Abstract
A unique subfamily of calmodulin-dependent Ca2+-ATPases was recently identified in plants. In contrast to the most closely related pumps in animals, plasma membrane-type Ca2+-ATPases, members of this new subfamily are distinguished by a calmodulin-regulated autoinhibitor located at the N-terminal instead of a C-terminal end. In addition, at least some isoforms appear to reside in non-plasma membrane locations. To begin delineating their functions, we investigated the subcellular localization of isoform ACA2p (Arabidopsis Ca2+-ATPase, isoform 2 protein) in Arabidopsis. Here we provide evidence that ACA2p resides in the endoplasmic reticulum (ER). In buoyant density sucrose gradients performed with and without Mg2+, ACA2p cofractionated with an ER membrane marker and a typical “ER-type” Ca2+-ATPase, ACA3p/ECA1p. To visualize its subcellular localization, ACA2p was tagged with a green fluorescence protein at its C terminus (ACA2-GFPp) and expressed in transgenic Arabidopsis. We collected fluorescence images from live root cells using confocal and computational optical-sectioning microscopy. ACA2-GFPp appeared as a fluorescent reticulum, consistent with an ER location. In addition, we observed strong fluorescence around the nuclei of mature epidermal cells, which is consistent with the hypothesis that ACA2p may also function in the nuclear envelope. An ER location makes ACA2p distinct from all other calmodulin-regulated pumps identified in plants or animals.
Ca2+ is thought to function as an important second messenger in all eukaryotes (Bootman and Berridge, 1995; Clapham, 1995). In addition, Ca2+ is required for the stability and activity of many proteins and appears to play a critical role in protein processing in the secretory pathway (Rudolph et al., 1989; Gill et al., 1996). To control Ca2+ concentrations in different compartments, cells commonly use two active transport systems, Ca2+-ATPases (pumps) and H+- or Na+-coupled antiporters. In plants there is evidence for multiple Ca2+ pumps (Evans and Williams, 1998) and low-affinity Ca2+/H+ antiporters (Hirschi et al., 1996).
Ca2+ pumps belong to a large superfamily of P-type ATPases that include the Na+/K+-ATPase of animals and the H+-ATPase of plants and fungi. Axelsen and Palmgren (1998) have proposed two distinct families of Ca2+ pumps, type IIA and IIB, based on protein sequence homologies. Type IIA and IIB pumps include the “ER-type” and the “PM-type” Ca2+ pumps, respectively, first characterized in animal cells. Previously, homologs of ER- or PM-type pumps were distinguished by three criteria: (a) localization to either the ER or PM, respectively, (b) differential sensitivity to inhibitors (e.g. ER-type inhibition by cyclopiazonic acid and thapsigargin), and (c) direct activation of PM-type pumps by calmodulin. However, not all plant homologs conform to these criteria (Bush, 1995; Evans and Williams, 1998).
In plants several genes encoding type IIA pumps (ER-type homologs) have been cloned, including LCA1 from tomato (Wimmers et al., 1992), OsCA from rice (Chen et al., 1997), and ACA3/ECA1 (Arabidopsis Ca2+-ATPase, isoform 3/ER-Ca2+-ATPase isoform 1) from Arabidopsis (Liang et al., 1997). Consistent with the criteria for a typical ER-type pump, ACA3p (ACA isoform 3 protein) appears to reside in the ER (Liang et al., 1997). However, non-ER locations have been suggested for other isoforms. For example, Ferrol and Bennett (1996) obtained evidence for tonoplast and PM isoforms from membrane fractionation and immunodetection of pumps cross-reacting with an anti-LCA1 antibody.
Three plant genes encoding type IIB pumps (PM-type homologs) have also been identified: ACA1 and ACA2 from Arabidopsis (Huang et al., 1993; Harper et al., 1998) and BCA1 from Brassica oleracea pimprivate (Malmstrom et al., 1997). These plant homologs are distinguished from animal PM-type pumps by a unique structural arrangement with putative autoinhibitory sequences located in the N-terminal instead of C-terminal domain and proposed non-PM locations (Harper et al., 1998). Nevertheless, as expected for type IIB pumps, at least some plant homologs have calmodulin-dependent Ca2+-ATPase activity. For isoform ACA2p, this contention is supported by three lines of evidence (Harper et al., 1998). First, ACA2p was expressed in yeast and shown to have a Ca2+ and calmodulin-stimulated ATPase activity. Second, a calmodulin-binding sequence was mapped within 36 residues of the N terminus, indicating that the pump could interact directly with calmodulin. Third, a partial deletion of the N-terminal domain resulted in a constitutively active Ca2+-ATPase (i.e. calmodulin independent). Together, these results support the hypothesis that the N-terminal domain functions as a calmodulin-regulated autoinhibitor.
Harper et al. (1998) previously obtained evidence that ACA2p is targeted to a non-PM location from aqueous two-phase partitioning of microsomal membranes. However, these studies did not determine a specific endomembrane location for ACA2p. Non-PM locations have also been proposed for ACA1p and BCA1p. ACA1p is thought to target to a plastid inner membrane, based on membrane fractionation and immunodetection with an anti-ACA1 polyclonal antibody (Huang et al., 1993). BCA1p is thought to target to the tonoplast, based on correspondence to the peptide sequence obtained from a purified tonoplast ATPase (Askerlund, 1996; Malmstrom et al., 1997). However, none of these studies used an isoform-specific probe, nor were they confirmed by cytological evidence.
Here we show that ACA2p is most abundant in the ER, as indicated by membrane fractionation and corroborated by fluorescence imaging in live cells of ACA2p tagged with a GFP. An ER location establishes ACA2p as the first calmodulin-regulated Ca2+-ATPase to be identified in the ER of any organism. The ER in Arabidopsis also contains a typical ER-type Ca2+ pump. Thus, to our knowledge, our results provide the first example of an organism in which the ER has been found to function with two different types of Ca2+ pumps.
MATERIALS AND METHODS
Plant experiments were conducted with Arabidopsis cv Columbia. Yeast experiments were conducted with the Saccharomyces cerevisiae strain K616 (MATa pmr1::HIS3 pmc1::TRP1 cnb1::LEU2, ura3 [Cunningham and Fink, 1994]). DNA cloning was done in Escherichia coli strain XL1-Blue (Stratagene) or DH10α (a derivative of DH5α; Stratagene). Unless noted otherwise, we used standard molecular techniques according to the method of Sambrook et al. (1989).
