length. β-Carbolines are structurally related to MPP+ and possess a pyridinium-like structure attached to an aromatic heterocyclic indole ring (category 3). They are considered as natural or environmental analogs of MPP+ and have been implicated as environmental risk factors for Parkinson's diseases (Nagatsu, 1997; Collins and Neafsey, 2000; Storch et al., 2004). On the basis of the current data and previously reported PMAT inhibitors and noninhibitors, we used computer-aided modeling to generate three-dimensional qualitative and quantitative pharmacophore models with the aim of characterizing the substrate-inhibitor interaction site in PMAT.
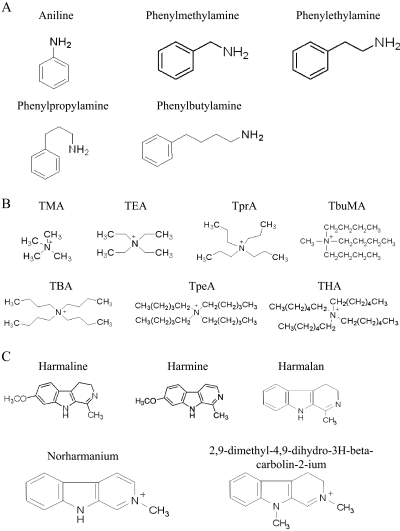
Fig. 1.
Structures of phenylalkylamines (A), n-TAA compounds (B), and β-carbolines (C) tested for interactions with PMAT.
Materials and Methods
Materials.
[3H]MPP+ (85 Ci/mmol) was obtained from American Radiolabeled Chemicals, Inc. (St. Louis, MO). The n-TAA compounds [tetramethylammonium (TMA), TEA, tetrapropylammonium (TPrA), tetrabutylammonium (TBA), tetrampentylammonium (TPeA), and tetrahexylammonium (THA)], tributylmethylammonium (TbuMA), aniline and phenylalkylamine derivatives (phenylmethylamine, phenylethylamine, phenylpropylamine, and phenylbutylamine), and β-carbolines [harmaline (4,9-dihydro-7-methoxy-1-methyl-3H-pyrido[3,4-b]indole), harmine (7-methoxy-1-methyl-9H-pyrido[3,4-b]indole), harmalan methosulfate (1-methyl-4,9-dihydro-3H-β-carboline, methosulfate), norharmanium methiodide (2-methyl-9H-β-carboline-2-ium, iodide), and 2,9-dimethyl-4,9-dihydro-3H-β-carbolin-2-ium, iodide] were obtained from Sigma-Aldrich (St. Louis, MO).
Cloning of PMAT and Expression in MDCK and HEK293 Cells.
PMAT cDNA was obtained and expressed in MDCK cells as described previously (Engel et al., 2004). In brief, PMAT cDNA, isolated from human kidney, was subcloned into the HindIII and XbaI restriction sites of the pcDNA3 vector (Invitrogen, Carlsbad, CA) and transfected into MDCK cells by liposome-mediated transfection (Lipofectamine; Invitrogen). A stably transfected cell line was obtained by G418 selection. PMAT- and vector-transfected MDCK cells were cultured in minimal essential medium containing 10% fetal bovine serum and 200 μg of G418/ml medium.
Because MDCK cells are resistant to MPP+ and β-carboline toxicity (Wang et al., unpublished data), we used a sensitive cell line, HEK293, as the expression system for the analyses of β-carbolines. The generation of HEK293 cell lines stably transfected with human PMAT and the empty vector pcDNA5 was described previously (Duan and Wang 2010). In brief, human PMAT cDNA was subcloned into the pcDNA5/FRT vector (Invitrogen). The pcDNA5/FRT empty vector or PMAT expression vector was cotransfected with pOG44 expressing the Flp-recombinase into the Flp-in HEK293 cell line. Flp-in HEK293 cells stably transfected with human PMAT were maintained in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 150 μg/ml hygromycin. Cells were cultured in a 37°C humidified incubator with 5% CO2. For better attachment of cells, all cell culture plastic surfaces were pretreated with 0.01% poly l-ornithine (molecular weight 30,000–70,000)/phosphate-buffered saline solution before plating.
Inhibition Studies.
For inhibition studies, cells were plated in 24-well plates and allowed to grow at 37°C for 2 to 3 days until confluent. Growth medium was aspirated, and each well was rinsed with Krebs-Ringer-Henseleit (KRH) buffer (5.6 mM glucose, 125 mM NaCl, 4.8 mM KCl, 1.2 mM KH2PO4, 1.2 mM CaCl2, 1.2 mM MgSO4, and 25 mM HEPES, pH 7.4) and then preincubated in KRH buffer for 15 min at 37°C. Cells were then incubated at 37°C for 1 min in KRH buffer containing [3H]MPP+ (1 μM) in the absence or presence of an inhibitor at various concentrations. Earlier studies from our laboratory have shown that 1 min is within the linear range for PMAT-mediated MPP+ uptake in transfected MDCK and HEK293 cells (Engel et al., 2004; Duan and Wang, 2010). This time point was also used in our previous studies to obtain IC50 values for various inhibitors (Engel and Wang, 2005; Zhou et al., 2007a,c). After incubation, uptake was terminated by aspirating the reaction mixture and washing the cells three times with ice-cold KRH buffer. Cells were then solubilized with 0.5 ml of 1 N NaOH. After 2 h, the solubilized cells were neutralized with 0.5 ml of 1 N HCl. This solution (0.5 ml) was quantified by liquid scintillation counting, and 25 μl was used for the protein assay using a BCA protein assay kit (Pierce, Waltham, MA). The amount of protein was calculated from the standard curve generated by the use of known amounts of bovine serum albumin, and the uptake in each well was normalized to its protein content. All inhibition experiments were performed in triplicate in three different wells on the same plate and repeated two to four times. Results from a representative experiment are shown. Data are expressed as the mean ± S.D. Statistical significance was determined by Student's t test, and kinetic parameters were determined by nonlinear least-squares regression fitting as described previously (Wang et al., 1997). The IC50 was determined by fitting the data to the equation V = V0 + (Vmax − V0)/[1 + (I/IC50)nH], where V is the rate of uptake of MPP+ in the presence of the inhibitor, V0 is the residual noninhibitable baseline value, Vmax is the rate of uptake of MPP+ in the absence of inhibitor, I is the inhibitor concentration, and nH is the Hill coefficient.
