Abstract
Myogenesis, the formation of skeletal muscle, is a multistep event that commences with myoblast proliferation, followed by cell-cycle arrest, and finally the formation of multinucleated myotubes via fusion of mononucleated myoblasts. Each step is orchestrated by well-documented intracellular factors, such as cytoplasmic signalling molecules and nuclear transcription factors. Regardless, the key step in getting a more comprehensive understanding of the regulation of myogenesis is to explore the extracellular factors that are capable of eliciting the downstream intracellular factors. This could further provide valuable insight into the acute cellular response to extrinsic cues in maintaining normal muscle development. In this paper, we survey the intracellular factors that respond to extracellular cues that are responsible for the cascades of events during myogenesis: myoblast proliferation, cell-cycle arrest of myoblasts, and differentiation of myoblasts into myotubes. This focus on extracellular perspective of muscle development illustrates our mass spectrometry-based proteomic approaches to identify differentially expressed secreted factors during skeletal myogenesis.
1. Introduction
Myogenesis, the formation of skeletal muscle, has been recognized as a hierarchical cellular event, commencing with myogenic lineage specification and followed by iterative proliferation of the muscle precursor cells called myoblasts in which cell-cell contact is initiated. This triggers withdrawal of myoblasts from the proliferation cycle (i.e., cell-cycle arrest) and in turn switches on the differentiation program in which mononucleated myoblasts are fused to each other and give rise to multinucleated myotubes (i.e., building blocks for contractile muscle fibres in the mature animal). Each step is orchestrated by groups of intracellular factors, such as cytoplasmic signalling molecules and nuclear transcription factors, which are described in further detail below.
1.1. Myogenic Lineage Specification
Skeletal muscle originates from the paraxial mesoderm, epithelialization and segmentation of which gives rise to the somites in a cranio-caudal manner (i.e., somites are generated and specified from head to tail) (Figure 1). Various compartments of the somite are committed to distinct cell lineages: myotome (muscle), dermatome (skin), and sclerotome (bone and cartilage), according to their relative orientations to the surrounding tissue, such as ectoderm, neural tube, notochord, and lateral mesoderm [1]. The ventral medial portion of the somite is specified as the sclerotome, whereas the double-layered structure remaining is called the dermomyotome which gives rise to the dermatome and myotome. The latter is subdivided into two compartments: dorsal medial lip (DML) and ventral lateral lip (VLL). The former compartment gives rise to the epaxial myotome that becomes the back muscle, whereas the latter gives the hypaxial myotome that generates the muscles of the body wall, limbs, and tongue [2–5].
Figure 1.
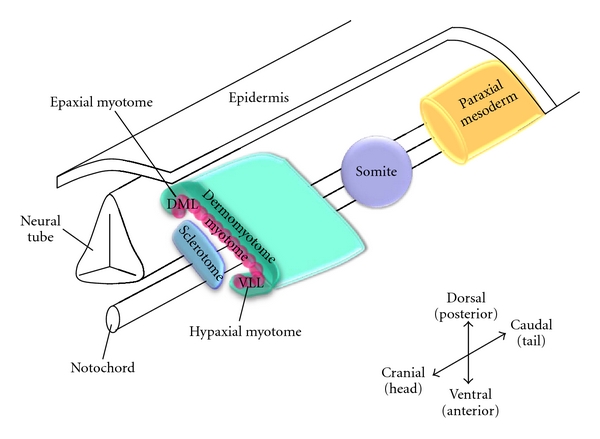
Myogenic lineage specification. Dorsal medial lip and ventral lateral lip were denoted as DML and VLL, respectively. Redrawn from Buckingham et al. [6].
1.2. Myoblast Proliferation with Simultaneous Repression of Muscle Differentiation
After the primary wave of myoblasts is generated from the somite, they enter the cell cycle and undergo iterative propagation to expand the cell population, eventually cell-cell contact occurs. This step has been shown to be essential to withdraw the myoblasts from the proliferation cycle and initiate the differentiation program (Figure 2(a)) [7–9]. Thus, the proliferation and differentiation of myoblasts are mutually exclusive events; the tipping point between the two is governed by a master regulator: the retinoblastoma protein (pRb) [10–12].
Figure 2.
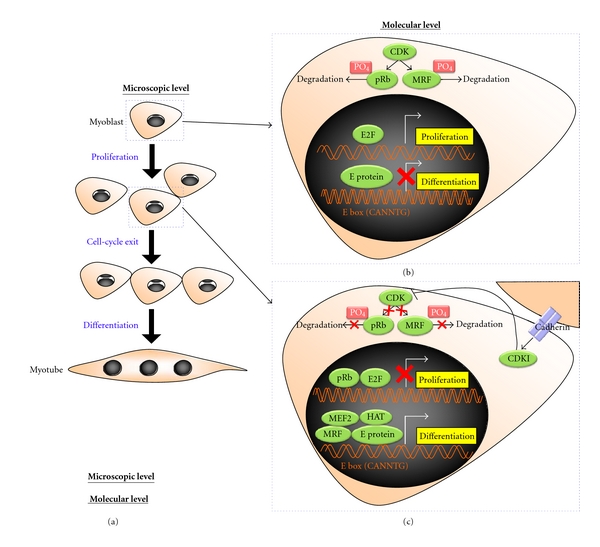
Skeletal muscle differentiation at the microscopic and molecular level. (a) During myogenesis, mononucleated myoblast proliferate, followed by cell-cycle exit, and fusion to form multinucleated myotube; (b) during proliferation, at the molecular level, active CDK could trigger myoblast proliferation by phosphorylating and subjecting pRb to degradation, in which E2F transcription factor is free from the inhibitory effect of pRb and elicits the proliferation of myoblasts. Simultaneously, CDK can also block myoblasts from differentiation via the phosphorylation-induced degradation of MRF. As a consequence, E protein by itself cannot drive the differentiation program; (c) upon cell-cell contact, m-cadherin is activated, by which CDKI is induced. This in turn inhibits CDK from phosphorylating its downstream substrates: pRb and MRF. Hence, both pRb and MRF are exempted from degradation, in which the former can withdraw the myoblasts from the cell cycle by inhibiting E2F transcription factor from activating the proliferation-associated events, whereas the latter complexes with E protein, myogenic co-activator MEF2, and the chromatin remodeling molecule HATs, in an effort to evoke the differentiation program of myoblasts synergistically. Phosphate groups were indicated as “PO4”.
During proliferation, cyclin/cyclin-dependent kinases (CDKs), such as cyclin D/cdk4, cyclin D/cdk6, cyclin E/cdk2, and cyclin A/cdk2, are active. These kinases phosphorylate pRb, holding it inactive [13–18]. As a result, pRb is unable to bind to the E2F transcription factor complex and inhibit its activation of downstream proliferation-associated cellular events, including chromosome segregation, mitotic spindle formation, and chromatin remodelling [19] (Figure 2(b)).
Notably, the differentiation of these myoblasts is critically dependent upon a family of myogenic transcription factors: the myogenic regulatory factors (MRFs), including myogenic differentiation factor (MyoD) [20, 21] and myogenic factor 5 (Myf5) [22, 23]. The MRFs confer on the myoblasts a potent ability to differentiate. By contrast, mitogenic myoblasts may be prohibited from differentiation by myogenic repressors, including Id [24, 25], twist [26–28], MyoR [29, 30], Mist 1 [31], and I-mf [32]. In the absence of myogenic repressors, MRFs, which are members of the class II basic helix-loop-helix (bHLH) superfamily, can dimerize with members of the class I bHLH family, the E proteins. The E protein: MRF heterodimer thus resulted recognizes and binds to the consensus DNA sequence (CANNTG) named the E-box, which lies upstream of most muscle-specific genes, for example, the myosin heavy chain and muscle creatine kinase [33]. Conversely, in the presence of myogenic repressors, the dimerization between MRF and the E protein inside the nucleus is negated either by (1) competitive binding to MRFs or the E proteins by means of Id, twist, MyoR, and Mist 1, or (2) sequestering MRFs in the cytoplasm by means of I-mf. Additional control can come via other interactions, including those of pRb and CDKs which can also phosphorylate MRFs and subject them to degradation [34–36] (Figure 2(b)). The initial repression of muscle differentiation is essential for ensuring a sufficiently large number of myoblasts are attained prior to differentiation to populate the vast amount of skeletal musculature in the metazoan species.
