Abstract
![]()
The ChEMBL database was mined to efficiently assemble an ion channel-focused screening library. The compiled library consists of 3241 compounds representing 123 templates across nine ion channel categories. Compounds in the screening library are annotated with their respective ion channel category to facilitate back-tracing of prospective molecular targets from phenotypic screening results. The established workflow is adaptable to the construction of focused screening libraries for other therapeutic target classes with diverse recognition motifs.
Introduction
Ion channels are integral membrane proteins that govern the passage of ions across cell membranes. Encoded by approximately 400 ion channel genes in the human genome, this superfamily of membrane proteins is involved in many important physiological functions such as the regulation of blood pressure, neurotransmission, and hormonal secretion.1,2 Ion channels are implicated in a wide range of diseases including hypertension, neuromuscular disorders and Parkinson’s disease. Consequently, they constitute the third largest class of targets in drug discovery after protein kinases and G-protein coupled receptors.2,3 Despite its wide implication in disease conditions, the development of drugs targeting this membrane protein superfamily has remained underexploited, with only about 10% of drugs on the current market known to bind to ion channels.1,4
Since the advent of high-throughput screening (HTS) for drug discovery in the 1980s, HTS has become an important tool for hit identification in pharmaceutical research. In the past decade, strategies have evolved from traditional diverse HTS to the screening of focused libraries for a particular class of biological targets.5−7 For example, protein kinase-focused screening libraries have been reported by us and other research groups using well-documented structural motifs to select compounds satisfying a defined pharmacophore.8,9 However, in comparison to protein kinases, the assembly of a focused screening library targeting ion channels is more challenging owing to limited structural information about the targets, their structural diversity, and the absence of well-defined pharmacophores required for binding to ion channels.(10)
The release of the ChEMBL database(11) has facilitated open-access to a large volume of small-molecule bioactivity data for various therapeutic target families, including ion channels. Here, we describe an efficient workflow for compiling a focused screening library for ion channel targets using a combination of database mining and various chemoinformatics analytical tools for structural class generation and compound selection. The established workflow can easily be adopted to assemble focused screening libraries for other therapeutic target classes with diverse recognition motifs.
Results
To enable an efficient assembly of the ion channel-focused screening library, the procedure was divided into five stages (Figure 1).
Figure 1.
Workflow for the assembly of the ion channel-focused screening library.
Data Retrieval and Analysis
Bioactivity data of compounds active against ion channel targets were retrieved from the ChEMBL database.(11) This data set (ChEMBLinitial data set) contained 25150 compounds reported to be active against 337 different molecular targets. These compounds were subsequently grouped under 14 ion channel categories (Figure 2). The majority of the classification of ion channel categories followed those as defined in ChEMBL, apart from the groups of ligand- (LGIC) and voltage-gated ion channels (VGC) which were further divided to facilitate data handling. For LGIC, individual categories were constructed for glutamate-activated receptors and purinoceptors. Acetylcholine and serotoninergic 5HT3 receptors were grouped together to form the cationic Cys-loop channels (CationicCysLoop), whereas GABAA and glycine receptors were categorized as anionic Cys-loop channels (AnionicCysLoop). For VGC, each of sodium (Na+), calcium (Ca2+), potassium (K+), and cGMP-gated channels formed individual categories. The viral category was excluded as there was no bioactive compounds reported.
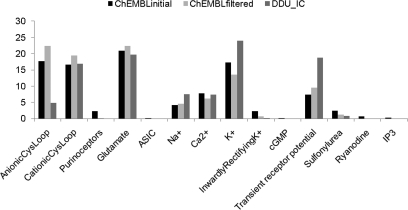
Figure 2.
Percentage composition per ion channel category in the ChEMBLinitial, ChEMBLfiltered, and DDU_IC data sets. No desirable bioactive compounds were found for four categories of ion channels (amiloride-sensitive sodium channels (ASIC), cGMP-gated channels, ryanodine receptors, and IP3 receptors).