Antibodies
Anti-ACA2 (no. 1371) rabbit polyclonal antiserum against a GST fusion protein with ACA2 residues Val-119 to Pro-161 was described by Harper et al. (1998). Anti-CTF2 was made against a GST fusion protein with the C-terminal domain of a PM H+-ATPase (AHA2) (DeWitt et al., 1996). Anti-ACA3 (no. 1374) was generated against a GST fusion protein containing the last 27 residues of ECA1p/ACA3p, an ER-type Ca2+-ATPase from Arabidopsis (Liang et al., 1997). Dr. M. Chrispeels (University of California, San Diego) kindly provided the anti-BiP and anti-γ-tonoplast intrinsic protein. We purchased anti-GFP from Clontech (Palo Alto, CA).
For immunocytochemistry, we used affinity-purified anti-ACA2 antibodies. Serum was first precipitated with 50% ammonium sulfate and resuspended in PBS. Anti-GST antibodies were removed using a column coupled with GST protein. Anti-ACA2 antibodies were then allowed to bind to a column containing the fusion protein encoded by pACA2-Ns (Harper et al., 1998). Columns were made using cyanogen bromide-activated Sepharose 4B according to the manufacturer's instructions (Pharmacia). Anti-ACA2 antibodies were eluted with 0.1 m Gly, pH 2.7, immediately neutralized with 0.1 volume of 1 m Tris-HCl, pH 8.0, and dialyzed against PBS. Control preimmune serum was purified over a protein-A Sepharose CL-4B column (Sigma).
Plasmid Constructs
We used standard PCR reactions and subcloning procedures to engineer ACA2 sequences into the clones described below. We used Ampli-Taq (Perkin-Elmer) or Taka Ra Ex Taq (PanVera, Madison, WI) to perform PCR. All PCR-derived sequences were sequenced to ensure the absence of PCR mistakes. DNA sequencing was done at The Scripps Biochemistry Core Facility using an automated sequencer (Prism 373XL, ABI, Foster City, CA).
pYX-ACA2–1 and pYX-ACA2–2 encode full-length and N-terminally truncated versions, respectively, of ACA2p for expression in yeast (Harper et al., 1998). The parent vector used for all yeast constructs in this study was pYX-112-TEV, which contains a URA3 gene for selection in yeast.
pYX-ACA2–7 (mp11) encodes an ACA2-GFPp identical to that diagrammed in Figure 3 but engineered for expression in yeast. The construct was engineered as an insertion between the SalI and NheI sites of pYX-112-TEV. The sequence at the 5′ end is GTCGACATG (start codon is underlined). The coding sequence contains a BstB1 and SacII site as shown for pACA2-wt (Harper et al., 1998).
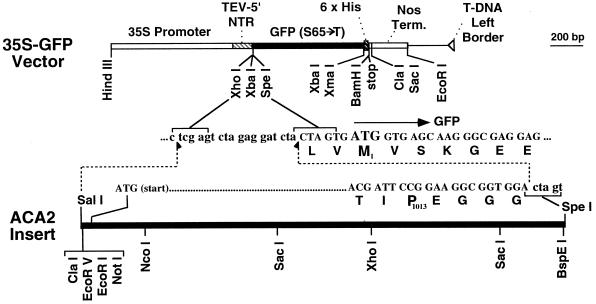
Figure 3.
Diagram of p35S-ACA2-GFP used for expression of a GFP-tagged pump in transgenic plants. ACA2-GFP was placed under the control of a CaMV-35S promoter. A translational enhancer sequence derived from the tobacco etch virus was used as a 5′-untranslated leader sequence (Harper et al., 1998). For the junction between ACA2 and the C-terminal GFP, the nucleotide and amino acid sequences are shown. Pro-1013 is the last amino acid contributed by ACA2p. The downstream M1 in bold corresponds to the “start” Met for GFP. A six-His motif present at the C-terminal end of the GFP domain, encoded by the following sequence, starts with the XbaI site: T CTA GAC CCG GGA ATG CAT CAC CAT CAC CAT CAC GGA TCC TGA). Some useful enzyme sites are shown for diagnostic and subcloning purposes.
pYX-ACA2–8 (mp20) encodes an N-terminally truncated ACA2p (Harper et al., 1998) fused to a C-terminal GFP, as described for pYX-ACA2–7. The sequence at the modified 5′ end is GTCGACATGAGT (start codon for M1 is underlined, followed by the codon for “S81”).
p35S-ACA2-GFP1 encodes a full-length ACA2-GFPp fusion as shown in Figure 3. The 35S-GFP vector used was p35S-GFP-JFH1 (mp16). This vector was derived from pBIN19 (accession no. U09365; Frisch et al., 1995), modified to include a CaMV-35S promoter from a pRT-derived vector (Topfer et al., 1987), a translational enhancer derived from a tobacco etch virus (sequence shown from 20 to 155 [Harper et al., 1998]), a synthetic GFP sequence with an S65-T mutation (derived from a clone provided by J. Sheen; Sheen, 1996), and a six-His affinity tag. The coding sequence for ACA2p was cut from clone pACA2-BSX-3 (mp 62) as an SalI-Spe1 insert and subcloned into the XhoI-Spe1 site of p35S-GFP-JFH1. The 3′ end of pACA2-BSX-3 had been modified by site-specific mutagenesis to include three Gly residues followed by an Spe1.
Yeast Transformations
For transformations, K616 was grown in standard yeast extract, peptone, and dextrose medium supplemented with 10 mm CaCl2. Yeast were transformed by lithium acetate/PEG methods (Elble, 1992) and selected for uracil prototrophy by plating on synthetic medium minus uracil supplemented with 2% Suc as a carbon source and 1.5% agar (Rose et al., 1990). For complementation studies, Ura+ colonies were streaked on plates containing 10 mm EGTA (pH 6.0) and incubated for 2 to 3 d at 30°C as described by Liang et al. (1997).
Plant Transformation
Transgenic plants were generated by a vacuum-infiltration protocol (Bechtold et al., 1993) using Agrobacterium tumefaciens strain GV3101. Infiltration was performed a few days after clipping primary bolts. Dry seeds were harvested, sterilized in 20% bleach for 20 min, and plated on Gamborg's B5 medium containing kanamycin (50 mg L−1), and carbenicillin (300 mg L−1). Kanamycin-resistant plants (T0) were identified and grown from seed. We conducted experiments with T1 or T2 plants and selected two independent transgenic plant lines for detailed analysis of ACA2-GFPp expression and localization (identification nos. 1284 and 1289).