1.3. Cell-Cycle Arrest of Myoblasts with Simultaneous Activation of Muscle Differentiation
Under growth conditions, myoblasts proliferate until they reach confluency and cell-cell contact provokes growth arrest. The switch between cell-cell contact and cell-cycle arrest is mediated by transmembrane proteins, such as m-cadherin [37–42]. Upon cell-cell contact, m-cadherin is activated and induces CDK inhibitors (CDKIs), for example, p21 and p57 [43, 44]. As the name suggests, CDKIs inhibit CDK from phosphorylating its respective substrates, such as pRb and MRF [45, 46]. As a result, both pRb and MyoD are spared from degradation. The corollary to that is twofold: (1) nonphosphorylated pRb can bind and inhibit E2F from activating the downstream proliferation events, by which cell-cycle arrest of myoblasts is achieved [47, 48]; (2) nonphosphorylated MyoD can dimerize with the E protein and cooperatively bind to the E box to activate the expression of muscle-specific gene, thus triggering the differentiation program. Furthermore, with the recruitment of myogenic coactivators, such as myocyte enhancer factor 2 (MEF2) [49–52] as well as the chromatin remodelling factors, the histone acetyltransferases (HATs), for example, p300 and p300/CBP-associated factor (PCAF) [53–62], the differentiation program is initiated (Figure 2(c)). In addition, activated cadherin interacts and triggers a cell adhesion molecule of the Ig superfamily called CAM-related/downregulated by oncogenes (CDO) [63, 64]. The CDO complex promotes myogenesis by activating the p38 MAPK signalling pathway [65–68], which is a well-known promyogenic signal acting at various steps [69–71]. p38, for example, enhances the activity of MyoD [72], and its co-activator MEF2 [73], favouring MyoD/E protein heterodimerization by phosphorylating E protein [74], recruiting SWI-SNF chromatin-remodelling complex to the promoter of muscle-specific genes to enhance accessibility to transcriptional regulators required for subsequent gene expression [75]. Intriguingly, CDO is a target of MyoD, establishing positive feedback loop which reinforces the muscle differentiation program [64, 76].
1.4. From Intra- to Extracellular Perspective of Myogenesis
Irrespective of well-documented intracellular factors entailed in myogenesis, the key step in developing a more comprehensive picture of the regulation of muscle development is to investigate the extracellular factors that prime these downstream intracellular events. This, in turn, may provide valuable insight into the acute cellular response as a result of extrinsic cues in normal muscle development and regeneration. Intriguingly, the effects exerted by the “conditioned” media (CM) on the development of muscle cells have been documented some time ago [77, 78], illustrating the phenomena that myogenic cells modify their own extracellular milieu by secreting factors that exert autocrine and paracrine effects on the differentiation program. Furthermore, the skeletal muscle has been recognized as the largest endocrine organ in humans for secreting extracellular factors, the myokines that orchestrate muscle development in an autocrine fashion [79, 80]. Apart from the well-known myokines, such as members of the insulin-like growth factor-1 (IGF1) [81–90] and transforming growth factor (TGF) families [91–99], which have potent, but opposing effects on myogenesis, there were individual studies investigating other myokines, such as plasminogen activator [100], collagenase [101], decorin [102], glial growth factor [103], neurocrescin [104], meltrin alpha [105], musculin [79, 106], interleukin-1 beta [107], interleukin-7 [108], ADAMTS-like 2 [109], follistatin-like 1 [110], secreted protein acidic and rich in cysteine (SPARC) [111–113]. To make progress on the characterization of the “secretome” in an unbiased manner, we implemented an initial mass spectrometry-based proteomics study to identify secreted proteins in the mouse skeletal muscle cell line C2C12 [114]. Furthermore, a more quantitative approach using stable-isotope labelling by amino acids in cell culture (SILAC) in conjunction with online reverse phase liquid chromatography tandem mass spectrometry (RPLC-MS/MS), has now been implemented to identify differentially expressed secreted proteins during myogenesis.
2. Workflow of SILAC Quantification
In differential proteomics, stable-isotope labelling, for example, 2H versus 1H, 13C versus 12C, and 15N versus 14N, is employed to introduce a signature mass difference between the samples of interest (e.g., treatment versus control). After enzymatic protein digest, the ratios of the labelled peptide peak intensities reveal the relative protein expression. There are two general ways to introduce the stable-isotope label into the sample: (1) chemical labelling, typically achieved via the isotope-coded affinity tag (ICAT) or the isobaric tag for relative and absolute quantitation (iTRAQ); (2) metabolic labelling, conveniently performed via SILAC. ICAT targets the sulfhydryl group on the cysteine residue [115], whereas iTRAQ modifies the amino group on the N-terminus and the lysine residue [116]. For SILAC, stable-isotope labelled amino acids are metabolically incorporated into the living cells as they grow. Irrespective of the labelling methodology, the tagged samples are then combined and processed as one in subsequent treatment, separation, and analysis. This minimizes the impact of nonquantitative recovery of the proteins and peptides in these steps on the accuracy of the quantification [117, 118].
In recent years, SILAC has been widely applied to various biological models and cell types, including immune B cells [119], fibroblasts [120], neuronal cells [121], blood cells [122], lung cells [123], chondrocytes [124], prostate cancer [125], ovarian cancer [126], liver cancer [127, 128], breast cancer [129, 130], esophageal cancer [131, 132], and embryonic stem cells [133–135]. In addition, it has also been successfully implemented in tissues [136, 137] and living organisms [122, 138–140].
We employed SILAC labelling in an attempt to identify differentially expressed secreted factors at the myotube- versus myoblast-stage (i.e., differentiation versus proliferation) in C2C12 cells. As illustrated in Figure 3, CM proteins derived from [12C6]-lysine labelled myoblasts (light) and [13C6]-lysine labelled myotubes (heavy) were mixed in equal amounts and subjected to one-dimensional gel electrophoresis (1D-SDS PAGE), followed by trypsin digestion. The resulting tryptic peptides were analyzed by online RPLC-MS/MS. The ratio of the heavy- versus light-labelled peptide peak intensities in the MS mass spectrum mirrored the relative expression level of that particular protein during myogenesis.
Figure 3.
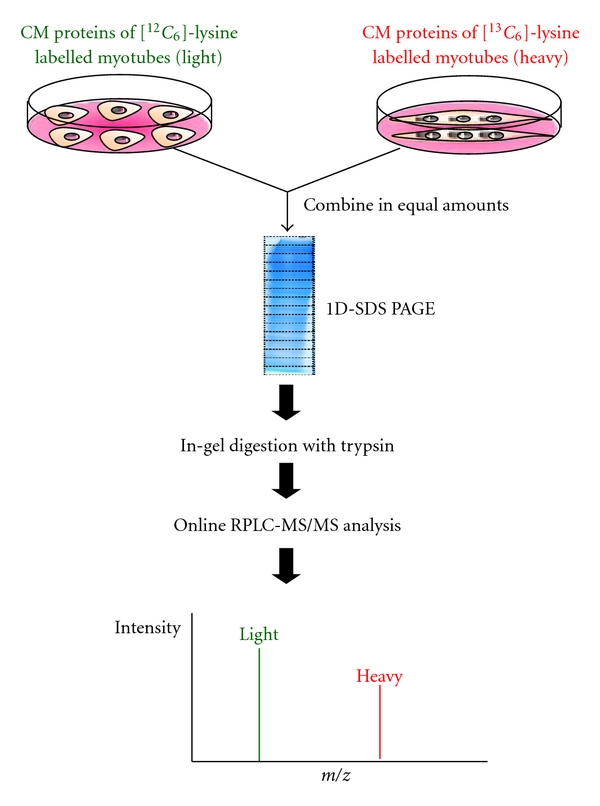
The workflow of using SILAC to identify differentially expressed secreted factors during skeletal myogenesis.
3. Implications of the Secreted Proteins Identified in Myogenesis
As previously discussed, myogenesis is a multistep process, beginning with myogenic lineage specification, followed by cell proliferation, cell-cycle arrest, and ultimately the differentiation of myoblasts into myotubes. We postulated that each of these steps is regulated by secreted factor(s). According to our preliminary data, novel secreted proteins, such as osteoglycin (OGN), peroxiredoxin 1 (Prx1), and cytokine-induced apoptosis inhibitor 1 (CIAPIN1), were identified as differentially expressed proteins. Their respective role(s) in myogenesis were proposed as follows.