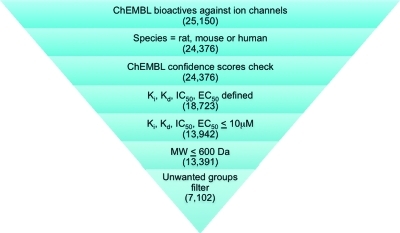
Various filters were applied to the ChEMBLinitial data set for the selection of compounds to form the ChEMBLfiltered data set (Figure 3). First, filters were introduced to exclude compounds for which bioactivity data was reported only for species other than rat, mouse or human. Next, the ChEMBL confidence scores for all bioactivity data were checked to ensure there was no experimental data representing activity against nonmolecular or nonprotein targets (ChEMBL confidence score between 1 and 3 inclusive). As expected, no compounds were removed as a result. Further, only compounds with reported bioactivity (Ki, Kd, IC50, or EC50) of ≤10 μM were kept. Compounds were then filtered using a molecular weight cutoff of 600 Da. Although this cutoff value would allow compounds that violate Lipinski’s rule-of-five,(12) it was considered appropriate at this stage of the analysis to minimize loss of core structural information when generating structural classes (see below). Finally, compounds containing unwanted groups(8) were excluded. This led to a collection of 7102 compounds representing 10 ion channel categories (Figure 2).
Figure 3.
Compilation of the ChEMBLfiltered data set containing bioactive ion channel compounds. Number of compounds retained at each filter step is shown in parentheses.
Structural Class Generation
Commercial availability search for compounds of the ChEMBLfiltered data set using our in-house database(8) revealed only 329 available compounds. In light of this, bioactive templates representing the different structural classes of ion channel modulators were generated for subsequent substructure searches. Bioactive templates were identified by searching for maximum common substructures (MCS) of compounds within each ion channel category in the ChEMBLfiltered data set. During this process, singletons and under-represented classes (see Experimental Procedures) were excluded. Afterward, the structures of the bioactive templates were visually inspected, and any synthetically intractable structures were rejected to avoid the presence of synthetically challenging compounds in the final screening library that would not be proceeded as hit or lead candidates. 307 bioactive templates out of 548 generated templates were selected in the final collection and annotated against their respective categories of ion channels (Table 1).
Table 1. Number of Compounds, Bioactive Templates, and Singletons within Each Category of Ion Channels in the ChEMBLfiltered Data Set.
| ion channel category | total no. of compounds | total no. of templates | no. of templates selected | no. of singletons |
|---|---|---|---|---|
| AnionicCysLoop | 1589 | 94 | 36 | 288 |
| CationicCysLoop | 1383 | 94 | 60 | 856 |
| purinoceptors | 14 | 2 | 2 | 3 |
| glutamate | 1586 | 106 | 59 | 368 |
| Na+ | 326 | 30 | 17 | 63 |
| Ca2+ | 437 | 46 | 21 | 108 |
| K+ | 959 | 112 | 71 | 119 |
| InwardlyRectifyingK+ | 51 | 9 | 3 | 13 |
| transient receptor potential | 671 | 42 | 34 | 51 |
| sulfonylurea | 86 | 13 | 4 | 11 |
| total | 7102 | 548 | 307 | 1880 |
Commercial Availability Search
After merging identical templates present in multiple categories, 297 unique bioactive templates were used as substructures to search for commercially available lead-like compounds in our in-house database.(8) This search identified 92340 compounds representing 149 bioactive templates, forming the CommAvail data set which contained on average ∼620 compounds per template.
Diversity Analysis and Compound Selection
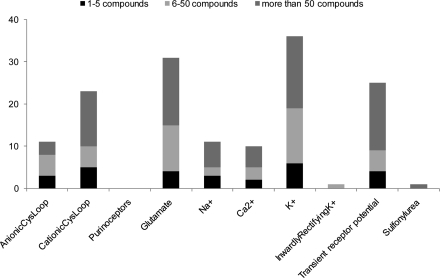
To avoid over-representation of certain templates and to keep the library at an affordable size, a maximum of 50 compounds per bioactive template was imposed.(8) In the 77 templates which were represented by more than 50 compounds (Figure 4), molecular diversity of compounds was analyzed using molecular fingerprints and the 50 structurally most diverse compounds were retained in the data set. Subsequently, the data set was visually inspected to remove any compounds containing synthetically intractable structures attached to the templates. Finally, all bioactive templates which had five or fewer examples remaining were considered under-represented, and therefore these compounds were excluded from the data set.