β-Carboline Toxicity Assay.
The cytotoxicity of various β-carbolines toward PMAT- and pcDNA5-transfected HEK293 cells was measured by MTT assay. Cells were seeded in Dulbecco's modified Eagle's medium with 10% fetal bovine serum on 96-well plates at a density of ∼5000 cells/well. After an ∼48-h incubation (∼40–50% confluence), cells were changed to fresh growth medium containing a β-carboline at graded concentrations. After a 72-h incubation in a 95% O2 incubator at 37°C, cells were washed, and cell viability in each well was determined using the thiazolyl blue tetrazolium bromide assay according to the manufacturer's instructions (Sigma-Aldrich). Cell viability was quantified by measuring light absorbance at 570 nm of the reduced MTT formazan. Five to six determinations were performed within each experiment, and three independent experiments were performed. Because we observed a systematic increase (10–25%) in optical intensity in cells treated with all β-carbolines at low and nontoxic concentrations, which is probably due to additional optical absorbance by residual β-carbolines, the survival rate in cells treated with the lowest drug concentration (1 μM) was set to 100% and used as a control to normalize cell viability across the entire concentration range (1–1000 μM). The EC50, defined as the β-carboline concentration leading to half-maximal cell survival, was determined by fitting the cell growth data to the following model using nonlinear regression (WinNonlin version 3.2; Pharsight, Mountain View, CA): S = Smax − [Smax − S0] × [Cγ/(Cγ + EC50γ)], where S is the cell survival expressed as percentage of the optical density to control cells, Smax is the maximal cell survival, S0 is the lowest residual cell survival at the high drug concentration, C is the β-carboline concentration, and γ is the Hill coefficient.
Physiochemical Descriptors.
The octanol/water partition coefficient (log P) values were calculated using Advanced Chemistry Development Software ChemSketch (version 12.01; ACD/Labs, Toronto, ON, Canada). The distance between the amine nitrogen atom and the middle of the nearest aromatic ring was determined with Marvin-Space (version 5.3.6; ChemAxon, Budapest, Hungary).
Qualitative and Quantitative Pharmacophore Modeling.
Molecular modeling studies were performed using Discovery Studio (DS) 3.0 (Accelrys, San Diego, CA). A qualitative pharmacophore model was developed on the basis of 23 previously reported and currently identified PMAT inhibitors and noninhibitors (Table 3) using the Common Feature Pharmacophore Generation protocol embedded in DS3.0. The 3D molecular structures of compounds used for both qualitative and quantitative model generation were obtained from either PubChem Compound (http://www.ncbi.nlm.nih.gov/ pccompound) or by sketching and subsequent energy minimization within SYBYL-X 1.2 (Tripos, St. Louis, MO). Hydrophobic, hydrophobic-aromatic, hydrogen bond acceptor and donor, and positive charge features were used for mapping functional groups of molecules in both qualitative and quantitative models. The molecular conformations for both models were generated by the FAST algorithm (Larger and Hoffmann, 2006) with maximum conformer number 255. The compounds fluvoxamine, rhodamine123, and decynium-22 were defined as active compounds, whereas other inhibitors were considered moderately active compounds during common feature model generation. A test set comprising nine PMAT inhibitors and noninhibitors (Table 3) were used for common feature pharmacophore validation with the Ligand Profiler protocol of DS3.0. Their conformers were generated with FAST and their fitting algorithm was RIGID (Kabsch, 1978). The maximum number of features that were allowed to be missed when these ligands were mapped to the pharmacophore model was set to 1.
TABLE 3.
Training and test set compounds for the qualitative pharmacophore model
| No. | Name | Observed Activity |
Fit Valuea | Fit Scoreb | |
|---|---|---|---|---|---|
| IC50 | Ki | ||||
| μM | |||||
| 1 | Fluvoxaminec | 11.00 | 3.00 | ||
| 2 | Sertralinec | 13.54 | 2.64 | ||
| 3 | Phenylbutylamine | 27.70 | 2.14 | ||
| 4 | Fluoxetined | 28.39 | 22.7 | 2.92 | |
| 5 | Harmaline | 39.10 | 1.67 | ||
| 6 | Norharmanium | 44.20 | 1.28 | ||
| 7 | Harmalan | 46.00 | 1.41 | ||
| 8 | Harmine | 48.60 | 1.59 | ||
| 9 | Aniline | >10,000 | 0.99 | ||
| 10 | Decynium-22d | 0.10 | 1.99 | ||
| 11 | Rhodamine123d | 1.02 | 2.29 | ||
| 12 | Quinidined | 25.30 | 2.87 | ||
| 13 | Desipramined | 32.60 | 2.87 | ||
| 14 | Tryptamined | 62.90 | 1.81 | ||
| 15 | Corticosteroned | 450.50 | 2.47 | ||
| 16 | Clonidined | <500 | 2.44 | ||
| 17 | Amantadined | <500 | 1.63 | ||
| 18 | Cimetidined | >500 | 2.69 | ||
| 19 | Procainamided | >500 | 2.57 | ||
| 20 | Cholined | >2000 | 1.00 | ||
| 21 | Creatinined | NI | 1.19 | ||
| 22 | l-Dopad | NI | 1.98 | ||
| 23 | Thiamined | NI | 1.96 | ||
| 24 | Phenylpropylamined,e | 9.80 | 0.67 | ||
| 25 | Paroxetineb,e | 22.46 | 0.87 | ||
| 26 | Phenyethylaminee | 23.40 | 0.66 | ||
| 27 | Phenymethylaminee | 170.60 | 0.59 | ||
| 28 | Citalopramb,e | 116.67 | 0.67 | ||
| 29 | Phenformine,f | 245.00 | 0.67 | ||
| 30 | Buformine,f | 1500.00 | 0.25 | ||
| 31 | PAHd,e | NI | 0.38 | ||
| 32 | Agmatined,e | NI | 0.00 | ||
NI, no interaction with PMAT.