3.1. OGN
OGN is also known as mimecan. It belongs to the small leucine-rich repeat proteoglycan (SLRP) family of proteins [141–147]. This protein was found to be essential in maintaining the integrity of the extracellular matrix (ECM) of the cornea [148, 149] and the vascular smooth muscle [150, 151] by inhibiting the ECM-cleaving enzyme gelatinase [152]. This anti-ECM cleaving property contributed to OGN's tumour suppressor role in hepatocarcinoma cells by attenuating tumour cell migration [153]. Given OGN's differential expression in myogenesis, we hypothesized that OGN may play an inhibitory role by hindering myoblast migration and the subsequent cell-cell contact. As result, cell-cycle arrest is inhibited and hence the muscle differentiation program is sabotaged. Interestingly, the E box has been identified in the promoter region of OGN [154]. This projects a compelling regulation mechanism of OGN during myogenesis in which binding of the MRF and E protein heterodimer to the E box may function as a docking site to recruit a chromatin remodelling molecule, such as histone deacetyltransferases (HDACs); as consequence, the transcription and subsequent expression of OGN decrease.
Furthermore, OGN may also play a role in myogenic lineage commitment, where the protein was initially identified as a bone-inductive factor [155–159]. Intriguingly, we have demonstrated the possibility that C2C12 myoblasts could be recommitted to the osteoblast lineage by overexpressing a bone-inductive gene called menin1 [160]. With this taken into account, it is tempting for us to speculate a plausible link between OGN and menin1 in which downregulation of OGN may be essential in directing the myoblasts to myogenic lineage.
3.2. Prx1
Prx1, also known as Pag [161] or MSP23 [162], belongs to the antioxidant protein family for cellular defence against reactive oxygen species (ROS) [163]. Prx1 was revealed to be upregulated in various cancer types, such as oral cancer [164], lung cancer [165–172], pancreatic cancer [173], and esophageal cancer [174]. Expression level of Prx1 was shown to positively correlate with cancer progression; knocking down Prx1 not only attenuated malignancy, but also sensitized the cancer cells to chemotherapy and improved survival [175–177]. Given the role of Prx1 as a prosurvival factor by blocking apoptosis signal-regulating kinase (ASK)- induced cell death [178–180], we hypothesized that Prx1 may function as a mitogen that promotes the proliferation of myoblasts. As proliferation and differentiation are mutually exclusive events, the down-regulation of Prx1 (unpublished data) may be essential for the withdrawal of myoblasts from the proliferation cycle and subsequent differentiation.
3.3. CIAPIN1
CIAPIN1 has been characterized as an antiproliferation molecule in cell division and angiogenesis [181–183]. CIAPIN1 was shown to be a suppressor of various cancers, for instance gastric cancer [184], renal carcinoma [185], esophageal cancer [186], and colorectal cancer [187]. The antiproliferation effect of CIAPIN1 was found to be mediated by upregulating CDKI, which in turn allows pRb to inhibit the E2F transcription factor from activating downstream proliferation events; as a result, cell-cycle arrest prevails [185, 188]. We postulated that CIAPIN1 may function as a positive regulator of myogenesis, in which the upregulation of CIAPIN1 (unpublished data) may be essential in triggering cell-cycle arrest of myoblasts for subsequent differentiation to take place.
4. Conclusion
We have demonstrated the fidelity of applying SILAC to identify secreted factors during skeletal myogenesis in an unbiased proteomics approach. OGN, Prx1, and CIAPIN1 were identified as novel differentially expressed extracellular factors that are proposed to play a role in the myogenic program (Figure 4). Based on the findings of this “discovery” approach, gain and loss of function studies are now in progress to further dissect these proteins' individual and combinatorial roles in myogenesis. The identification of secretome factors that regulate myogenesis will enhance our knowledge of extracellular regulation of differentiation as well as identify biomarkers of potential therapeutic value in muscle regeneration and stem cell programming.
Figure 4.
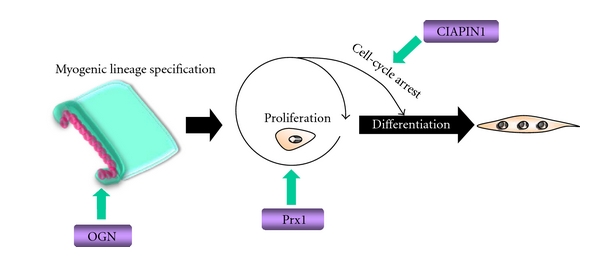
Overview of the implications of OGN, Prx1, and CIAPIN1 in myogenesis.
Acknowledgments
This work was made possible by support from the Natural Sciences and Engineering Research Council (NSERC) of Canada and from the Canadian Institutes of Health Research (CIHR) to JCM and KWMS.
Abbreviations
- 1D-SDS PAGE:
One-dimensional gel electrophoresis
- ASK:
Apoptosis signal-regulating kinase
- bHLH:
Basic helix-loop-helix
- CDKIs:
CDK inhibitors
- CDKs:
Cyclin-dependent kinases
- CDO:
CAM-related/downregulated by oncogenes
- CIAPIN1:
Cytokine-induced apoptosis inhibitor 1
- CM:
Conditioned media
- DML:
Dorsal medial lip
- ECM:
Extracellular matrix
- HATs:
Histone acetyltransferases
- HDACs:
Histone deacetyltransferases
- ICAT:
Isotope-coded affinity tag
- IGF1:
Insulin-like growth factor-1
- iTRAQ:
Isobaric tag for relative and absolute quantitation
- MEF2:
Myocyte enhancer factor 2
- MRFs:
Myogenic regulatory factors
- Myf5:
Myogenic factor 5
- MyoD:
Myogenic differentiation factor
- OGN:
Osteoglycin
- PCAF:
p300/CBP-associated factor
- pRb:
Retinoblastoma protein
- Prx1:
Peroxiredoxin 1
- ROS:
Reactive oxygen species
- RPLC-MS/MS:
Reversed phase liquid chromatography tandem mass spectrometry
- SILAC:
Stable isotope labelling by amino acids in cell culture
- SLRP:
Small leucine-rich repeat proteoglycan
- SPARC:
Secreted protein acidic and rich in cysteine
- TGF:
Transforming growth factor
- VLL:
Ventral lateral lip.
References
- 1.Youn BW, Malacinski GM. A comparative analysis of amphibian somite morhphogenesis: cell rearrangement patterns during rosette formation and myoblast fusion. Journal of Embryology and Experimental Morphology. 1981;66:1–26. [PubMed] [Google Scholar]
- 2.Ordahl CP, Le Douarin NM. Two myogenic lineages within the developing somite. Development. 1992;114(2):339–353. doi: 10.1242/dev.114.2.339. [DOI] [PubMed] [Google Scholar]
- 3.Pourquie O, Coltey M, Teillet MA, Ordahl C, Le Douarin NM. Control of dorsoventral patterning of somitic derivatives by notochord and floor plate. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(11):5242–5246. doi: 10.1073/pnas.90.11.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christ B, Ordahl CP. Early stages of chick somite development. Anatomy and Embryology. 1995;191(5):381–396. doi: 10.1007/BF00304424. [DOI] [PubMed] [Google Scholar]
- 5.Kato N, Aoyama H. Dermomyotomal origin of the ribs as revealed by extirpation and transplantation experiments in chick and quail embryos. Development. 1998;125(17):3437–3443. doi: 10.1242/dev.125.17.3437. [DOI] [PubMed] [Google Scholar]
- 6.Buckingham M, Bajard L, Chang T, et al. The formation of skeletal muscle: from somite to limb. Journal of Anatomy. 2003;202(1):59–86. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurdon JB. A community effect in animal development. Nature. 1988;336(6201):772–774. doi: 10.1038/336772a0. [DOI] [PubMed] [Google Scholar]
- 8.Gurdon JB, Tiller E, Roberts J, Kato K. A community effect in muscle development. Current Biology. 1993;3(1):1–11. doi: 10.1016/0960-9822(93)90139-f. [DOI] [PubMed] [Google Scholar]
- 9.Cossu G, Kelly R, Di Donna S, Vivarelli E, Buckingham M. Myoblast differentiation during mammalian somitogenesis is dependent upon a community effect. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(6):2254–2258. doi: 10.1073/pnas.92.6.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graña X, Garriga J, Mayol X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene. 1998;17(25):3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 11.Tamrakar S, Rubin E, Ludlow JW. Role of pRB dephosphorylation in cell cycle regulation. Front Biosci. 2000;5:D121–137. doi: 10.2741/tamrakar. [DOI] [PubMed] [Google Scholar]
- 12.De Falco G, Comes F, Simone C. pRb: master of differentiation. Coupling irreversible cell cycle withdrawal with induction of muscle-specific transcription. Oncogene. 2006;25(38):5244–5249. doi: 10.1038/sj.onc.1209623. [DOI] [PubMed] [Google Scholar]