Figure 4.
Number of compounds per bioactive templates across different ion channel categories in the CommAvail data set.
The 3241 compounds that passed all filter steps were purchased to form the final focused screening library (DDU_IC data set) (Figure 2). Covering 123 bioactive templates across nine ion channel categories, these compounds were annotated with their respective ion channel category to assist back-tracing of prospective molecular targets from phenotypic screening results.
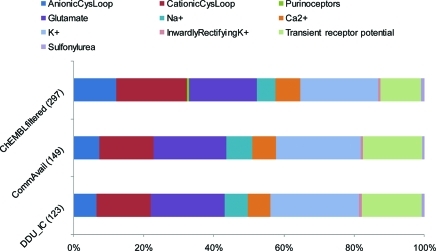
Despite the number of templates progressively decreasing from 297 templates in the ChEMBLfiltered data set to only 123 templates in the DDU_IC data set, the percentage composition of each ion channel category remained approximately constant throughout the process (Figure 5). On average, 38% of the templates per category in ChEMBLfiltered were represented in DDU_IC. Out of the nine ion channel categories represented, only the AnionicCysLoop category showed a considerable reduction, with eight out of the 36 templates (22%) in ChEMBLfiltered represented in DDU_IC. In contrast, 61% of the 34 templates for transient receptor potential channels were represented in the final library.
Figure 5.
Percentage composition of each ion channel category by number of templates in the ChEMBLfiltered, CommAvail, and DDU_IC data sets. Total number of represented templates in each data set in parentheses.
Discussion
Screening of focused libraries is considered to be a cost-effective strategy for hit discovery.5−7 Compiling focused libraries requires analysis of relevant chemical space in order to enrich compounds that are likely to interact with the desired target class.(5) This is commonly achieved by defining pharmacophoric or structural motifs satisfying specific binding interactions for the desired target class, which consequently requires a thorough understanding of the structural features and patterns of the protein–ligand interactions.8,9 Therefore, focused library construction is more challenging when the recognition motifs of the target class are less established.(10) With the first crystal structures of ion channel targets just emerging(13) and due to their heterogeneous nature involving 16 subfamilies,(14) the assembly of focused screening libraries for ion channels clearly represents a demanding task.
The workflow described here (Figure 1) provides an efficient protocol to analyze relevant bioactive data for ion channels and to use this information for compiling a focused library. The ChEMBL database offers direct access to chemical compounds associated with ion channel activity. However, since most of the compounds in ChEMBL are not commercially available,(15) the identification of bioactive templates using MCS represents an effective method to derive substructures which can subsequently be used to retrieve commercially available compounds that are likely to modulate ion channel activity. In addition to improving compound availability, these bioactive templates, unlike many descriptors derived to predict bioactivity of chemical compounds,16,17 are easy to interpret and do not require expert chemoinformatics knowledge, hence synthetically intractable templates can be rejected at an early stage by visual inspection. Besides, this MCS approach also allows the identification of promiscuous templates which appear across multiple ion channel categories. Indeed, we identified eight bioactive templates which are common to multiple ion channel categories using this workflow. Such observation would have been much more difficult if using more traditional similarity-based approaches such as molecular fingerprints comparison. However, promiscuous inhibitors(18) in ChEMBL erroneously reported to be active against certain targets are difficult to be detected using this workflow. The selected compounds can be annotated with the respective ion channel category they were derived from, which facilitates target identification when using the library for phenotypic screening. This workflow is not limited to ion channels but can be adapted to any target family for which chemical information is available in ChEMBL or other related databases.