Training set compounds fit values.
Test set compounds fit scores when mapped to the qualitative pharmacophore.
IC5 = 0 data are from Zhou et al. (2007a).
Ki data are from Engel and Wang (2005).
IC50 data are from Zhou et al. (2007c).
Test set compounds.
A quantitative pharmacophore model was generated with the 3D quantitative structure-activity relationship Pharmacophore Generation protocol of DS3.0. Ten hypotheses were produced on the basis of 13 PMAT inhibitors with IC50 values (Table 4). The hypothesis with the largest difference between the total energy cost and null cost, which indicates a lower risk of chance correlation reflected by the hypothesis (Chang et al., 2006a; Zheng et al., 2009), was selected for further analysis.
TABLE 4.
Observed and predicted IC50 values of training set compounds for the quantitative pharmacophore model
| No. | Name | Activity (IC50) |
Errora | Fit Value | |
|---|---|---|---|---|---|
| Observed | Predicted | ||||
| μM | |||||
| 1 | Fluvoxamineb | 11.00 | 9.17 | 1.20 | 5.24 |
| 2 | Sertralineb | 13.54 | 16.60 | 1.23 | 4.98 |
| 3 | Paroxetineb | 22.46 | 35.46 | 1.58 | 4.65 |
| 4 | Phenylbutylamine | 27.65 | 22.86 | 1.21 | 4.85 |
| 5 | Fluoxetineb | 28.39 | 22.35 | 1.27 | 4.86 |
| 6 | Harmaline | 39.10 | 18.36 | 2.13 | 4.94 |
| 7 | Norharmanium | 44.20 | 105.78 | 2.39 | 4.18 |
| 8 | Harmalan | 46.00 | 64.21 | 1.40 | 4.40 |
| 9 | Harmine | 48.60 | 19.15 | 2.54 | 4.92 |
| 10 | Citalopramb | 116.67 | 583.18 | 5.00 | 3.44 |
| 11 | Phenforminc | 245.00 | 583.52 | 2.38 | 3.44 |
| 12 | Buforminc | 1500.00 | 366.03 | 4.10 | 3.64 |
| 13 | Aniline | >10,000d | 11,198.90 | 1.79 | 2.16 |
The error represents the ratio of the predicted activity to the observed activity or its inverse if the ratio is less than 1.
IC50 data are from Zhou et al. (2007a).
IC50 data are from Zhou et al. (2007c).
Used a putative IC50 of 20,000 μM in the pharmacophore generation.
Results
cis-Inhibition Studies with Phenylalkylamines.
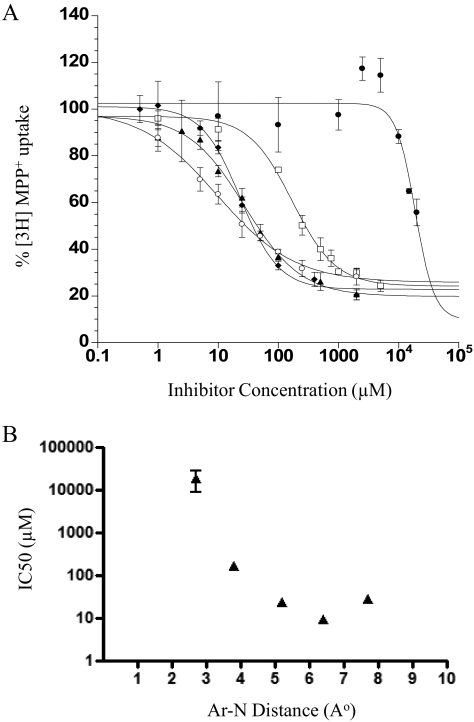
Phenylalkylamine is the core structure shared by many PMAT substrates such as tyramine, dopamine, and serotonin (Engel et al., 2004; Engel and Wang, 2005). We determined the inhibition potency of a series of phenylalkylamine analogs, which vary in the distance between the aromatic ring (Ar) and the positively charged amine nitrogen atom (N). The sensitivity of PMAT to these phenylalkylamines (Fig. 1A) was analyzed, and their IC50 values were determined (Table 1). The dose-dependent inhibition of PMAT by the phenylalkylamines is shown in Fig. 2A. The inhibition potencies of these amines differed by more than 17-fold, and the inhibition was greatest for phenylpropylamine (IC50 = 9.8 μM) with an Ar-N distance of 6.4 Å. Phenylethylamine and phenylbutylamine, with Ar-N distances of 5.2 and 7.7 Å, respectively, are also potent inhibitors of PMAT. On the other hand, analogs with shorter spacer distance did not interact with PMAT well. Aniline, which bears an Ar-N distance of 2.8 Å, does not interact with PMAT, whereas phenylmethylamine (Ar-N distance of 3.8 Å) is 6- to 17-fold less potent in inhibiting PMAT compared with amines with Ar-N distances >4 Å (Fig. 2B).
TABLE 1.
IC50 values of various compounds toward PMAT
Values are given as mean ± S.D. (n = 3).
| Inhibitors | IC50 |
|---|---|
| μM | |
| Phenylalkylamines (Ar-N distance) | |
| Aniline (2.8 Å) | >10,000 |
| Phenylmethylamine (3.8 Å) | 170.6 ± 19.5 |
| Phenylethylamine (5.2 Å) | 23.4 ± 1.9 |
| Phenylpropylamine (6.4 Å) | 9.8 ± 1.0 |
| Phenylbutylamine (7.7 Å) | 27.7 ± 4.7 |
| n-TAA | |
| TMA | >10,000 |
| TEA | 3555.8 ± 817.4 |
| TPrA | 1854.8 ± 458.4 |
| TBuMA | 405.3 ± 93.8 |
| TBA | 199.3 ± 39.6 |
| TPeA | 7.4 ± 1.8 |
| THA | 0.5 ± 0.1 |
| β-Carbolines | |
| Harmaline | 39.1 ± 6.1 |
| Harmine | 48.6 ± 6.3 |
| Harmalan | 46.0 ± 9.4 |
| Norharmanium | 44.2 ± 6.4 |
| 2,9-Dimethyl-4,9-dihydro-3H-β-carbolin-2-ium | 65.5 ± 7.5 |
Fig. 2.