- 13.Buchkovich K, Duffy LA, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell. 1989;58(6):1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 14.Chen PL, Scully P, Shew JY, Wang JYJ, Lee WH. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell. 1989;58(6):1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 15.DeCaprio JA, Ludlow JW, Lynch D, et al. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell. 1989;58(6):1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- 16.Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes and Development. 1993;7(3):331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 17.Obeyesekere MN, Herbert JR, Zimmerman SO. A model of the G1 phase of the cell cycle incorporating cyclin E/cdk2 complex and retinoblastoma protein. Oncogene. 1995;11(6):1199–1205. [PubMed] [Google Scholar]
- 18.Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98(6):859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 19.Ren B, Cam H, Takahashi Y, et al. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes and Development. 2002;16(2):245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 21.Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(20):7988–7992. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braun T, Buschhausen-Denker G, Bober E, Tannich E, Arnold HH. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. The EMBO Journal. 1989;8(3):701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tajbakhsh S, Rocancourt D, Buckingham M. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature. 1996;384(6606):266–270. doi: 10.1038/384266a0. [DOI] [PubMed] [Google Scholar]
- 24.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61(1):49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 25.Neuhold LA, Wold B. HLH forced dimers: tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell. 1993;74(6):1033–1042. doi: 10.1016/0092-8674(93)90725-6. [DOI] [PubMed] [Google Scholar]
- 26.Hebrok M, Wertz K, Fuchtbauer EM. M-twist is an inhibitor of muscle differentiation. Developmental Biology. 1994;165(2):537–544. doi: 10.1006/dbio.1994.1273. [DOI] [PubMed] [Google Scholar]
- 27.Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein twist. Science. 1996;272(5267):1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- 28.Hamamori Y, Wu HY, Sartorelli V, Kedes L. The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, twist. Molecular and Cellular Biology. 1997;17(11):6563–6573. doi: 10.1128/mcb.17.11.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu J, Webb R, Richardson JA, Olson EN. MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(2):552–557. doi: 10.1073/pnas.96.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu L, Sangster N, Perez A, McCormick PJ. The bHLH protein MyoR inhibits the differentiation of early embryonic endoderm. Differentiation. 2004;72(7):341–347. doi: 10.1111/j.1432-0436.2004.07207005.x. [DOI] [PubMed] [Google Scholar]
- 31.Lemercier C, To RQ, Carrasco RA, Konieczny SF. The basic helix-loop-helix transcription factor Mist1 functions as a transcriptional repressor of MyoD. The EMBO Journal. 1998;17(5):1412–1422. doi: 10.1093/emboj/17.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CMA, Kraut N, Groudine M, Weintraub H. I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell. 1996;86(5):731–741. doi: 10.1016/s0092-8674(00)80148-8. [DOI] [PubMed] [Google Scholar]
- 33.Lassar AB, Davis RL, Wright WE, et al. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66(2):305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 34.Song AN, Wang QI, Goebl MG, Harrington MA. Phosphorylation of nuclear MyoD is required for its rapid degradation. Molecular and Cellular Biology. 1998;18(9):4994–4999. doi: 10.1128/mcb.18.9.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitzmann M, Vandromme M, Schaeffer V, et al. cdk1- and cdk2-mediated phosphorylation of MyoD Ser200 in growing C2 myoblasts: role in modulating MyoD half-life and myogenic activity. Molecular and Cellular Biology. 1999;19(4):3167–3176. doi: 10.1128/mcb.19.4.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Floyd ZE, Trausch-Azar JS, Reinstein E, Ciechanover A, Schwartz AL. The nuclear ubiquitin-proteasome system degrades MyoD. Journal of Biological Chemistry. 2001;276(25):22468–22475. doi: 10.1074/jbc.M009388200. [DOI] [PubMed] [Google Scholar]
- 37.Hatta K, Takagi S, Fujisawa H, Takeichi M. Spatial and temporal expression pattern of N-cadherin cell adhesion molecules correlated with morphogenetic processes of chicken embryos. Developmental Biology. 1987;120(1):215–227. doi: 10.1016/0012-1606(87)90119-9. [DOI] [PubMed] [Google Scholar]
- 38.Knudsen KA, Myers L, McElwee SA. A role for the Ca2+-dependent adhesion molecular, N-cadherin, in myoblast interaction during myogenesis. Experimental Cell Research. 1990;188(2):175–184. doi: 10.1016/0014-4827(90)90157-6. [DOI] [PubMed] [Google Scholar]
- 39.Holt CE, Lemaire P, Gurdon JB. Cadherin-mediated cell interactions are necessary for the activation of MyoD in Xenopus mesoderm. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(23):10844–10848. doi: 10.1073/pnas.91.23.10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeschnigk M, Kozian D, Kuch C, Schmoll M, Starzinski-Powitz A. Involvement of M-cadherin in terminal differentiation of skeletal muscle cells. Journal of Cell Science. 1995;108(9):2973–2981. doi: 10.1242/jcs.108.9.2973. [DOI] [PubMed] [Google Scholar]
- 41.George-Weinstein M, Gerhart J, Blitz J, Simak E, Knudsen KA. N-cadherin promotes the commitment and differentiation of skeletal muscle precursor cells. Developmental Biology. 1997;185(1):14–24. doi: 10.1006/dbio.1997.8542. [DOI] [PubMed] [Google Scholar]
- 42.Goichberg P, Geiger B. Direct involvement of N-cadherin-mediated signaling in muscle differentiation. Molecular Biology of the Cell. 1998;9(11):3119–3131. doi: 10.1091/mbc.9.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gavard J, Marthiens V, Monnet C, Lambert M, Mège RM. N-cadherin activation substitutes for the cell contact control in cell cycle arrest and myogenic differentiation: involvement of p120 and β-catenin. Journal of Biological Chemistry. 2004;279(35):36795–36802. doi: 10.1074/jbc.M401705200. [DOI] [PubMed] [Google Scholar]
- 44.Messina G, Blasi C, La Rocca SA, Pompili M, Calconi A, Grossi M. p27 acts downstream of N-cadherin-mediated cell adhesion to promote myogenesis beyond cell cycle regulation. Molecular Biology of the Cell. 2005;16(3):1469–1480. doi: 10.1091/mbc.E04-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynaud EG, Leibovitch MP, Tintignac LAJ, Pelpel K, Guillier M, Leibovitch SA. Stabilization of MyoD by direct binding to p57Kip2 . Journal of Biological Chemistry. 2000;275(25):18767–18776. doi: 10.1074/jbc.M907412199. [DOI] [PubMed] [Google Scholar]
- 46.Tintignac LAJ, Sirri V, Leibovitch MP, et al. Mutant MyoD lacking Cdc2 phosphorylation sites delays M-phase entry. Molecular and Cellular Biology. 2004;24(4):1809–1821. doi: 10.1128/MCB.24.4.1809-1821.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flemington EK, Speck SH, Kaelin WG. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(15):6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. Journal of Cell Science. 2004;117(11):2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- 49.Gossett LA, Kelvin DJ, Sternberg EA, Olson EN. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Molecular and Cellular Biology. 1989;9(11):5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu YT, Breitbart RE, Smoot LB, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes and Development. 1992;6(9):1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 51.Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83(7):1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 52.Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brownell JE, Allis CD. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Current Opinion in Genetics and Development. 1996;6(2):176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 54.Yuan W, Condorelli G, Caruso M, Felsani A, Giordanoi A. Human p300 protein is a coactivator for the transcription factor MyoD. Journal of Biological Chemistry. 1996;271(15):9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 55.Puri PL, Sartorelli V, Yang XJ, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Molecular Cell. 1997;1(1):35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 56.Puri PL, Avantaggiati ML, Balsano C, et al. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. The EMBO Journal. 1997;16(2):369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Molecular and Cellular Biology. 1997;17(2):1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sartorelli V, Puri PL, Hamamori Y, et al. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Molecular Cell. 1999;4(5):725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 59.Polesskaya A, Duquet A, Naguibneva I, et al. CREB-binding protein/p300 activates MyoD by acetylation. Journal of Biological Chemistry. 2000;275(44):34359–34364. doi: 10.1074/jbc.M003815200. [DOI] [PubMed] [Google Scholar]