Membrane Fractionation
We prepared microsomal membranes by modifying a procedure previously described by Serrano (1984). All manipulations were conducted on ice or in a cold room with prechilled buffers. Membranes came from plants grown in liquid Gamborg's B5 medium with 0.05% (w/v) Mes (pH 5.7) on a shaker at about 125 rpm. Fresh tissues were cut into small pieces under an extraction buffer (290 mm Suc, 2 mm EDTA, 250 mm Tris-HCl, pH 8.5, 2 mm PMSF, and 76 mm β-mercaptoethanol) and homogenized with a mortar and pestle. For some preparations, the homogenization buffer contained 5 mm MgCl2. Homogenates were filtered through cheesecloth to remove large debris and then centrifuged at 5,000g to remove intact organelles and cell walls. Supernatants were spun at 100,000g for 1 h to pellet microsomal membranes. Membrane pellets were resuspended in homogenization buffer (1 mL/10 g starting material) using a glass homogenizer. Microsomes (0.5 mL) were then layered onto a gradient of 15% to 45% (w/w) Suc in a centrifugation buffer (10 mm Tris, pH 7.5, 1 mm EDTA, 1 mm DTT, 2 mm benzamidine, and 0.1 mm PMSF). For all the plus Mg2+ Suc gradients, 5 mm MgCl2 was added to the homogenization and centrifugation buffers. Gradients were centrifuged at 110,000g for 16 h and 1-mL fractions were collected, frozen in liquid nitrogen, and stored at −80°C. A refractometer was used to measure the Suc concentration of each fraction.
Western Blots
For SDS-PAGE, samples were mixed with 3× loading buffer (100 mm Tris, pH 6.8, 3.7% [w/v] SDS, 5% [w/v] DTT, 20% [w/v] Suc or glycerol, and 0.3% [w/v] bromphenol blue) and incubated for 15 min at 37°C. After the sample was electrophoresed, a transfer apparatus (Bio-Rad) transferred proteins to nitrocellulose. The transfer buffer consisted of 192 mm Gly, 25 mm Tris-HCl (pH 8.3), 20% (v/v) methanol, and 0.02% (w/v) SDS.
Blots were incubated for 2 h in a blocking buffer (20 mm Tris, [pH 7.6], 137 mm NaCl, and 0.5% [w/v] Tween 20 [TBS-T] with 5% [w/v] nonfat dry milk). We diluted the primary antisera accordingly in the blocking buffer. The secondary antibody used for immunodetection on western blots was a donkey anti-rabbit IgG conjugated with horseradish peroxidase (Amersham), and it was diluted at 1:5000 in a blocking buffer. Two-hour primary and secondary antibody incubations at room temperature were followed by four 15-min washes in TBS-T. Secondary antibodies were detected using ECL (Amersham).
Marker Enzyme Assays
Chlorophyll a and b concentrations were determined spectrophotometrically by mixing 10-μL samples with 750 μL of 95% ethanol, and the A648.6 and A664.2 were measured. Chlorophyll a and b content was determined using the equation Ca+b = 5.24A664.2 + 22.24A648.6 (Lichtenthaler, 1987).
Triton-stimulated UDPase activity was assayed by adding 10-μL samples of membrane fractions to 100 μL of assay buffer consisting of 3 mm UDP, 3 mm MnSO4, and 30 mm Mes-Tris, pH 6.5 (Nagahashi and Kane, 1982). A parallel reaction was conducted with the addition of 0.03% (w/v) Triton X-100. Reactions were incubated at 37°C for 20 min, and we used the Malachite Green method (Lanzetta et al., 1979) to determine the released phosphate. We calculated latent activity as the difference in activity between the presence and absence of detergent.
Immunocytochemistry
Roots were dissected from 1-week-old seedlings grown vertically on Gamborg's B5 medium with 1% agar and fixed for 1 h at 25°C in freshly prepared 4% paraformaldehyde with 5% Suc in 100 mm phosphate, pH 7.3. Fixed tissues were washed three times at 10-min intervals in 100 mm phosphate and dehydrated in a graded ethanol series. Dehydrated tissues were infiltrated and embedded in London Resin White at 50°C for 16 h. Sections of 1-μm thickness were cut with a glass knife on a microtome (JB-4, Sorvall) and placed on glass slides.
For cyrosectioning, roots were fixed for 2 h in freshly prepared periodate-Lys-paraformaldehyde (McClean and Nakane, 1974). Fixed tissues were washed three times at 10-min intervals in 100 mm phosphate and embedded with frozen tissue medium (Electron Microscopy Sciences, Fort Washington, PA) at −70°C. Sections were cut 8 μm thick at −20°C on a cryostat (Cryocut 1800, Reichert-Jung, Heidelberg, Germany). Frozen sections were affixed to Poly-l-Lys-coated glass slides (Sigma) and allowed to soak for 15 min in PBS-T (PBS with 0.1% Tween 20).
Tissue sections were treated with blocking solution (PBS-T containing 10% normal goat serum and 1% BSA) for 1 h at room temperature and then incubated for 2 h with 10 μg mL−1 affinity-purified anti-ACA2 antibodies or protein A-purified preimmune IgG in the blocking solution. After the sections were washed in three changes of PBS-T (10 min each), they were incubated for 2 h at room temperature with goat anti-rabbit IgG coupled to fluorescein isothiocyanate (Molecular Probes, Eugene, OR) diluted 1:100 in a blocking solution. The sections were washed with three changes of PBS-T (10 min each) and mounted with SlowFade (Molecular Probes).
Fluorescence Confocal Microscopy and Computational Optical-Sectioning Microscopy
For imaging GFP fluorescence, root tips were excised from young seedlings and mounted in Gamborg's B5 medium under glass coverslips. Confocal images were collected on a confocal laser-scanning microscope (MRC-600, Bio-Rad) attached to an inverted microscope (Nikon) equipped with a fluorescein filter. For computational optical sectioning, specimens were sometimes irrigated with 0.1% azide to slow or stop cytoplasmic streaming to generate three-dimensional stacks. The wide-field computational optical-sectioning microscope system was described by Gens et al. (1996). The objective lens used was an X60, 1.4 NA Plan apo lens (Nikon). CCD (charge-coupled device) wells of 6.8 μm width were binned 2 × 2 or 4 × 4, with the cubic voxel size set at either 0.23 × 0.23 × 0.23 or 0.45 × 0.45 × 0.45 μm. Restoration of Figure 7, E through G, was by the maximum-likelihood algorithm with 1000 iterations and with an intensity penalty of 5 × 10−6 for F and G, and these were also subjected to light-Gaussian filtering. The algorithm and user-friendly interfaces are available at http://ibc.wustl.edu:80/bcl/.