A, concentration-dependent inhibition of MPP+ uptake by phenylalkylamines. Aniline (●), phenylmethylamine (□), phenylethylamine (♦), phenylpropylamine (○), and phenylbutylamine (▴) were tested in PMAT-expressing MDCK cells. Cells were incubated at 37°C with 1 μM [3H]MPP+ for 1 min in the presence of varying concentrations of inhibitors. B, relationships between IC50 values of phenylalkylamines and their Ar-N distance (angstroms). Each IC50 value represents the mean ± S.D. from three separate experiments.
cis-Inhibition Studies with n-TAA Compounds.
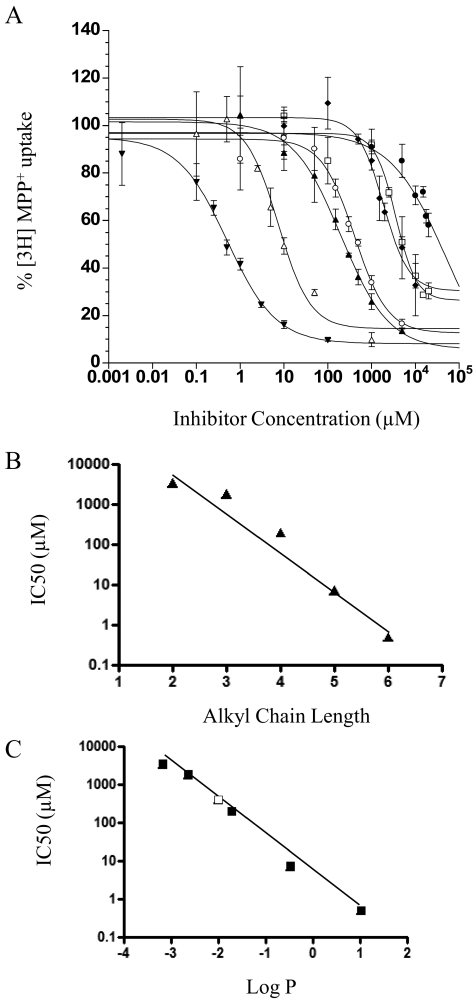
TEA is a classic substrate for OCTs (Gorboulev et al., 1997; Kekuda et al., 1998). We previously showed that TEA is also transported by PMAT (Engel and Wang, 2005). Here, we analyzed the interaction of PMAT with a series of TEA analogs (n-TAA compounds) that share similarity in backbone structure with TEA but differ in their hydrocarbon chain length (Fig. 1B). The dose-dependent inhibition of PMAT by the n-TAA compounds is shown in Fig. 3A, and their IC50 values are summarized in Table 1. The IC50 values of the n-TAA toward PMAT decreased drastically with increasing alkyl chain lengths (Table 1). A semilogarithmic plot of IC50 values versus alkyl chain length (N) revealed a linear relationship [log(IC50) = −1.01 × N + 5.98, r2 = 0.958] (Fig. 3B), suggesting that there is a good correlation between IC50 values and alkyl chain length. Because alkyl chain length is related to hydrophobicity of these compounds, we determined whether there is a correlation between IC50 and hydrophobicity (expressed as calculated log P) and observed a strong correlation [log(IC50) = −0.96 × log P + 0.606, r2 = 0.993] between these two parameters (Fig. 3C). The IC50 of TbuMA, which has a different side chain structure (Fig. 1B) and was not included in the analysis in Fig. 3B, also fitted well against its log P value (Fig. 3C), suggesting that hydrophobicity is a major determinant of the potency of the aliphatic organic cations to interact with PMAT.
Fig. 3.
A, concentration-dependent inhibition of MPP+ uptake by n-TAAs. TMA (●), TEA (□), TprA (♦), TbuMA (○), TBA (▴), TPeA (▵), and THA (▾) were tested in PMAT-expressing MDCK cells. Cells were incubated at 37°C with 1 μM [3H]MPP+ for 1 min in the presence of varying concentrations of inhibitors. B, relationships between IC50 values of n-TAA compounds and their alkyl chain length (N). Each IC50 value represents the mean ± S.D. fitted with data from three separate experiments by nonlinear regression. The fitted equation is log(IC50) = −1.01 × N + 5.98, r2 = 0.958. C, relationships between IC50 of n-TAA compounds and their Log P values. The fitted equation is log(IC50) = −0.96 × log P + 0.606, r2 = 0.993. ■, data for TEA, TprA, TBA, TpeA, and THA; □, TbuMA.
cis-Inhibition Studies with β-Carbolines.
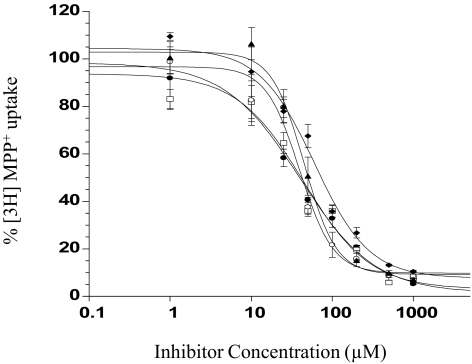
β-Carbolines (Fig. 1C) are structurally related to MPP+, which is efficiently transported by PMAT as well as by the OCTs. MPP+ differs from the other two substrate categories in that the positively charged nitrogen atom is imbedded into the pyridinium ring. Five commercially available β-carbolines possessing a pyridine or pyridinium-like structure and an indole ring were tested for their dose-dependent inhibition of PMAT (Fig. 4). Their IC50 values toward PMAT are summarized in Table 1. All five β-carbolines are relatively potent inhibitors of PMAT and showed similar IC50 values in the range of 39.1 to 65.5 μM. Comparison among the β-carboline structures indicated that 3,4-carbon saturation on the pyridine ring (harmaline versus harmine), and the methoxyl substitution on position 7 of the indole ring (harmaline versus harmalan) does not significantly affect the β-carboline inhibition against PMAT.
Fig. 4.