- 60.Polesskaya A, Naguibneva I, Fritsch L, et al. CBP/p300 and muscle differentiation: no HAT, no muscle. The EMBO Journal. 2001;20(23):6816–6825. doi: 10.1093/emboj/20.23.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polesskaya A, Naguibneva I, Duquet A, Bengal E, Robin P, Harel-Bellan A. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Molecular and Cellular Biology. 2001;21(16):5312–5320. doi: 10.1128/MCB.21.16.5312-5320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(32):11593–11598. doi: 10.1073/pnas.0404192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang JS, Gao M, Feinleib JL, Cotter PD, Guadagno SN, Krauss RS. CDO: an oncogene-, serum-, and anchorage-regulated member of the Ig/fibronectin type III repeat family. Journal of Cell Biology. 1997;138(1):203–213. doi: 10.1083/jcb.138.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang JS, Mulieri PJ, Miller C, Sassoon DA, Krauss RS. CDO, a Robo-related cell surface protein that mediates myogenic differentiation. Journal of Cell Biology. 1998;143(2):403–413. doi: 10.1083/jcb.143.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takaesu G, Kang JS, Bae GU, et al. Activation of p38α/β MAPK in myogenesis via binding of the scaffold protein JLP to the cell surface protein Cdo. Journal of Cell Biology. 2006;175(3):383–388. doi: 10.1083/jcb.200608031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang JS, Bae GU, Yi MJ, et al. A Cdo-Bnip-2-Cdc42 signaling pathway regulates p38α/β MAPK activity and myogenic differentiation. Journal of Cell Biology. 2008;182(3):497–507. doi: 10.1083/jcb.200801119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bae GU, Kim BG, Lee HJ, et al. Cdo binds Abl to promote p38α/β mitogen-activated protein kinase activity and myogenic differentiation. Molecular and Cellular Biology. 2009;29(15):4130–4143. doi: 10.1128/MCB.00199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu M, Krauss RS. N-cadherin ligation, but not Sonic hedgehog binding, initiates Cdo-dependent p38α/β MAPK signaling in skeletal myoblasts. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(9):4212–4217. doi: 10.1073/pnas.0908883107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cuenda A, Cohen P. Stress-activated protein kinase-2/p38 and a rapamycin-sensitive pathway are required for C2C12 myogenesis. Journal of Biological Chemistry. 1999;274(7):4341–4346. doi: 10.1074/jbc.274.7.4341. [DOI] [PubMed] [Google Scholar]
- 70.Lluís F, Perdiguero E, Nebreda AR, Muñoz-Cánoves P. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends in Cell Biology. 2006;16(1):36–44. doi: 10.1016/j.tcb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 71.Perdiguero E, Ruiz-Bonilla V, Gresh L, et al. Genetic analysis of p38 MAP kinases in myogenesis: fundamental role of p38α in abrogating myoblast proliferation. The EMBO Journal. 2007;26(5):1245–1256. doi: 10.1038/sj.emboj.7601587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zetser A, Gredinger E, Bengal E. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation: participation of the MEF2C transcription factor. Journal of Biological Chemistry. 1999;274(8):5193–5200. doi: 10.1074/jbc.274.8.5193. [DOI] [PubMed] [Google Scholar]
- 73.Wu Z, Woodring PJ, Bhakta KS, et al. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Molecular and Cellular Biology. 2000;20(11):3951–3964. doi: 10.1128/mcb.20.11.3951-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lluís F, Ballestar E, Suelves M, Esteller M, Muñoz-Cánoves P. E47 phosphorylation by p38 MAPK promotes MyoD/E47 association and muscle-specific gene transcription. The EMBO Journal. 2005;24(5):974–984. doi: 10.1038/sj.emboj.7600528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nature Genetics. 2004;36(7):738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 76.Cole F, Zhang W, Geyra A, Kang JS, Krauss RS. Positive regulation of myogenic bHLH factors and skeletal muscle development by the cell surface receptor CDO. Developmental Cell. 2004;7(6):843–854. doi: 10.1016/j.devcel.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 77.Bischoff R. A satellite cell mitogen from crushed adult muscle. Developmental Biology. 1986;115(1):140–147. doi: 10.1016/0012-1606(86)90235-6. [DOI] [PubMed] [Google Scholar]
- 78.Bischoff R. Interaction between satellite cells and skeletal muscle fibers. Development. 1990;109(4):943–952. doi: 10.1242/dev.109.4.943. [DOI] [PubMed] [Google Scholar]
- 79.Engler D. Hypothesis: musculin is a hormone secreted by skeletal muscle, the body's largest endocrine organ. Evidence for actions on the endocrine pancreas to restrain the beta-cell mass and to inhibit insulin secretion and on the hypothalamus to co-ordinate the neuroendocrine and appetite responses to exercise. Acta Bio-Medica: Atenei Parmensis. 2007;78(supplement 1):156–206. [PubMed] [Google Scholar]
- 80.Pedersen BK, Febbraio MA. Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiological Reviews. 2008;88(4):1379–1406. doi: 10.1152/physrev.90100.2007. [DOI] [PubMed] [Google Scholar]
- 81.Florini JR, Ewton DZ, Magri KA, Mangiacapra FJ. IGFs and muscle differentiation. Advances in Experimental Medicine and Biology. 1993;343:319–326. doi: 10.1007/978-1-4615-2988-0_31. [DOI] [PubMed] [Google Scholar]
- 82.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75(1):59–72. [PubMed] [Google Scholar]
- 83.Powell-Braxton L, Hollingshead P, Warburton C, et al. IGF-I is required for normal embryonic growth in mice. Genes and Development. 1993;7(12 B):2609–2617. doi: 10.1101/gad.7.12b.2609. [DOI] [PubMed] [Google Scholar]
- 84.Coleman ME, DeMayo F, Yin KC, et al. Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. Journal of Biological Chemistry. 1995;270(20):12109–12116. doi: 10.1074/jbc.270.20.12109. [DOI] [PubMed] [Google Scholar]
- 85.Barton-Davis ER, Shoturma DI, Musaro A, Rosenthal N, Lee Sweeney H. Viral mediated expression of insulin-like growth factor I blocks the aging-related loss of skeletal muscle function. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(26):15603–15607. doi: 10.1073/pnas.95.26.15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawlor MA, Rotwein P. Coordinate control of muscle cell survival by distinct insulin-like growth factor activated signaling pathways. Journal of Cell Biology. 2000;151(6):1131–1140. doi: 10.1083/jcb.151.6.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lawlor MA, Rotwein P. Insulin-like growth factor-mediated muscle cell survival: central roles for Akt and cyclin-dependent kinase inhibitor p21. Molecular and Cellular Biology. 2000;20(23):8983–8995. doi: 10.1128/mcb.20.23.8983-8995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lawlor MA, Feng X, Everding DR, Sieger K, Stewart CEH, Rotwein P. Dual control of muscle cell survival by distinct growth factor-regulated signaling pathways. Molecular and Cellular Biology. 2000;20(9):3256–3265. doi: 10.1128/mcb.20.9.3256-3265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barton ER, Morris L, Musaro A, Rosenthal N, Sweeney HL. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. Journal of Cell Biology. 2002;157(1):137–147. doi: 10.1083/jcb.200108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Musaró A, Giacinti C, Borsellino G, et al. Stem cell-mediated muscle regeneration is enhanced by local isoform of insulin-like growth factor 1. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(5):1206–1210. doi: 10.1073/pnas.0303792101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Florini JR, Roberts AB, Ewton DZ. Transforming growth factor-β. A very potent inhibitor of myoblast differentiation, identical to the differentiation inhibitor secreted by Buffalo rat liver cells. Journal of Biological Chemistry. 1986;261(35):16509–16513. [PubMed] [Google Scholar]