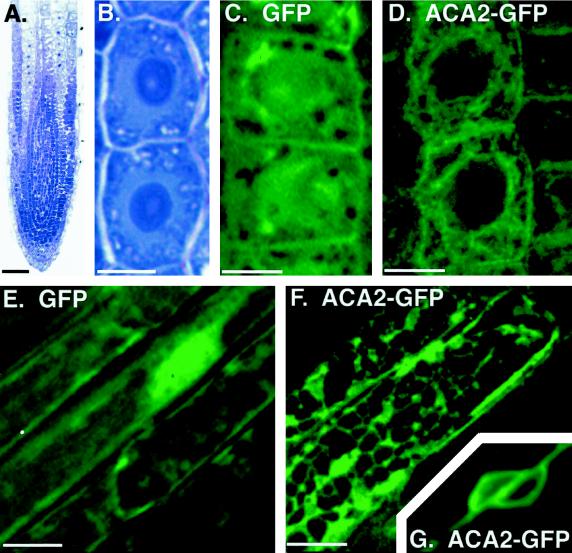
Figure 7.
Imaging of ACA2-GFPp fluorescence reveals ER-like structures in root cells. A, Longitudinal section from a primary root shows undifferentiated cells at the root tip. Cells are stained with toluidine blue to emphasize densely stained nuclei. Toluidine blue-stained sections were photographed on an inverted microscope (model IMT2, Olympus) using Kodak Ektachrome P1600. B, Two undifferentiated cells from A are shown at higher magnification to emphasize the position of the large central nuclei. These cells are representative of live cells imaged for GFP fluorescence in C and D. C and D, Confocal images of green fluorescence from two undifferentiated cells expressing a GFPp-only control (C) or ACA2-GFPp (D). E and F, Green fluorescence from mature root epidermal cells expressing a GFPp-only control (E) or ACA2-GFPp (F), as imaged by computational optical-sectioning microscopy. G, The nuclear region from two neighboring epidermal cells expressing ACA2-GFPp. Imaging of the nuclear region required shorter exposure times, which left the image of the surrounding network only faintly fluorescent. The thickness and angle of this section does not show the cell wall, which lies at an oblique angle between two nuclei in two separate epidermal cells. Scale bars = 50 μm in A; 5 μm in B, C, and D; and 10 μm in E, F, and G. Images were arranged using Adobe PhotoShop (Adobe Systems, Mountain View, CA).
RESULTS
Immunocytology Indicates That ACA2p Is an Endomembrane Pump
Harper et al. (1998) previously showed ACA2p to be most abundant in the roots, as determined by a western-blot analysis. As a first approach to determine the subcellular localization of ACA2p, root sections were examined by immunofluorescence microscopy (Fig. 1). Cryosections were immunodecorated with an affinity-purified anti-ACA2 polyclonal antibody. The antiserum was generated against residues Val-119 to Pro-161 within the N-terminal domain of ACA2p (Harper et al., 1998). Affinity-purified antibodies detected a single protein band of 110 kD in a western blot of total cellular protein (Harper et al., 1998). These antibodies appeared to be highly specific for ACA2p, as indicated by their failure to detect cross-reacting proteins in tobacco or onion (data not shown). Nevertheless, the possibility that these antibodies cross-react with a second, closely related isoform in Arabidopsis has not been excluded.
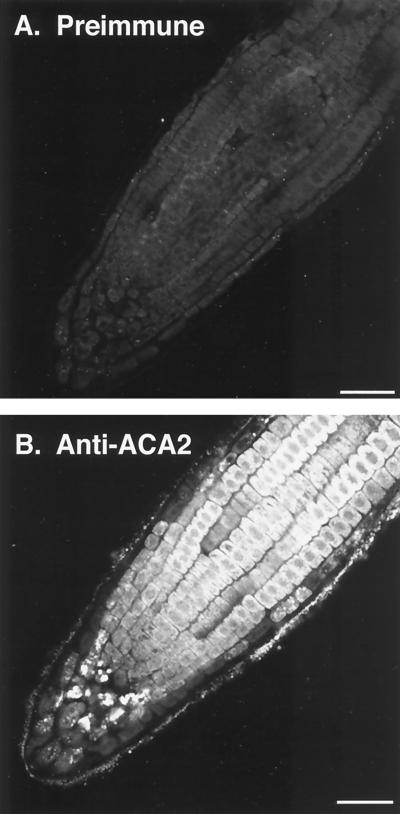
Figure 1.
Immunofluorescence microscopy of root cells showing ACA2p in an endomembrane system. A, Longitudinal section of a root tip labeled with preimmune IgG and fluorescein isothiocyanate-conjugated secondary antibody. B, A similar section labeled with affinity-purified anti-ACA2p and fluorescein isothiocyanate-conjugated secondary antibody. Figure 7A shows a similar section stained with toluidine to show structural preservation of root tip cells and the presence of large, densely stained nuclei in the center of most cells. Bar = 50 μm.
Confocal microscopy revealed anti-ACA2p staining of all cell types within roots. Immunodecorated proteins were observed inside the cell boundary, with an apparent exclusion from the nucleoplasm. These results are consistent with a non-PM location, as previously suggested, based on membrane-fractionation studies using aqueous two-phase partitioning (Harper et al., 1998). However, our immunocytology did not provide sufficient resolution to identify specific subcellular structures.