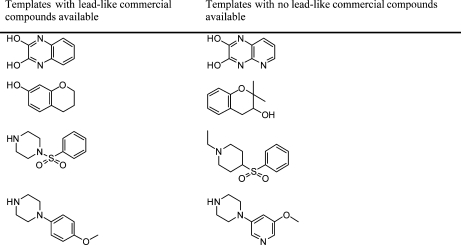
The presence of gaps for ion channel-active compounds in lead-like commercial chemical space became apparent when assembling the ion channel library. Only 329 compounds (<5%) in the ChEMBLfiltered data set were commercially available based on our in-house database. This is perhaps not surprising, since many compounds in ChEMBL are retrieved from medicinal chemistry literature, which often describes hit or lead optimization efforts. When searching for lead-like commercially available compounds(8) using the 297 bioactive templates derived from MCS (Table 1), compound availability was vastly improved, although a significant proportion of the selected templates remained unrepresented (Figure 5). These unavailable templates are present across all ion channel categories (Table S1 in the Supporting Information), and many of them do not appear to be synthetically more challenging than the available ones (Table 2). A similar observation was noted before when we assembled a focused kinase screening library.(8) The observed absence of ion channel templates in commercial chemical space is also supported by a recent publication by Chuprina et al. who estimated a generally low occurrence of prospective ion channel modulators relative to other target classes from a collection of commercial chemical suppliers similar to those in our in-house database.(19) Although the collection of suppliers in our database may not represent a comprehensive coverage of the entire commercial chemical space, our observations here, together with that of Chuprina et al., indicate there is still scope for compound vendors to increase the diversity of lead-like compounds on offer in their libraries.
Table 2. Comparison of Example Structures of Bioactive Templates with/without Lead-like Commercial Compounds.
Conclusions
We have demonstrated an efficient workflow to assemble a focused screening library for ion channel targets using bioactivity data retrieved from ChEMBL. The workflow is based on the efficient mining of an open-access database containing bioactivity data for structurally diverse therapeutic target families and demonstrates an effective solution using bioactive templates to overcome the problem associated with limited compound availability. The final screening library contained 3241 compounds representing 123 templates across nine ion channel categories. These compounds were annotated with their respective ion channel category to enable efficient back-tracing of prospective molecular targets from phenotypic screening results. The screening library is currently being used in campaigns to identify new chemical starting points toward ion channel targets for various neglected disease programs within the Drug Discovery Unit at Dundee. These results will offer valuable data to evaluate the quality of this newly assembled screening library.
Experimental Procedures
ChEMBLfiltered Data Set
Bioactivity data of compounds annotated with associated ion channel targets (337 molecular targets in 14 categories) were retrieved from the ChEMBL database (accessed Feb 16–Mar 4, 2010). All filters applied were carried out using Pipeline Pilot professional client 7.5 (Accelrys, Inc.). Unwanted groups were described as SMARTS strings,(8) and compounds were matched against the SMARTS description using substructure mapping.
CommAvail Data Set
Structural classes of the ChEMBLfiltered data set were generated using ClassPharmer 4.7 (SimulationsPlus, Inc.). The criteria of structural classes were
a minimum of 2 rings (either 2 single rings linked together or 1 fused ring),
1–3 functional groups attached to any ring system,
a maximum of 5 bonds between a ring and a functional group, and
a maximum of 5 bonds between ring-connected fragments.
Under-represented classes were defined as those containing fewer than five compounds of which no bioactivities were better than 5 μM. Bioactive templates selected were annotated with their respective ion channel categories. These templates were used as substructures to search for commercially available compounds in our in-house database containing ∼5.9 million unique compounds from 20 commercial chemical suppliers (as of June 2010). Compounds in the CommAvail data set were chosen according to the lead-like criteria as previously described.(8)
DDU_IC Data Set
Compound clustering and the selection of representative examples followed the same procedure as previously published.(8)
Acknowledgments
This work was funded by the Wellcome Trust (WT077705 and WT083481) and the Drugs for Neglected Diseases initiative (DNDi). We thank the Scottish Universities Life Sciences Alliance (SULSA) for financial support of Pipeline Pilot software (Grant HR07019) and Claire Mackenzie and Michael Thomas for help with triaging compounds and selecting synthetically tractable structures.