- 92.Massague J, Cheifetz S, Endo T, Nadal-Ginard B. Type β transforming growth factor is an inhibitor of myogenic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(21):8206–8210. doi: 10.1073/pnas.83.21.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Olson EN, Sternberg E, Hu JS. Regulation of myogenic differentiation by type β transforming growth factor. Journal of Cell Biology. 1986;103(5):1799–1805. doi: 10.1083/jcb.103.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brennan TJ, Edmondson DG, Li L, Olson EN. Transforming growth factor β represses the actions of myogenin through a mechanism independent of DNA binding. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(9):3822–3826. doi: 10.1073/pnas.88.9.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martin JF, Li L, Olson EN. Repression of myogenin function by TGF-β1 is targeted at the basic helix-loop-helix motif and is independent of E2A products. Journal of Biological Chemistry. 1992;267(16):10956–10960. [PubMed] [Google Scholar]
- 96.De Angelis L, Borghi S, Melchionna R, et al. Inhibition of myogenesis by transforming growth factor β is density-dependent and related to the translocation of transcription factor MEF2 to the cytoplasm. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(21):12358–12363. doi: 10.1073/pnas.95.21.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu D, Black BL, Derynck R. TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes and Development. 2001;15(22):2950–2966. doi: 10.1101/gad.925901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu D, Kang JS, Derynck R. TGF-β-activated Smad3 represses MEF2-dependent transcription in myogenic differentiation. The EMBO Journal. 2004;23(7):1557–1566. doi: 10.1038/sj.emboj.7600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cohn RD, Van Erp C, Habashi JP, et al. Angiotensin II type 1 receptor blockade attenuates TGF-β-induced failure of muscle regeneration in multiple myopathic states. Nature Medicine. 2007;13(2):204–210. doi: 10.1038/nm1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Festoff BW, Patterson MR, Romstedt K. Plasminogen activator: the major secreted neutral protease of cultured skeletal muscle cells. Journal of Cellular Physiology. 1982;110(2):190–195. doi: 10.1002/jcp.1041100213. [DOI] [PubMed] [Google Scholar]
- 101.Beach RL, Rao JS, Festoff BW. Extracellular-matrix synthesis by skeletal muscle in culture. Major secreted collagenous protein of clonal myoblasts. Biochemical Journal. 1985;225(3):619–627. doi: 10.1042/bj2250619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brandan E, Fuentes ME, Andrade W. The proteoglycan decorin is synthesized and secreted by differentiated myotubes. European Journal of Cell Biology. 1991;55(2):209–216. [PubMed] [Google Scholar]
- 103.Florini JR, Samuel DS, Ewton DZ, Kirk C, Sklar RM. Stimulation of myogenic differentiation by a neuregulin, glial growth factor 2: are neuregulins the long-sought muscle trophic factors secreted by nerves? Journal of Biological Chemistry. 1996;271(22):12699–12702. doi: 10.1074/jbc.271.22.12699. [DOI] [PubMed] [Google Scholar]
- 104.Nishimune H, Uyeda A, Nogawa M, Fujimori K, Taguchi T. Neurocrescin: a novel neurite-outgrowth factor secreted by muscle after denervation. NeuroReport. 1997;8(16):3649–3654. doi: 10.1097/00001756-199711100-00045. [DOI] [PubMed] [Google Scholar]
- 105.Gilpin BJ, Loechel F, Mattei MG, Engvall E, Albrechtsen R, Wewer UM. A novel secreted form of human ADAM 12 (meltrin α) provokes myogenesis in vivo. Journal of Biological Chemistry. 1998;273(1):157–166. doi: 10.1074/jbc.273.1.157. [DOI] [PubMed] [Google Scholar]
- 106.Nishizawa H, Matsuda M, Yamada Y, et al. Musclin, a novel skeletal muscle-derived secretory factor. Journal of Biological Chemistry. 2004;279(19):19391–19395. doi: 10.1074/jbc.C400066200. [DOI] [PubMed] [Google Scholar]
- 107.Tateno K, Minamino T, Toko H, et al. Critical roles of muscle-secreted angiogenic factors in therapeutic neovascularization. Circulation Research. 2006;98(9):1194–1202. doi: 10.1161/01.RES.0000219901.13974.15. [DOI] [PubMed] [Google Scholar]
- 108.Haugen F, Norheim F, Lian H, et al. IL-7 is expressed and secreted by human skeletal muscle cells. American Journal of Physiology. 2010;298(4):C807–C816. doi: 10.1152/ajpcell.00094.2009. [DOI] [PubMed] [Google Scholar]
- 109.Koo BH, Goff CL, Jungers KA, et al. ADAMTS-like 2 (ADAMTSL2) is a secreted glycoprotein that is widely expressed during mouse embryogenesis and is regulated during skeletal myogenesis. Matrix Biology. 2007;26(6):431–441. doi: 10.1016/j.matbio.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 110.Ouchi N, Oshima Y, Ohashi K, et al. Follistatin-like 1, a secreted muscle protein, promotes endothelial cell function and revascularization in ischemic tissue through a nitric-oxide synthase-dependent mechanism. Journal of Biological Chemistry. 2008;283(47):32802–32811. doi: 10.1074/jbc.M803440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bassuk JA, Iruela-Arispe ML, Lane TF, Benson JM, Berg RA, Sage EH. Molecular analysis of chicken embryo SPARC (osteonectin) European Journal of Biochemistry. 1993;218(1):117–127. doi: 10.1111/j.1432-1033.1993.tb18358.x. [DOI] [PubMed] [Google Scholar]
- 112.Cho WJ, Kim EJ, Lee SJ, Kim HD, Shin HJ, Lim WK. Involvement of SPARC in in vitro differentiation of skeletal myoblasts. Biochemical and Biophysical Research Communications. 2000;271(3):630–634. doi: 10.1006/bbrc.2000.2682. [DOI] [PubMed] [Google Scholar]
- 113.Jørgensen LH, Petersson SJ, Sellathurai J, et al. Secreted protein acidic and rich in cysteine (SPARC) in human skeletal muscle. Journal of Histochemistry and Cytochemistry. 2009;57(1):29–39. doi: 10.1369/jhc.2008.951954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chan XCY, McDermott JC, Siu KWM. Identification of secreted proteins during skeletal muscle development. Journal of Proteome Research. 2007;6(2):698–710. doi: 10.1021/pr060448k. [DOI] [PubMed] [Google Scholar]
- 115.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature Biotechnology. 1999;17(10):994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 116.Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & Cellular Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 117.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & Cellular Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 118.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nature chemical biology. 2005;1(5):252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 119.Romijn EP, Christis C, Wieffer M, et al. Expression clustering reveals detailed co-expression patterns of functionally related proteins during B cell differentiation: a proteomic study using a combination of one-dimensional gel electrophoresis, LC-MS/MS, and stable isotope labeling by amino acids in cell culture (SILAC) Molecular & Cellular Proteomics. 2005;4(9):1297–1310. doi: 10.1074/mcp.M500123-MCP200. [DOI] [PubMed] [Google Scholar]
- 120.Pinto AFM, Ma L, Dragulev B, Guimaraes JA, Fox JW. Use of SILAC for exploring sheddase and matrix degradation of fibroblasts in culture by the PIII SVMP atrolysin A: identification of two novel substrates with functional relevance. Archives of Biochemistry and Biophysics. 2007;465(1):11–15. doi: 10.1016/j.abb.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 121.Klegeris A, Li J, Bammler TK, et al. Prolyl endopeptidase is revealed following SILAC analysis to be a novel mediator of human microglial and THP-1 cell neurotoxicity. GLIA. 2008;56(6):675–685. doi: 10.1002/glia.20645. [DOI] [PubMed] [Google Scholar]
- 122.Krüger M, Moser M, Ussar S, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134(2):353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 123.Duan X, Kelsen SG, Clarkson AB, Ji R, Merali S. SILAC analysis of oxidative stress-mediated proteins in human pneumocytes: new role for treacle. Proteomics. 2010;10(11):2165–2174. doi: 10.1002/pmic.201000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Polacek M, Bruun J-A, Johansen O, Martinez I. Differences in the secretome of cartilage explants and cultured chondrocytes unveiled by SILAC technology. Journal of Orthopaedic Research. 2010;28(8):1040–1049. doi: 10.1002/jor.21067. [DOI] [PubMed] [Google Scholar]