ACA2p Cofractionates with ER
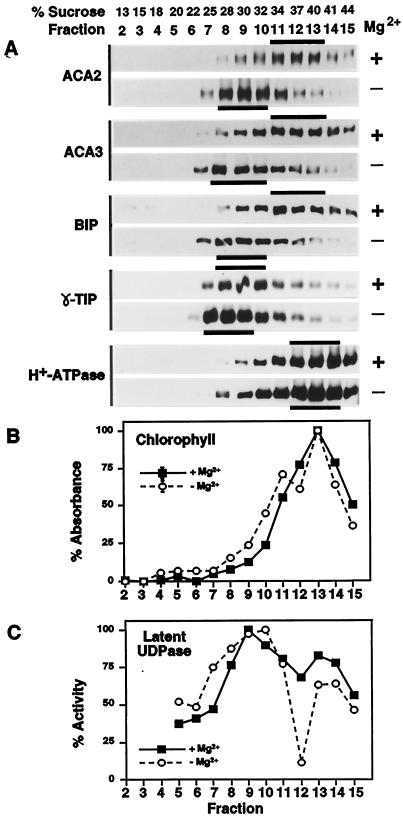
To further define the endomembrane location of ACA2p, microsomal membranes were fractionated on Suc gradients and characterized by western blots and marker enzyme assays (Fig. 2). ACA2p cofractionated with ER membranes, as indicated by sedimentation profiles overlapping with two other ER resident proteins, ACA3p and BiP. BiP is a commonly used ER marker (Haas, 1994) and ACA3p (ECA1p) is a typical ER-type Ca2+ pump in Arabidopsis (Liang et al., 1997). We analyzed two independent sets of gradients with equivalent results. In gradients prepared with 5 mm Mg2+, ACA2p was most abundant at a Suc density of 34% to 40% (w/v), as detected in western blots using anti-ACA2p antibodies. In gradients prepared without Mg2+ (i.e. + 5 mm EDTA), the ACA2p peak shifted to a lighter Suc density between 28% and 32%. A large Mg2+-dependent density shift is characteristic of ER membranes and it occurs when Mg2+ is chelated and polyribosomes are dissociated from the ER. Controls also identified peak fractions for marker enzymes, detecting the PM (H+-ATPase marker at 40%–44% Suc), tonoplast (γ-tonoplast intrinsic protein marker at 25%–32% Suc), chloroplast thylakoid membranes (chlorophyll marker at 40% Suc), and Golgi (latent UDPase marker at 32%–34% Suc). None of these markers revealed a fractionation profile comparable to ACA2p. Thus, our fractionation effectively separated the ER from other major membrane systems and supported an ER localization for ACA2p.
Figure 2.
Suc-gradient membrane fractionation showing cofractionation of ACA2p with ER markers. Microsomal membranes were fractionated over 15% to 45% Suc gradients, and 1-mL fractions were collected. Mg2+ + or − indicate parallel gradients run with 5 mm MgCl2 (+) to stabilize membrane-associated proteins or with 5 mm EDTA (−) to dissociate membrane-associated proteins. A, Western-blot analysis of membrane fractions probed with anti-ACA2, ACA3 (ER marker), BiP (ER marker; Denecke et al., 1991; Hofte and Chrispeels, 1992), γ-TIP (tonoplast marker; Johnson et al., 1990), and H+-ATPase antibodies (PM marker; DeWitt et al., 1996). Black bars highlight peak fractions. Samples (10 μL) from each fraction were separated by SDS-PAGE, blotted, and probed with anti-ACA2 (1:2000), anti-ACA3 (1:2000), anti-BiP (1 2000), anti-γ-TIP (1:500), or anti-CTF2 (H+-ATPase) (1:7500). B, Spectrophotometric analysis of membrane fractions (10-μL samples) for chlorophyll a and b (chloroplast marker). C, Marker enzyme analysis of membrane fractions (10-μL samples) for Triton-stimulated UDPase activity (Golgi marker; Nagahashi and Kane, 1982). The maximal activity in the −Mg2+ gradient was 0.2 μmol min−1 mL−1, and in the +Mg2+ gradient it was 0.3 μmol min−1 mL−1.
Engineering Plants with ACA2-GFPp
To provide cytological corroboration for an ER localization, we tagged ACA2p with a GFP from jellyfish (Aequorea victoria) and visualized its subcellular location in transgenic plants. A GFP was fused to the C-terminal end of ACA2p immediately following the penultimate Pro (Fig. 3). Three Gly residues were included in the linker sequence to provide a flexible attachment. The GFP sequence contained an S65-T mutation that provided an optimal excitation of approximately 490 nm and an emission at 510 nm (Chiu et al., 1996). In addition, a six-His motif was included at the C terminus to permit nickel-resin affinity purification. Figure 3 diagrams the engineering of ACA2-GFP into a plant-expression vector, with transcription under the control of a CaMV-35S promoter.
There were two rationales for choosing a C-terminal location for the GFP tag. First, DeWitt et al. (1996) previously showed that adding epitopes to the C-terminal end of H+-ATPase isoforms AHA2p and AHA3p did not alter membrane insertion or PM-targeting properties. Second, the predicted C-terminal domain of ACA2p had no proposed function and contained only five residues (KTIPV). We considered an N-terminal location to be a risky alternative because the N-terminal domain functions as an autoinhibitor and a tag in this location could easily disrupt the autoinhibitor and generate a hyperactive pump. This was a serious concern because a “deregulated” pump might result in pleiotropic cellular dysfunctions and thereby raise uncertainties about whether normal targeting pathways were being observed.
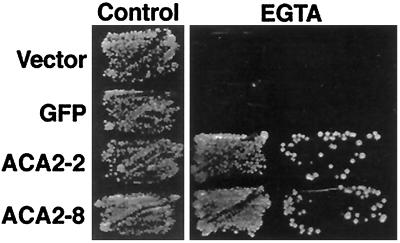
To confirm that our engineered ACA2-GFPp retained the expected functional properties, we genetically tested its activity in a yeast host, K616. This yeast strain harbors a disruption of both endogenous Ca2+ pumps and does not grow on Ca2+-depleted media (e.g. with 10 mm EGTA). Previous studies of an untagged ACA2p demonstrated that only a deregulated mutant (ACA2–2p) provided complementation and allowed K616 to grow on Ca2+-depleted media (Harper et al., 1998). Complementation is thought to require the activity of a high-affinity Ca2+ pump to transport Ca2+ into the ER/Golgi. Enzyme assays established that, in contrast to the Ca2+/calmodulin-regulated activity of the full-length pump (ACA2–1p), the mutant ACA2–2p was constitutively active. This calmodulin-independent activity is proposed to allow ACA2–2p to restore growth to the yeast K616 on Ca2+-depleted media. Under these same growth conditions, the full-length pump probably remains inactive, since the yeast calmodulin would likely be in an apo state because of the expected low levels of cytosolic Ca2+. Consistent with these earlier studies, the complementation properties of ACA2p were not altered by the addition of a GFP tag to either the full-length or the deregulated versions (Fig. 4 shows complementation by the “deregulated ACA2–2 + GFP” = ACA2–8). These results indicated that a C-terminal GFP did not disrupt the potential of ACA2p to function as a Ca2+ pump or for the pump to be regulated by its autoinhibitor. Therefore, the C terminus of ACA2p appeared to be a suitable location for a GFP tag.
Figure 4.