Supporting Information Available
Tables listing the 297 shortlisted bioactive templates used for commercial availability search and their corresponding SMARTS definitions. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Terstappen G. C.; Roncarati R.; Dunlop J.; Peri R. Screening technologies for ion channel drug discovery. Future Med. Chem. 2010, 2, 715–730. [DOI] [PubMed] [Google Scholar]
- Wible B. A.; Kuryshev Y. A.; Smith S. S.; Liu Z. Q.; Brown A. M. An ion channel library for drug discovery and safety screening on automated platforms. Assay Drug Dev. Technol. 2008, 6, 765–780. [DOI] [PubMed] [Google Scholar]
- Wulff H.; Castle N. A.; Pardo L. A. Voltage-gated potassium channels as therapeutic targets. Nat. Rev. Drug Discovery 2009, 8, 982–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry Y.; Gies J. P. Drugs and their molecular targets: an updated overview. Fundam. Clin. Pharmacol. 2008, 22, 1–18. [DOI] [PubMed] [Google Scholar]
- Mayr L. M.; Bojanic D. Novel trends in high-throughput screening. Curr. Opin. Pharmacol. 2009, 9, 580–588. [DOI] [PubMed] [Google Scholar]
- Miller J. L. Recent developments in focused library design: Targeting gene-families. Curr. Top. Med. Chem. 2006, 6, 19–29. [DOI] [PubMed] [Google Scholar]
- Fox S.; Farr-Jones S.; Sopchak L.; Boggs A.; Nicely H. W.; Khoury R.; Biros M. High-throughput screening: Update on practices and success. J. Biomol. Screening 2006, 11, 864–869. [DOI] [PubMed] [Google Scholar]
- Brenk R.; Schipani A.; James D.; Krasowski A.; Gilbert I. H.; Frearson J.; Wyatt P. G. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem 2008, 3, 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ahsen O.; Bomer U. High-throughput screening for kinase inhibitors. ChemBioChem 2005, 6, 481–490. [DOI] [PubMed] [Google Scholar]
- Harris C. J.; Hill R. D.; Sheppard D. W.; Slater M. J.; Stouten P. F. The design and application of target-focused compound libraries. Comb. Chem. High Throughput Screening 2011, 14, 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChEMBL database, European Bioinformatics Institute(EBI): Cambridge, U.K., 2010. http://www.ebi.ac.uk/chembl/ (accessed 16 Feb 2010–4 Mar 2010).
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 1997, 23, 3–25. [DOI] [PubMed] [Google Scholar]
- Corringer P. J.; Baaden M.; Bocquet N.; Delarue M.; Dufresne V.; Nury H.; Prevost M.; Van Renterghem C. Atomic structure and dynamics of pentameric ligand-gated ion channels: new insight from bacterial homologues. J. Physiol. (Lond). 2010, 588, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharman J. L.; Mpamhanga C. P.; Spedding M.; Germain P.; Staels B.; Dacquet C.; Laudet V.; Harmar A. J. NC-IUPHAR IUPHAR-DB: new receptors and tools for easy searching and visualization of pharmacological data. Nucleic Acids Res. 2011, 39, D534–D538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.1% of all compounds in ChEMBL were available when searched in our in-house database (as of May 2011).
- Rogers D.; Hahn M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [DOI] [PubMed] [Google Scholar]
- Nidhi; Glick M.; Davies J. W.; Jenkins J. L. Prediction of biological targets for compounds using multiple-category Bayesian models trained on chemogenomics databases. J. Chem. Inf. Model. 2006, 46, 1124–1133. [DOI] [PubMed] [Google Scholar]
- McGovern S. L.; Caselli E.; Grigorieff N.; Shoichet B. K. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 2002, 45, 1712–1722. [DOI] [PubMed] [Google Scholar]
- Chuprina A.; Lukin O.; Demoiseaux R.; Buzko A.; Shivanyuk A. drug- and lead-likeness, target class, and molecular diversity analysis of 7.9 million commercially available organic compounds provided by 29 suppliers. J. Chem. Inf. Model. 2010, 50, 470–479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.