Concentration-dependent inhibition of MPP+ uptake by tested β-carbolines. Harmaline (●), harmine (□), 2,9-dimethyl-4,9-dihydro-3H-β-carbolin-2-ium (♦), harmalan (○), and norharmanium (▴) were analyzed in PMAT-expressing HEK293 cells. Cells were incubated at 37°C with 1 μM [3H]MPP+ for 1 min in the presence of varying concentrations of inhibitors.
Toxicity Studies with β-Carbolines.
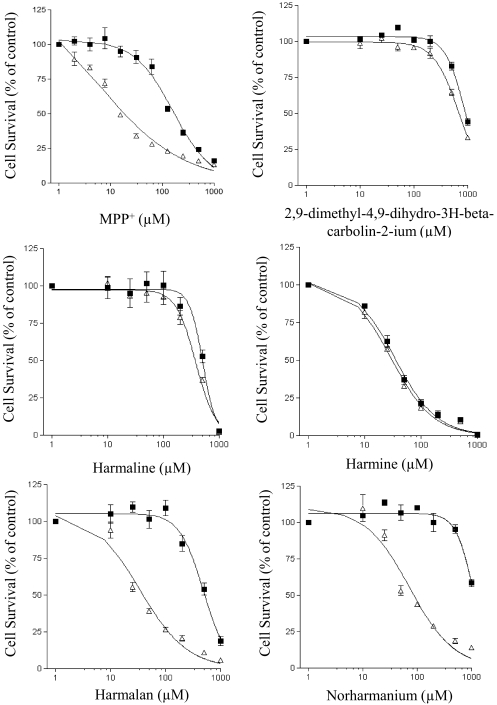
To further determine whether the tested β-carbolines are transportable substrates for PMAT, we took advantage of the observation that HEK293 cells are sensitive to the toxicity of MPP+. Because MPP+ exerts its toxicity by interfering with oxidative phosphorylation in mitochondria, PMAT-mediated cellular uptake of MPP+ would increase cell sensitivity to this toxin. Indeed, HEK293 cells expressing PMAT are much more sensitive to MPP+ cytotoxicity than vector-transfected control cells (Fig. 5). On the basis of the EC50 values obtained from the dose-response curves, PMAT-expressing cells are approximately 12-fold more sensitive to MPP+ than cells that do not harbor the transporter (Table 2). Of importance, our result showed that expression of PMAT also significantly sensitized cells to harmalan and norharmanium by approximately 14- and 15-fold, respectively (Table 2; Fig. 5). These data suggest that harmalan and norharmanium are indeed substrates of PMAT.
Fig. 5.
Effect of PMAT expression on HEK293 cell sensitivity to MPP+ and β-carboline toxicity. Concentration-dependent cell survival curves of PMAT-transfected (▵) and pcDNA5 vector-transfected (■) HEK293 cells after treatment with MPP+, harmaline, harmalan, 2,9-dimethyl-4,9-dihydro-3H-β-carbolin-2-ium, harmine, and norharmanium. Cells were incubated with various concentrations of above compounds for 72 h at 37°C.
TABLE 2.
MPP+ and β-carboline sensitivity in HEK293 cells stably transfected with pcDNA5 and PMAT
Each compound was tested in both pcDNA5- and PMAT-transfected HEK293 cell lines. EC50 values represent the effective concentration inducing a 50% decrease in cell viability of pcDNA5- and PMAT-expressing HEK293 cells, measured with the MTT assay after 72 h of drug exposure. Values are given as mean ± S.D. (n = 3). Fold increase in drug sensitivity was calculated by dividing the EC50 values of pcDNA5-transfected cells by those of PMAT-expressing cells.
| Compounds | EC50 in HEK293 Cells |
Fold Increase | |
|---|---|---|---|
| PMAT | pcDNA5 | ||
| μM | |||
| MPP+ | 10.2 ± 0.56* | 125 ± 27 | 12.2 |
| Harmaline | 382.6 ± 42.6 | 509.0 ± 61.1 | 1.33 |
| Harmine | 29.54 ± 3.90 | 34.60 ± 4.90 | 1.17 |
| Harmalan | 34.31 ± 8.90* | 489.7 ± 68.4 | 14.3 |
| Norharmanium | 73.18 ± 22.70* | 1081 ± 117 | 14.8 |
| 2,9-Dimethyl-4,9-dihydro-3H-β-carbolin-2-ium | 694.9 ± 70.5 | 887.4 ± 75.2 | 1.28 |
P < 0.01 versus pcDNA5 values.
Qualitative Pharmacophore Modeling.
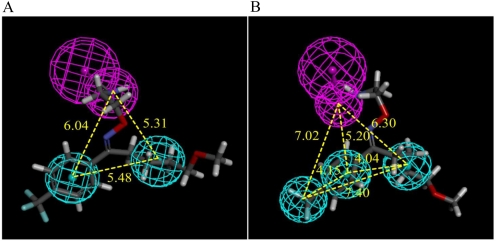
The 23 PMAT inhibitors and noninhibitors published previously and reported here (Table 3) were used as training set to develop a common feature pharmacophore model. Category 2 (n-TAA) compounds were excluded from both qualitative and quantitative analyses because of their limited structural features. In general, molecules with few molecular features can negatively affect model generation because of the intrinsic difficulty in detecting features that affect and correlate with their activities. Figure 6A illustrates the pertinent PMAT pharmacophore features characterized by a hydrogen bond donor and two hydrophobic groups. The inhibitor with the most favorable IC50 value, fluvoxamine, which had the highest fit value to the pharmacophore model, is shown in Fig. 6A. The distances between the hydrogen bond donor and the hydrophobic features range between 5.31 and 6.04 Å, whereas the distances between the hydrophobic groups were 5.48 Å.
Fig. 6.
Qualitative (A) and quantitative (B) pharmacophores for PMAT inhibition. Hydrophobic features are indicated with cyan spheres. The magenta spheres represent hydrogen bond donor with a vector pointing to a putative hydrogen bond donor atom. The figure show fluvoxamine, the most active drug in the quantitative pharmacophore training set, mapped to both models. Yellow numbers indicate the distances between the pharmacophore features.