Genetic evidence that ACA2-GFPp retains normal functional properties when expressed in yeast. The yeast mutant K616 was transformed with different constructs and tested for its ability to grow on synthetic medium supplemented with 10 mm CaCl2 (control) or on synthetic medium supplemented with 10 mm EGTA. Growth of K616 harboring different constructs is shown. Both tagged and untagged versions of a deregulated pump (ACA2–8 and ACA2–2, respectively) were able to restore growth of the K616 mutant on EGTA. The constructs shown are: Vector, the parent vector pYX-112; GFP, pYX-GFP-JFH1, which encodes a GFP(S65/T) (J.F. Harper, unpublished data); ACA2–2, pYX-ACA2–2, which encodes a deregulated ACA2p (Harper et al., 1998); and ACA2–8, pYX-ACA2–8, which encodes the same deregulated ACA2–2p with an additional C-terminal GFP tag. The tagged and untagged full-length versions of ACA2p (respectively encoded by pYX-ACA2–7 and pYX-ACA2–1) did not complement and are not shown.
In working with plant cells, our initial evaluation of ACA2-GFPp expression was conducted by electroporating the 35S-ACA2-GFP plasmid into tobacco cv BY2 protoplasts (not shown). Transient expression was easily detected by fluorescence microscopy. The fluorescence was concentrated in subcellular structures similar to those observed in stable, transformed Arabidopsis cells (see below).
We pursued subcellular localization further in transgenic Arabidopsis. The rationale was to examine ACA2-GFPp localization in cell types in which the endogenous ACA2p was known to be expressed. Of 20 independent transgenic plant lines showing ACA2-GFPp expression, all grew into normal, healthy plants, indicating that ACA2-GFPp expression did not adversely alter cellular functions. Expression levels and subcellular localization were examined in detail for two transgenic plant lines, and both expressed ACA2GFPp at approximately 5-fold higher levels than observed for the endogenous ACA2p in wild-type plants (see below). Thus, in the following subcellular localization study healthy cells with only modest levels of transgene overexpression were used.
ACA2-GFPp Cofractionates with ER
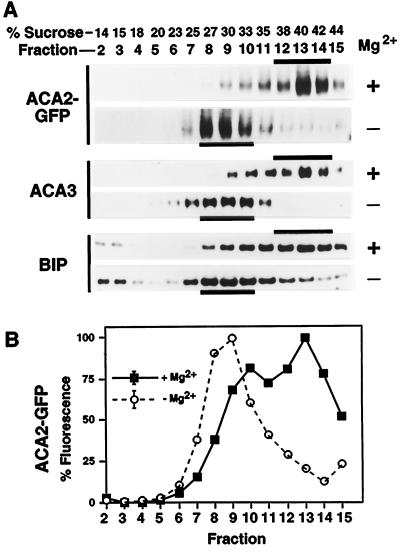
To examine whether the targeting of tagged ACA2-GFPp was comparable to the endogenous ACA2p, we performed parallel membrane-fractionation analyses. We obtained equivalent cofractionation results for the two independent transgenic plants lines that we analyzed. As observed for the endogenous ACA2p (Fig. 2) the tagged ACA2-GFPp cofractionated with two ER markers, BiP and ACA3p, in Suc gradients performed with and without Mg2+ (Fig. 5). Not shown are additional marker assays for the PM, tonoplast, chloroplast, and Golgi, which also showed fractionation profiles equivalent to those shown in Figure 2. ACA2-GFPp was detected in these gradients using two different assays: a western-blot analysis using anti-GFP to immunodetect the chimeric protein at 136 kD (Fig. 5A) and fluorescence spectroscopy to detect the GFP fluorescence (Fig. 5B).
Figure 5.
Suc-gradient fractionation of membranes from 35S-ACA2-GFP transgenic plants showing cofractionation of ACA2-GFPp with ER markers. Microsomal membranes were fractionated through a 15% to 45% (w/w) Suc gradient and analyzed as described in Figure 2. A, Western-blot analysis of membrane fractions probed for ACA2-GFPp, ACA3p, and BIP. Western blots were probed with anti-GFP (1:2000) to specifically detect the ACA2-GFP fusion. ACA3p and BIP were detected as described in Figure 2. B, Fractionation profile of ACA2-GFP as detected by GFP fluorescence. Fluorescence was measured at 510 nm (±5 nm) with excitation at 480 nm. The values shown were derived by subtracting the background fluorescence detected in parallel gradients from a wild-type control (normalized per microgram of protein). Each point is the mean from the analysis of two gradients.
ACA2p Expression Is Silenced in ACA2-GFPp Plants
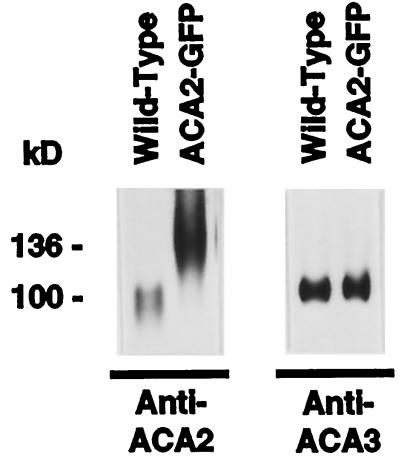
To more directly assess whether ACA2-GFPp targets the same membrane as ACA2p, we attempted to simultaneously detect the endogenous ACA2p in the same Suc gradients. Figure 6 shows that both pumps can be separated by SDS-PAGE and detected on a western blot by anti-ACA2 antibodies as bands of 136 and 110 kD, respectively. However, in both transgenic plant lines analyzed, the endogenous ACA2p was not detectable in any fraction, even after long overexposures (Fig. 6 shows a representative comparison). We estimated that expression of the endogenous ACA2p was suppressed more than 10-fold, based on western-blot analyses conducted with wild-type and two transgenic plant lines. We also examined, as a control, the expression levels of two other resident ER proteins, ACA3p and BiP. In contrast to ACA2p, normal expression levels were observed for ACA3p (Fig. 6) and BiP (not shown). These results indicate that ACA2p suppression was specific and not caused by a general suppression of ER resident proteins.
Figure 6.
Western-blot analysis showing suppression of endogenous ACA2p in transgenic plants expressing ACA2-GFPp. Suc gradients from two independent 35S-ACA2-GFP plants were simultaneously analyzed for endogenous and GFP-tagged ACA2p using anti-ACA2 or anti-GFP antibodies, respectively. Shown here is a representative comparison of two 10-μg protein samples taken from fraction 9 of the +Mg2+ gradients derived from wild-type and 35S-ACA2-GFP transgenic plants (see Figs. 2 and 5). Parallel samples were analyzed for the expression levels of ACA3p using anti-ACA3 antibodies. Proteins were separated by SDS-PAGE, blotted, and probed with anti-ACA2 (1:2000) or anti-ACA3 (1:2000).