- 125.Everley PA, Krijgsveld J, Zetter BR, Gygi SP. Quantitative cancer proteomics: stable isotope labeling with amino acids in cell culture (SILAC) as a tool for prostate cancer research. Molecular & Cellular Proteomics. 2004;3(7):729–735. doi: 10.1074/mcp.M400021-MCP200. [DOI] [PubMed] [Google Scholar]
- 126.Amanchy R, Kalume DE, Iwahori A, Zhong J, Pandey A. Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC) Journal of Proteome Research. 2005;4(5):1661–1671. doi: 10.1021/pr050134h. [DOI] [PubMed] [Google Scholar]
- 127.Chen N, Sun W, Deng X, et al. Quantitative proteome analysis of HCC cell lines with different metastatic potentials by SILAC. Proteomics. 2008;8(23-24):5108–5118. doi: 10.1002/pmic.200800280. [DOI] [PubMed] [Google Scholar]
- 128.Zhang G, Fenyo D, Neubert TA. Screening for EphB signaling effectors using SILAC with a linear ion trap-orbitrap mass spectrometer. Journal of Proteome Research. 2008;7(11):4715–4726. doi: 10.1021/pr800255a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cuomo A, Moretti S, Minucci S, Bonaldi T. SILAC-based proteomic analysis to dissect the “histone modification signature" of human breast cancer cells. doi: 10.1007/s00726-010-0668-2. Amino Acids, http://www.ncbi.nlm.nih.gov/pubmed/20617350. In press. [DOI] [PubMed] [Google Scholar]
- 130.Zhou Q, Chaerkady R, Shaw PG, Kensler TW, Pandey A, Davidson NE. Screening for therapeutic targets of vorinostat by SILAC-based proteomic analysis in human breast cancer cells. Proteomics. 2010;10(5):1029–1039. doi: 10.1002/pmic.200900602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kashyap MK, Harsha HC, Renuse S, et al. SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome. Cancer Biology and Therapy. 2010;10(8):796–810. doi: 10.4161/cbt.10.8.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee KK, Todorova K, Mandinova A. Maximizing early detection of esophageal squamous cell carcinoma via SILAC-proteomics. Cancer Biology and Therapy. 2010;10(8):811–813. doi: 10.4161/cbt.10.8.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Prokhorova TA, Rigbolt KTG, Johansen PT, et al. Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Molecular & Cellular Proteomics. 2009;8(5):959–970. doi: 10.1074/mcp.M800287-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Collier TS, Sarkar P, Rao B, Muddiman DC. Quantitative top-down proteomics of SILAC labeled human embryonic stem cells. Journal of the American Society for Mass Spectrometry. 2010;21(6):879–889. doi: 10.1016/j.jasms.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 135.Tian R, Wang S, Elisma F, et al. Rare cell proteomic reactor applied to SILAC based quantitative proteomic study of human embryonic stem cell differentiation. Molecular & Cellular Proteomics. 2011;10(2) doi: 10.1074/mcp.M110.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xu Y, Liang S, Shen G, et al. Application of the SILAC (stable isotope labelling with amino acids in cell culture) technique in quantitative comparisons for tissue proteome expression. Biotechnology and Applied Biochemistry. 2009;54(1):11–20. doi: 10.1042/BA20090007. [DOI] [PubMed] [Google Scholar]
- 137.Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nature Methods. 2010;7(5):383–385. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- 138.Graumann J, Hubner NC, Kim JB, et al. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Molecular & Cellular Proteomics. 2008;7(4):672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- 139.Cuomo A, Bonaldi T. Systems biology "on-the-fly": SILAC-based quantitative proteomics and RNAi approach in Drosophila melanogaster. Methods in Molecular Biology. 2010;662:59–78. doi: 10.1007/978-1-60761-800-3_3. [DOI] [PubMed] [Google Scholar]
- 140.Sury MD, Chen J-X, Selbach M. The SILAC fly allows for accurate protein quantification in vivo. Molecular & Cellular Proteomics. 2010;9(10):2173–2183. doi: 10.1074/mcp.M110.000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Funderburgh JL, Corpuz LM, Roth MR, Funderburgh ML, Tasheva ES, Conrad GW. Mimecan, the 25-kDa corneal keratan sulfate proteoglycan, is a product of the gene producing osteoglycin. Journal of Biological Chemistry. 1997;272(44):28089–28095. doi: 10.1074/jbc.272.44.28089. [DOI] [PubMed] [Google Scholar]
- 142.Hocking AM, Shinomura T, McQuillan DJ. Leucine-rich repeat glycoproteins of the extracellular matrix. Matrix Biology. 1998;17(1):1–19. doi: 10.1016/s0945-053x(98)90121-4. [DOI] [PubMed] [Google Scholar]
- 143.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annual Review of Biochemistry. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 144.Iozzo RV. The biology of the small leucine-rich proteoglycans. Functional network of interactive proteins. Journal of Biological Chemistry. 1999;274(27):18843–18846. doi: 10.1074/jbc.274.27.18843. [DOI] [PubMed] [Google Scholar]
- 145.Matsushima N, Ohyanagi T, Tanaka T, Kretsinger RH. Super-motifs and evolution of tandem leucine-rich repeats within the small proteoglycans—biglycan, decorin, lumican, fibromodulin, PRELP, keratocan, osteoadherin, epiphycan, and osteoglycin. Proteins: Structure, Function and Genetics. 2000;38(2):210–225. doi: 10.1002/(sici)1097-0134(20000201)38:2<210::aid-prot9>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 146.Henry SP, Takanosu M, Boyd TC, et al. Expression pattern and gene characterization of asporin. A newly discovered member of the leucine-rich repeat protein family. Journal of Biological Chemistry. 2001;276(15):12212–12221. doi: 10.1074/jbc.M011290200. [DOI] [PubMed] [Google Scholar]
- 147.Lorenzo P, Aspberg A, Önnerfjord P, Bayliss MT, Neame PJ, Heinegård D. Identification and characterization of asporin. A novel member of the leucine-rich repeat protein family closely related to decorin and biglycan. Journal of Biological Chemistry. 2001;276(15):12201–12211. doi: 10.1074/jbc.M010932200. [DOI] [PubMed] [Google Scholar]
- 148.Tasheva ES, Koester A, Paulsen AQ, et al. Mimecan/osteoglycin-deficient mice have collagen fibril abnormalities. Molecular Vision. 2002;8:407–415. [PubMed] [Google Scholar]
- 149.Ge G, Seo NS, Liang X, Hopkins DR, Höök M, Greenspan DS. Bone morphogenetic protein-1/tolloid-related metalloproteinases process osteoglycin and enhance its ability to regulate collagen fibrillogenesis. Journal of Biological Chemistry. 2004;279(40):41626–41633. doi: 10.1074/jbc.M406630200. [DOI] [PubMed] [Google Scholar]
- 150.Shanahan CM, Cary NRB, Osbourn JK, Weissberg PL. Identification of osteoglycin as a component of the vascular matrix: differential expression by vascular smooth muscle cells during neointima formation and in atherosclerotic plaques. Arteriosclerosis, Thrombosis, and Vascular Biology. 1997;17(11):2437–2447. doi: 10.1161/01.atv.17.11.2437. [DOI] [PubMed] [Google Scholar]
- 151.Fernández B, Kampmann A, Pipp F, Zimmermann R, Schaper W. Osteoglycin expression and localization in rabbit tissues and atherosclerotic plaques. Molecular and Cellular Biochemistry. 2003;246(1-2):3–11. [PubMed] [Google Scholar]
- 152.Cui XN, Tang JW, Song B, Wang B, Chen SY, Hou L. High expression of osteoglycin decreases gelatinase activity of murine hepatocarcinoma Hca-F cells. World Journal of Gastroenterology. 2009;15(48):6117–6122. doi: 10.3748/wjg.15.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cui X, Song B, Hou L, Wei Z, Tang J. High expression of osteoglycin decreases the metastatic capability of mouse hepatocarcinoma Hca-F cells to lymph nodes. Acta Biochimica et Biophysica Sinica. 2008;40(4):349–355. doi: 10.1111/j.1745-7270.2008.00392.x. [DOI] [PubMed] [Google Scholar]
- 154.Tasheva ES, Conrad GW. Interferon-γ regulation of the human mimecan promoter. Molecular Vision. 2003;9:277–287. [PubMed] [Google Scholar]
- 155.Bentz H, Nathan RM, Rosen DM, et al. Purification and characterization of a unique osteoinductive factor from bovine bone. Journal of Biological Chemistry. 1989;264(34):20805–20810. [PubMed] [Google Scholar]
- 156.Madisen L, Neubauer M, Plowman G, et al. Molecular cloning of a novel bone-forming compound: osteoinductive factor. DNA and Cell Biology. 1990;9(5):303–309. doi: 10.1089/dna.1990.9.303. [DOI] [PubMed] [Google Scholar]