The test set with nine PMAT inhibitors and noninhibitors was virtually screened with the Ligand Profiler protocol of DS3.0 using the generated pharmacophore model (Table 3), with one missed feature allowed when a ligand was mapped to the pharmacophore. Among the test set compounds, six are relatively potent PMAT inhibitors (IC50 < 300 μM), one is a weak inhibitor (buformin, IC50 > 1000 μM), and two are noninhibitors (PAH and agmatine). The test set compounds in Table 3 are listed with their fit scores. A higher fit score indicates a better fit of a ligand to the model, indicating a higher probability of being a PMAT inhibitor. In Table 3, agmatine had a fit score of 0, representing its failure to match to more than one pharmacophore feature. Buformin, PAH, and agmatine, which are weak PMAT inhibitor and noninhibitors, had the lowest fit scores among all the test compounds, indicating they had less favorable matches to the pharmacophore model compared with potent PMAT inhibitors within the test set. The agreement between the fit scores and the observed PMAT affinity of test set drugs indicates that the qualitative pharmacophore model has a reasonable capacity to predict the inhibition of previously unrecorded compounds against PMAT. Because a linear correlation between fit values and compound activity is not expected to emerge from a qualitative model, we applied quantitative models to allow subsequent prediction of PMAT affinity.
Quantitative Pharmacophore Modeling.
A 3D quantitative structure-activity relationship model was developed from 13 PMAT inhibitors with IC50 values. Aniline, which has an observed IC50 > 10,000 μM, was included in the training set with a putative activity IC50 = 20,000 μM so that the training set compounds had a range of IC50 values over 1000-fold. Table 4 lists the predicted as well as observed IC50 values of the training set compounds. The resulting quantitative pharmacophore is illustrated in Fig. 6B. The pharmacophore was characterized by one hydrogen bond donor and three hydrophobic features. The distances between the hydrogen bond donor and the hydrophobic features ranged between 5.20 and 7.02 Å. The distances among the hydrophobes were 4.04, 4.15, and 7.40 Å, respectively. The statistical significance of the hypothesis generated was evaluated with its total energy cost relative to that of the null hypothesis. The total cost, null cost, and fixed cost were 59.91, 65.59, and 56.25, respectively. The magnitude of the difference between the total and null cost could be larger for an ideal generated hypothesis. The correlation coefficient for the training set was r = 0.92, reflecting a statistically acceptable model.
Discussion
In this study, we first systematically analyzed the interactions between PMAT and a series of structural analogs of known PMAT substrates. Previous analyses of a set of structurally diverse PMAT substrates/inhibitors showed that a positively charged nitrogen atom and a hydrophobic mass are the principal determinants for PMAT substrate-inhibitor interaction (Engel and Wang, 2005). However, the spatial requirement between these core elements was hereto unknown. Using a series of phenylalkylamine analogs, we determined the optimal distance between the positive charge feature and the aromatic feature for the most favorable interaction with PMAT. Our results showed that phenylalkylamines with an Ar-N distance in the range of 5.2 to 7.7 Å are more potent inhibitors of PMAT (Fig. 2; Table 1). β-Carbolines, which have similar IC50 values and are good inhibitors of PMAT (Fig. 4; Table 1), all have an Ar-N distance of approximately 5.5 Å. MPP+, a well established substrate of PMAT, has an Ar-N distance of 5.6 Å. These observations are in good agreement with our quantitative and qualitative pharmacophore models developed from a larger set of structurally diverse molecules, which revealed a distance range of 5.20 to 7.02 Å between the hydrogen bond donor (i.e., the positively charged nitrogen) and the hydrophobic features (Fig. 6). On the basis of these data, we predict that an Ar-N structure spaced by two to three carbon atoms is optimal for interacting with PMAT.
The n-TAA compounds are aliphatic amines that possess four symmetric carbon chains but lack an aromatic moiety, which permits evaluation of aliphatic interactions with PMAT. Inhibition studies with n-TAA compounds showed a good correlation between IC50 values and alkyl chain length and hydrophobicity (Fig. 3, B and C). Our results showed that apparent binding affinity increases drastically with increasing hydrophobicity, suggesting that for aliphatic organic cations hydrophobicity is a primary determinant of their binding to PMAT. A similar trend of interaction with n-TAA compounds has been reported for the OCTs (Zhang et al., 1999; Dresser et al., 2002; Bednarczyk et al., 2003). However, it should be noted that although hydrophobicity is important for binding, a balance between hydrophobicity and hydrophilicity seems to be critical for subsequent substrate translocation and dissociation (Engel and Wang, 2005). Highly hydrophobic cations may bind to the transporter too tightly to be translocated and/or released efficiently. In the present study we only examined inhibition but not transport properties of the n-TAA compounds. Because of their overly simple and symmetric structures (Fig. 1B), the n-TAA compounds were not included or further analyzed in the qualitative and quantitative SAR analyses to avoid potential model distortion and bias.
β-Carbolines are structurally related to MPP+ and possess a pyridinium-like structure attached to an aromatic indole ring. β-Carbolines are considered as natural or environmental analogs of MPP+ and have been implicated as environmental risk factors for Parkinson's disease (Nagatsu, 1997; Collins and Neafsey, 2000; Storch et al., 2004). These compounds produce neurotoxicity by inhibiting complex I of the mitochondrial respiratory chain. Here, five commercially available β-carbolines were tested. Among them, two compounds (norharmanium and 2,9-dimethyl-4,9-dihydro-3H-β-carbolin-2-ium) are carbolinium ions that are permanently charged (Fig. 1C). The other three, harmaline, harmine, and harmalan, coexist as uncharged neutral species and monovalent cations protonated at the nitrogen atom in the pyridine ring (Douglas et al., 1983). All five β-carbolines were found to be potent inhibitors of PMAT with similar IC50 values in the range of 39.1 to 65.5 μM. Of note, these IC50 values are comparable to the apparent binding affinity of MPP+, which was previously determined to be 33 μM. Using cell toxicity assays, we further demonstrated that harmalan and norharmanium are transportable substrates of PMAT. Because PMAT is strongly expressed in the choroid plexus (Dahlin et al., 2007, 2009), which forms the blood-cerebrospinal fluid barrier, it may play a role in brain disposition of these neurotoxins. It should be noted that cytotoxicity assays are surrogate assays that are also influenced by the intrinsic toxicity of the tested compounds and their passive membrane permeability. Thus, a negative observation of no difference in cytotoxic response between the PMAT- and vector-transfected cells does not necessarily preclude the tested compound as a PMAT substrate. Whether harmaline, harmine, and 2,9-dimethyl-4,9-dihydro-3H-β-carbolin-2-ium are actual PMAT substrates requires further analysis.