ACA2-GFPp Images as a Fluorescent Reticulum
To visualize the subcellular structures containing ACA2-GFPp, we imaged GFP fluorescence in living cells using confocal microscopy and computational optical-sectioning microscopy. We used both approaches to observe similar fluorescent images. No detectable signals were observed from nontransgenic plants (data not shown). Two separate transgenic plant lines were analyzed with equivalent results. Fluorescent images were evaluated from hundreds of cells showing both high and low levels of expression. Even within a single plant root tip there was considerable cell-to-cell variation in expression levels. The source of this variability is not known but may be related to the cosuppression phenomena discussed above (Fig. 6). This variability allowed us to make a direct comparison of strong and weak fluorescent patterns in neighboring cells, and the comparison revealed similar reticulate structures. Therefore, the images shown appear to be representative of a normal subcellular localization pattern and not an artifact produced by the overexpression and mislocalization of a GFP-tagged fusion protein.
Confocal images (Fig. 7, C and D) are shown for cells near the root tip from plants expressing ACA2-GFPp or a control GFP only. The root tip is shown as a toluidine blue-stained section in Figure 7A for reference. We chose the root tip for detailed analysis because the endogenous ACA2p was highly expressed in this tissue, as indicated by immunocytology with an anti-ACA2 antibody (Fig. 1). Toluidine blue-stained root tip cells appear at higher magnification in Figure 7B to illustrate the central location of a large, densely stained nucleus. Comparable cell types in Figure 7, C and D, show the fluorescent images of a GFP-only control and ACA2-GFPp. In the GFP control, the strongest fluorescent signal corresponded to the central nuclear region. This apparent nuclear accumulation is consistent with previous reports of imaging GFP expression in plants (Haseloff et al., 1997). In contrast, the signal from ACA2-GFPp was excluded from the nuclear region and showed a network of connecting strands throughout the cell, most similar to patterns seen for an actin/ER network (Boevink et al., 1998).
We also examined images from cells in more mature regions of the root. Figure 7, E and F, shows representative fluorescence images from mature root epidermal cells expressing a GFP-only control or the ACA2-GFP fusion. As with the subcellular localization in undifferentiated cells, the GFP-only control appeared to accumulate in the nucleus, whereas ACA2-GFPp was excluded from the nucleoplasm and formed a network of connecting strands. This network differed from that in undifferentiated cells, showing more evidence of membrane sheets in addition to interconnecting membrane strands. In addition, the perinuclear region of many epidermal cells showed strong fluorescence, as illustrated by an image of two nuclei in Figure 7G.
DISCUSSION
A PM-Type Ca2+ Pump in the ER
Our results indicate that ACA2p is localized to the ER, which makes ACA2p distinct from all other calmodulin-regulated Ca2+ pumps identified in any organism. Two lines of evidence support this novel, subcellular localization. First, membrane fractionation by Suc gradients showed that ACA2p and the tagged isoform ACA2-GFPp cofractionated with two ER markers, BiP and a second, more typical ER-type Ca2+ pump, ACA3p/ECA1p (Figs. 2 and 5). Second, fluorescence microscopy of cells expressing ACA2-GFPp revealed an interconnecting network of sheets and strands, forming a reticular pattern characteristic of plant ER (Fig. 7). Although both lines of evidence corroborated an ER localization, we cannot exclude the possibility that lesser amounts of ACA2p were targeted to other locations, such as the Golgi. These results identified ACA2-like pumps as a source of calmodulin-regulated Ca2+-ATPase activity previously reported in plant ER membrane fractions (Gavin et al., 1993; Hwang et al., 1997; Evans and Williams, 1998).
It is not clear whether the appearance of ACA2-GFPp around some nuclei reflects a preferential targeting of the pump to the nuclear envelope or to multiple layers of ER that can sometimes wrap around the nucleus. Ca2+ pumps are expected to be associated with the perinuclear region to control Ca2+ levels in this subcellular location. Nuclear-localized Ca2+ fluxes have been observed in plant and animal cells. For example, Ehrhardt et al. (1996) observed an oscillating Ca2+ signal around nuclei in alfalfa root epidermal cells exposed to a nodulation factor. In both plant and animal cells there is evidence for a typical ER-type Ca2+ pump in the nuclear envelope (Lanini et al., 1992; Santella and Carafoli, 1997; Downie et al., 1998). However, results here prompt speculation that plant cells differ from animal cells and use an additional calmodulin-regulated Ca2+ pump in the perinuclear region.
Use of a GFP-Tagged ACA2p
Tagging proteins with a GFP provides a powerful approach for imaging subcellular locations for proteins. In this study, imaging a GFP-tagged ACA2p provided important corroboration for our membrane-fractionation analysis for two reasons. First, the GFP tag provided isoform-specific information. This was important because immunocytology and membrane-fractionation studies both relied on detecting the endogenous pump with a polyclonal antibody. Although this antibody appeared to be highly specific, we could not exclude the possibility that it also detected a second, more abundant cross-reacting isoform. Second, Suc gradient fractionation protocols do not cleanly separate all membrane systems and the diversity of membrane systems in plants is still undefined. Thus, without cytological verification, membrane-fractionation studies may provide misleading information.
On the other hand, an investigation based solely on GFP tagging also requires a cautious interpretation. A protein tagged with GFP may not target the correct location for many reasons; e.g. the GFP tag may disrupt the normal targeting signals of the subject protein or overexpression of a tagged protein may overload a targeting pathway and cause significant missorting of membranes; and/or ectopic expression of a tagged protein may show an artifactual localization in a cell type where the organelles, interacting proteins, or modifying enzymes required for proper targeting are not present. With these concerns in mind, in this study we imaged ACA2-GFPp under conditions in which the transgene was expressed at low to moderate levels; we focused on cell types that showed expression of the endogenous gene. We further demonstrated, in a membrane-fractionation analysis (Fig. 5), that the behavior of the tagged pump was like that of the endogenous pump. Therefore, we believe that our imaging of ACA2-GFPp accurately reflects the subcellular distribution of the endogenous ACA2p.
Regulation of Ca2+-Pump Expression
We observed that ACA2p expression was suppressed in both of the transgenic plant lines analyzed in the present localization study (Fig. 6). This silencing may have two nonexclusive explanations. First, the endogenous ACA2p may be silenced by “transgene-cosuppression” (Baulcombe and English, 1996; Meyer and Saedler, 1996), or second and alternatively, it may be down-regulated by a feedback system that maintains an appropriate level of Ca2+-ATPase activity.