- 157.Dasch JR, Pace DR, Avis PD, Bentz H, Chu S. Characterization of monoclonal antibodies recognizing bovine bone osteoglycin. Connective Tissue Research. 1993;30(1):11–21. doi: 10.3109/03008209309032927. [DOI] [PubMed] [Google Scholar]
- 158.Xu T, Bianco P, Fisher LW, et al. Targeted disruption of the biglycan gene leads to an osteoporosis-like phenotype in mice. Nature Genetics. 1998;20(1):78–82. doi: 10.1038/1746. [DOI] [PubMed] [Google Scholar]
- 159.Hamajima S, Hiratsuka K, Kiyama-Kishikawa M, et al. Effect of low-level laser irradiation on osteoglycin gene expression in osteoblasts. Lasers in Medical Science. 2003;18(2):78–82. doi: 10.1007/s10103-003-0255-9. [DOI] [PubMed] [Google Scholar]
- 160.Aziz A, Miyake T, Engleka KA, Epstein JA, McDermott JC. Menin expression modulates mesenchymal cell commitment to the myogenic and osteogenic lineages. Developmental Biology. 2009;332(1):116–130. doi: 10.1016/j.ydbio.2009.05.555. [DOI] [PubMed] [Google Scholar]
- 161.Prosperi MT, Ferbus D, Karczinski I, Goubin G. A human cDNA corresponding to a gene overexpressed during cell proliferation encodes a product sharing homology with amoebic and bacterial proteins. Journal of Biological Chemistry. 1993;268(15):11050–11056. [PubMed] [Google Scholar]
- 162.Ishii T, Yamada M, Sato H, et al. Cloning and characterization of a 23-kDa stress-induced mouse peritoneal macrophage protein. Journal of Biological Chemistry. 1993;268(25):18633–18636. [PubMed] [Google Scholar]
- 163.Chae HZ, Robison K, Poole LB, Church G, Storz G, Rhee SG. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(15):7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yanagawa T, Iwasa S, Ishii T, et al. Peroxiredoxin I expression in oral cancer: a potential new tumor marker. Cancer Letters. 2000;156(1):27–35. doi: 10.1016/s0304-3835(00)00434-1. [DOI] [PubMed] [Google Scholar]
- 165.Chang JW, Jeon HB, Lee JH, et al. Augmented expression of peroxiredoxin I in lung cancer. Biochemical and Biophysical Research Communications. 2001;289(2):507–512. doi: 10.1006/bbrc.2001.5989. [DOI] [PubMed] [Google Scholar]
- 166.Kim HJ, Chae HZ, Kim YJ, et al. Preferential elevation of Prx I and Trx expression in lung cancer cells following hypoxia and in human lung cancer tissues. Cell Biology and Toxicology. 2003;19(5):285–298. doi: 10.1023/b:cbto.0000004952.07979.3d. [DOI] [PubMed] [Google Scholar]
- 167.Kinnula VL, Pääkkö P, Soini Y. Antioxidant enzymes and redox regulating thiol proteins in malignancies of human lung. FEBS Letters. 2004;569(1–3):1–6. doi: 10.1016/j.febslet.2004.05.045. [DOI] [PubMed] [Google Scholar]
- 168.Lehtonen ST, Svensk AM, Soini Y, et al. Peroxiredoxins, a novel protein family in lung cancer. International Journal of Cancer. 2004;111(4):514–521. doi: 10.1002/ijc.20294. [DOI] [PubMed] [Google Scholar]
- 169.Chang JW, Lee SH, Jeong JY, et al. Peroxiredoxin-I is an autoimmunogenic tumor antigen in non-small cell lung cancer. FEBS Letters. 2005;579(13):2873–2877. doi: 10.1016/j.febslet.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 170.Kim JH, Bogner PN, Ramnath N, Park Y, Yu J, Park YM. Elevated peroxiredoxin 1, but not NF-E2-related factor 2, is an independent prognostic factor for disease recurrence and reduced survival in stage I non-small cell lung cancer. Clinical Cancer Research. 2007;13(13):3875–3882. doi: 10.1158/1078-0432.CCR-06-2893. [DOI] [PubMed] [Google Scholar]
- 171.Kim JH, Bogner PN, Baek SH, et al. Up-regulation of peroxiredoxin 1 in lung cancer and its implication as a prognostic and therapeutic target. Clinical Cancer Research. 2008;14(8):2326–2333. doi: 10.1158/1078-0432.CCR-07-4457. [DOI] [PubMed] [Google Scholar]
- 172.Kim SY, Kim TJ, Lee KY. A novel function of peroxiredoxin 1 (Prx-1) in apoptosis signal-regulating kinase 1 (ASK1)-mediated signaling pathway. FEBS Letters. 2008;582(13):1913–1918. doi: 10.1016/j.febslet.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 173.Shen J, Person MD, Zhu J, Abbruzzese JL, Li D. Protein expression profiles in pancreatic adenocarcinoma compared with normal pancreatic tissue and tissue affected by pancreatitis as detected by two-dimensional gel electrophoresis and mass spectrometry. Cancer Research. 2004;64(24):9018–9026. doi: 10.1158/0008-5472.CAN-04-3262. [DOI] [PubMed] [Google Scholar]
- 174.Qi Y, Chiu JF, Wang L, Kwong DLW, He QY. Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics. 2005;5(11):2960–2971. doi: 10.1002/pmic.200401175. [DOI] [PubMed] [Google Scholar]
- 175.Chen MF, Chen WC, Wu CT, et al. p53 status is a major determinant of effects of decreasing peroxiredoxin I expression on tumor growth and response of lung cancer cells to treatment. International Journal of Radiation Oncology Biology Physics. 2006;66(5):1461–1472. doi: 10.1016/j.ijrobp.2006.07.1372. [DOI] [PubMed] [Google Scholar]
- 176.Chen MF, Keng PC, Shau H, et al. Inhibition of lung tumor growth and augmentation of radiosensitivity by decreasing peroxiredoxin I expression. International Journal of Radiation Oncology Biology Physics. 2006;64(2):581–591. doi: 10.1016/j.ijrobp.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 177.Kim Y-J, Lee W-S, Ip C, Chae H-Z, Park E-M, Park Y-M. Prx1 suppresses radiation-induced c-Jun NH2-terminal kinase signaling in lung cancer cells through interaction with the glutathione S-transferase Pi/c-Jun NH2-terminal kinase complex. Cancer Research. 2006;66(14):7136–7142. doi: 10.1158/0008-5472.CAN-05-4446. [DOI] [PubMed] [Google Scholar]
- 178.Manevich Y, Sweitzer T, Pak JH, Feinstein SI, Muzykantov V, Fisher AB. 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11599–11604. doi: 10.1073/pnas.182384499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pak JH, Manevich Y, Kim HS, Feinstein SI, Fisher AB. An antisense oligonucleotide to 1-cys peroxiredoxin causes lipid peroxidation and apoptosis in lung epithelial cells. Journal of Biological Chemistry. 2002;277(51):49927–49934. doi: 10.1074/jbc.M204222200. [DOI] [PubMed] [Google Scholar]
- 180.Shan SW, Tang MK, Cai DQ, et al. Comparative proteomic analysis identifies protein disulfide isomerase and peroxiredoxin 1 as new players involved in embryonic interdigital cell death. Developmental Dynamics. 2005;233(2):266–281. doi: 10.1002/dvdy.20404. [DOI] [PubMed] [Google Scholar]
- 181.Shibayama H, Takai E, Matsumura I, et al. Identification of a cytokine-induced antiapoptotic molecule anamorsin essential for definitive hematopoiesis. Journal of Experimental Medicine. 2004;199(4):581–592. doi: 10.1084/jem.20031858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hao Z, Qiao T, Jin X, Li X, Gao J, Fan D. Preparation and characterization of a specific monoclonal antibody against CIAPIN1. Hybridoma. 2005;24(3):141–145. doi: 10.1089/hyb.2005.24.141. [DOI] [PubMed] [Google Scholar]
- 183.Li X, Wu K, Fan D. CIAPIN1 as a therapeutic target in cancer. Expert Opinion on Therapeutic Targets. 2010;14(6):603–610. doi: 10.1517/14728221003774127. [DOI] [PubMed] [Google Scholar]
- 184.Hao Z, Li X, Qiao T, Li S, Lv Y, Fan D. Downregulated expression of CIAPIN1 may contribute to gastric carcinogenesis by accelerating cell proliferation and promoting cell cycle progression. Cancer Biology and Therapy. 2009;8(11):1064–1070. doi: 10.4161/cbt.8.11.8796. [DOI] [PubMed] [Google Scholar]
- 185.He L, Wang H, Jin H, et al. CIAPIN1 inhibits the growth and proliferation of clear cell renal cell carcinoma. Cancer Letters. 2009;276(1):88–94. doi: 10.1016/j.canlet.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 186.Zheng X, Zhao Y, Wang X, et al. Decreased expression of CIAPIN1 is correlated with poor prognosis in patients with esophageal squamous cell carcinoma. Digestive Diseases and Sciences. 2010;55(12):3408–3414. doi: 10.1007/s10620-010-1212-7. [DOI] [PubMed] [Google Scholar]
- 187.Shi H, Zhou Y, Liu H, et al. Expression of CIAPIN1 in human colorectal cancer and its correlation with prognosis. BMC Cancer. 2010;10, article 477 doi: 10.1186/1471-2407-10-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Li X, Hao Z, Fan R, et al. CIAPIN1 inhibits gastric cancer cell proliferation and cell cycle progression by downregulating CyclinD1 and upregulating P27. Cancer Biology and Therapy. 2007;6(10):1539–1545. doi: 10.4161/cbt.6.10.4684. [DOI] [PubMed] [Google Scholar]