Pharmacophore models attempt to describe ligand structural features that are responsible for their biological activities (Chang et al., 2006a,b; Zheng et al., 2009). In this study, one qualitative pharmacophore model for PMAT was derived with 23 PMAT inhibitors and noninhibitors, featuring one hydrogen bond donor and two hydrophobic functional groups. One quantitative pharmacophore hypothesis for PMAT based on 13 PMAT inhibitors suggested that a hydrogen bond donor and three hydrophobic features could be important for biological activity. The qualitative and quantitative models are in good agreement (despite their different training sets), and both feature a hydrogen bond donor and hydrophobic features. Figure 6, A and B, demonstrates that the features are oriented similarly in terms of their locations relative to the matched inhibitor fluvoxamine. Both hydrogen bond donors reside beside the –NH2 group; both models had a hydrophobic feature located at the aromatic ring of fluvoxamine, as well as a second one at the hydrocarbon chain connected to the –OCH3 group.
A test set consisting of nine PMAT inhibitors and noninhibitors was used to validate the qualitative pharmacophore model. The matching of a weak PMAT inhibitor and two PMAT noninhibitors was less favorable than that of all the potent PMAT inhibitors in terms of fit scores. The consistency between the mapping results and observed PMAT affinity of test set compounds indicates that the qualitative model has performed well in inhibition prediction against PMAT. Both models could be useful for future virtual screening of new PMAT inhibitors.
PMAT has been shown to share a range of inhibitors/substrates with OCTs from the SLC22 family (Engel and Wang, 2005). Of interest, molecular features that were identified to be important for PMAT interaction in this study, namely hydrophobicity and defined distance between the positive ionizable site and the hydrophobic aromatic site (Figs. 2 and 3), have also been found to be important descriptors for OCT1 and OCT2 affinity (Bednarczyk et al., 2003; Suhre et al., 2005; Zolk et al., 2009). Bednarczyk et al. (2003) developed a quantitative pharmacophore model based on 22 human OCT1 inhibitors and identified four pharmacophore features including three hydrophobes and one positive ionizable feature. Whereas they identified a positive ionizable feature residing near a nitrogen group as a pharmacophore feature, we detected a hydrogen bond donor at this nitrogen group. These two features agree with each other, because an amine group is both positively ionizable and a hydrogen bond acceptor. In another study, a qualitative OCT2 inhibition pharmacophore model featured a positive charge and a hydrogen bond donor/acceptor (Suhre et al., 2005). The similarities between the OCT and PMAT inhibition models provide further understanding into the functional parallels between transporters from two different solute carrier families (SLC29 and SLC22).
In summary, we systematically analyzed the interactions between PMAT and a series of structural analogs of known organic cation substrates and used computational modeling approaches to analyze the physicochemical descriptors that allow compound recognition at the substrate/inhibitor interaction site in PMAT. Our study provides new insights into the structure-activity relationship of this functionally unique transporter. The pharmacophore models generated from our study may serve as useful tools for future screening and development of novel PMAT substrates and inhibitors.
Acknowledgments
We thank Dr. Alex MacKerell in University of Maryland Baltimore for making Discovery Studio available to Y.P.
This work was supported by the National Institutes of Health National Institute of General Medicine Sciences [Grants GM066233, GM066233-07S1].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.111.184036.
- OCT
- organic cation transporter
- SLC
- solute carrier family
- PMAT
- plasma membrane monoamine transporter
- TM
- transmembrane domain
- MPP+
- 1-methyl-4-phenylpyridinium
- n-TAA
- n-tetraalkylammonium
- TMA
- tetramethylammonium
- TPrA
- tetrapropylammonium
- TBA
- tetrabutylammonium
- TPeA
- tetrampentylammonium
- THA
- tetrahexylammonium
- TbuMA
- tributylmethylammonium
- MDCK
- Madin-Darby canine kidney
- HEK
- human embryonic kidney
- KRH
- Krebs-Ringer Henseleit
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- DS
- Discovery Studio
- 3D
- three-dimensional
- Ar
- aromatic ring
- N
- positively charged amine nitrogen atom
- PAH
- p-aminohippuric acid.
Authorship Contributions
Participated in research design: Ho, Pan, Swaan, and Wang.
Conducted experiments: Ho, Pan, Cui, and Duan.
Performed data analysis: Ho, Pan, Swaan, and Wang.
Wrote or contributed to the writing of the manuscript: Ho, Pan, Swaan, and Wang.
References
- Bednarczyk D, Ekins S, Wikel JH, Wright SH. (2003) Influence of molecular structure on substrate binding to the human organic cation transporter, hOCT1. Mol Pharmacol 63:489–498 [DOI] [PubMed] [Google Scholar]
- Chang C, Bahadduri PM, Polli JE, Swaan PW, Ekins S. (2006a) Rapid identification of P-glycoprotein substrates and inhibitors. Drug Metab Dispos 34:1976–1984 [DOI] [PubMed] [Google Scholar]
- Chang C, Ekins S, Bahadduri P, Swaan PW. (2006b) Pharmacophore-based discovery of ligands for drug transporters. Adv Drug Deliv Rev 58:1431–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M, Neafsey E. (2000) β-Carboline analogues of MPP+ as environmental neurotoxins, in Neurotoxic Factors in Parkinson's Disease and Related Disorders (Storch A, Collins MA. eds) pp 115–130, Kluwer Academic Publishing/Plenum, New York [Google Scholar]
- Dahlin A, Royall J, Hohmann JG, Wang J. (2009) Expression profiling of the solute carrier gene family in the mouse brain. J Pharmacol Exp Ther 329:558–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin A, Xia L, Kong W, Hevner R, Wang J. (2007) Expression and immunolocalization of the plasma membrane monoamine transporter in the brain. Neuroscience 146:1193–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas KT, Sharma RK, Walmsley JF, Hider RC. (1983) Ionization processes of some harmala alkaloids. Mol Pharmacol 23:614–618 [PubMed] [Google Scholar]
- Dresser MJ, Xiao G, Leabman MK, Gray AT, Giacomini KM. (2002) Interactions of n-tetraalkylammonium compounds and biguanides with a human renal organic cation transporter (hOCT2). Pharm Res 19:1244–1247 [DOI] [PubMed] [Google Scholar]
- Duan H, Wang J. (2010) Selective transport of monoamine neurotransmitters by human plasma membrane monoamine transporter and organic cation transporter 3. J Pharmacol Exp Ther 335:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel K, Wang J. (2005) Interaction of organic cations with a newly identified plasma membrane monoamine transporter. Mol Pharmacol 68:1397–1407 [DOI] [PubMed] [Google Scholar]
- Engel K, Zhou M, Wang J. (2004) Identification and characterization of a novel monoamine transporter in the human brain. J Biol Chem 279:50042–50049 [DOI] [PubMed] [Google Scholar]
- Fujita T, Urban TJ, Leabman MK, Fujita K, Giacomini KM. (2006) Transport of drugs in the kidney by the human organic cation transporter, OCT2 and its genetic variants. J Pharm Sci 95:25–36 [DOI] [PubMed] [Google Scholar]
- Gorboulev V, Ulzheimer JC, Akhoundova A, Ulzheimer-Teuber I, Karbach U, Quester S, Baumann C, Lang F, Busch AE, Koepsell H. (1997) Cloning and characterization of two human polyspecific organic cation transporters. DNA Cell Biol 16:871–881 [DOI] [PubMed] [Google Scholar]
- Ho HT, Wang J. (2010) Tyrosine 112 is essential for organic cation transport by the plasma membrane monoamine transporter. Biochemistry 49:7839–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (1978) A discussion of the solution for the best rotation to relate two sets of vectors. Acta Cryst A 34:827–828 [Google Scholar]
- Kekuda R, Prasad PD, Wu X, Wang H, Fei YJ, Leibach FH, Ganapathy V. (1998) Cloning and functional characterization of a potential-sensitive, polyspecific organic cation transporter (OCT3) most abundantly expressed in placenta. J Biol Chem 273:15971–15979 [DOI] [PubMed] [Google Scholar]
- Koepsell H, Endou H. (2004) The SLC22 drug transporter family. Pflugers Arch 447:666–676 [DOI] [PubMed] [Google Scholar]
- Larger T, Hoffmann RD. (2006) Pharmacophore and Pharmacophore Searches, vol 32, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- Nagatsu T. (1997) Isoquinoline neurotoxins in the brain and Parkinson's disease. Neurosci Res 29:99–111 [DOI] [PubMed] [Google Scholar]
- Otsuka M, Matsumoto T, Morimoto R, Arioka S, Omote H, Moriyama Y. (2005) A human transporter protein that mediates the final excretion step for toxic organic cations. Proc Natl Acad Sci USA 102:17923–17928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch A, Hwang YI, Gearhart DA, Beach JW, Neafsey EJ, Collins MA, Schwarz J. (2004) Dopamine transporter-mediated cytotoxicity of β-carbolinium derivatives related to Parkinson's disease: relationship to transporter-dependent uptake. J Neurochem 89:685–694 [DOI] [PubMed] [Google Scholar]
- Suhre WM, Ekins S, Chang C, Swaan PW, Wright SH. (2005) Molecular determinants of substrate/inhibitor binding to the human and rabbit renal organic cation transporters hOCT2 and rbOCT2. Mol Pharmacol 67:1067–1077 [DOI] [PubMed] [Google Scholar]
- Wang J, Su SF, Dresser MJ, Schaner ME, Washington CB, Giacomini KM. (1997) Na+-dependent purine nucleoside transporter from human kidney: cloning and functional characterization. Am J Physiol 273:F1058–F1065 [DOI] [PubMed] [Google Scholar]
- Wright SH, Dantzler WH. (2004) Molecular and cellular physiology of renal organic cation and anion transport. Physiol Rev 84:987–1049 [DOI] [PubMed] [Google Scholar]
- Xia L, Engel K, Zhou M, Wang J. (2007) Membrane localization and pH-dependent transport of a newly cloned organic cation transporter (PMAT) in kidney cells. Am J Physiol Renal Physiol 292:F682–F690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Zhou M, Kalhorn TF, Ho HT, Wang J. (2009) Podocyte-specific expression of organic cation transporter PMAT: implication in puromycin aminonucleoside nephrotoxicity. Am J Physiol Renal Physiol 296:F1307–F1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gorset W, Dresser MJ, Giacomini KM. (1999) The interaction of n-tetraalkylammonium compounds with a human organic cation transporter, hOCT1. J Pharmacol Exp Ther 288:1192–1198 [PubMed] [Google Scholar]
- Zheng X, Ekins S, Raufman JP, Polli JE. (2009) Computational models for drug inhibition of the human apical sodium-dependent bile acid transporter. Mol Pharm 6:1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Duan H, Engel K, Xia L, Wang J. (2010) Adenosine transport by plasma membrane monoamine transporter: reinvestigation and comparison with organic cations. Drug Metab Dispos 38:1798–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Engel K, Wang J. (2007a) Evidence for significant contribution of a newly identified monoamine transporter (PMAT) to serotonin uptake in the human brain. Biochem Pharmacol 73:147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Xia L, Engel K, Wang J. (2007b) Molecular determinants of substrate selectivity of a novel organic cation transporter (PMAT) in the SLC29 family. J Biol Chem 282:3188–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Xia L, Wang J. (2007c) Metformin transport by a newly cloned proton-stimulated organic cation transporter (plasma membrane monoamine transporter) expressed in human intestine. Drug Metab Dispos 35:1956–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolk O, Solbach TF, König J, Fromm MF. (2009) Structural determinants of inhibitor interaction with the human organic cation transporter OCT2 (SLC22A2). Naunyn Schmiedebergs Arch Pharmacol 379:337–348 [DOI] [PubMed] [Google Scholar]