Although we cannot distinguish between these two mechanisms of down-regulation, there is precedence for an integrated feedback system that coordinates the expression of both PM- and ER-type Ca2+ pumps in animal cells (Guerini et al., 1995; Kuo et al., 1997). For example, overexpression of a PM-type Ca2+-ATPase in mammalian tissue-cultured cells correlated with the down-regulation of the ER-type pump (Kuo et al., 1997). In contrast to that example, we failed to observe any interdependence between the overexpression of ACA2-GFPp and the expression levels for the ER-type pump ACA3p (e.g. ACA3p levels were not down-regulated in response to ACA2-GFP overexpression). This result supports the hypothesis that, even though both the ACA2p and the ACA3p are located in the ER, they provide functionally separate Ca2+-efflux pathways that are not coordinately regulated at the protein expression level.
ER Targeting Signals
Exactly how P-type ATPases are targeted to different membrane systems in plants or animals is not known. A working hypothesis for ER localization of ACA3p (i.e. the more typical ER-type pump) is that a Lys-rich sequence (KQKEE) at the C-terminal end provides an ER-retention motif (Jackson et al., 1990; Townsley and Pelham, 1994). In animal cells many resident ER membrane proteins contain a C-terminal retention motif with the consensus (K/X)(K/X)KXX-stop (Jackson et al., 1993). Because the sequence KKXX can function as an ER retention signal in yeast (Townsley and Pelham, 1994), this retention motif appears to be conserved in distantly related phyla.
However, not all ER resident membrane proteins are localized by a common mechanism. A notable exception is an ER-type Ca2+ pump from chicken (SERCa1p), which does not contain a consensus C-terminal retention motif (its C-terminal sequence is DERRK; Karin and Settle, 1992). Instead, a targeting analysis of chimeras, made between animal ER-type- and PM-type Ca2+-pumps, suggests that the structure or sequence of the first transmembrane domain provides an ER-retention signal (Foletti et al., 1995). It is even less clear how ACA2p could be localized to the ER and how it could accumulate to high levels around the nucleus. A consensus ER-retention motif was not present in the C-terminal domain of ACA2p, and the sequence of the first transmembrane domain was not well conserved with the animal ER-type Ca2+-ATPase pumps.
In animals, calmodulin-regulated pumps are thought to be exclusively localized to the PM. By contrast, the identification of ACA2p in the plant ER and the proposed plastid and tonoplast locations of ACA1p and BCA1p, respectively, suggest a fundamental difference between the ways that plant and animal cells use calmodulin-regulated Ca2+ pumps.
Why Are There Two Ca2+ Pumps in the ER?
Our results indicate that both ACA3p and ACA2p cofractionate with ER isolated from Arabidopsis roots. It is likely that at least some cells in the root contain both pumps, because both ACA2p and ACA3p were predominantly expressed in the root, and immunocytology of the root tip suggested that ACA2p was expressed in every cell type. However, we have not investigated the issue of whether the colocalization of both types of Ca2+ pumps in the ER is a common feature of all plant cells.
We offer three hypotheses for considering the potential significance of two different Ca2+ pumps in the ER. First, the two pumps may be spatially separated into different functional domains. Separate domains of the ER have been reported in animal cells, each maintaining a distinct pool of Ca2+ (Golovina and Blaustein, 1997). Second, each pump may provide a unique enzymatic activity, perhaps the ability to pump a secondary cation such as Mn2+ (Liang et al., 1997). Third, the two pumps may act differently to control cytoplasmic/ER-Ca2+ levels in response to different stimuli. “Differential activation” is an attractive hypothesis because calmodulin directly activates ACA2p but not ACA3p. In any event, the observation that a plant ER contains both type IIA and IIB pumps highlights the complexity of Ca2+ transport into this organelle.
ACKNOWLEDGMENTS
The authors thank Catharine Conley for helpful discussions, Woo Sik Chung for discussions and assistance with subclonings, Maarten Chrispeels for providing anti-BiP and anti-γ-TIP antibodies, and Malcom Wood for assistance with confocal imaging. We thank Mrs. Frederick Garry and Mr. and Mrs. Kenneth E. Hill for general support to The Scripps Research Institute for operation of the laboratory for J.F.H.
Abbreviations:
- BiP
a homolog of the ER-resident immunoglobulin heavy chain-binding protein
- CaMV
cauliflower mosaic virus
- GFP
green fluorescence protein
- GST
glutathione S-transferase
- PM
plasma membrane
Footnotes
This research was supported by a U.S. Department of Energy grant (no. DE-FG03-94ER20152) to J.F.H. and a joint grant (no. IBN-9416038) from the National Aeronautics and Space Administration and the National Science Foundation for the Plant Sensory Systems Collaborative Research Network. Support for computational microscopy was provided by a National Institutes of Health grant (no. RR01380) to the Institute for Biomedical Computing at Washington University and to J.S.G. through a Monsanto predoctoral fellowship in plant biology.
LITERATURE CITED
- Askerlund P. Modulation of an intracellular calmodulin-stimulated Ca2+-pumping ATPase in cauliflower by trypsin. The use of calcium green-5N to measure Ca2+ transport in membrane vesicles. Plant Physiol. 1996;110:913–922. doi: 10.1104/pp.110.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. Mol Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- Baulcombe DC, English JJ. Ectopic pairing of homologous DNA and posttranscriptional gene silencing in transgenic plants. Curr Opin Biotechnol. 1996;7:173–180. [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci (Paris) 1993;316:1194–1199. [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;15:441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- Bootman MD, Berridge MJ. The elemental principles of calcium signaling. Cell. 1995;83:675–678. doi: 10.1016/0092-8674(95)90179-5. [DOI] [PubMed] [Google Scholar]
- Bush DS. Calcium regulation in plant cells and its role in signaling. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:95–122. [Google Scholar]
- Chen X, Chang M, Wang B, Wu B. Cloning of a Ca2+-ATPase gene and the role of cytosolic Ca2+ in the gibberellin-dependent signaling pathway in aleurone cells. Plant J. 1997;11:363–371. doi: 10.1046/j.1365-313x.1997.11030363.x. [DOI] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Clapham DE. Calcium signaling. Cell. 1995;80:259–268. doi: 10.1016/0092-8674(95)90408-5. [DOI] [PubMed] [Google Scholar]
- Cunningham KW, Fink GR. J Exp Biol. 1994;196:157–166. doi: 10.1242/jeb.196.1.157. [DOI] [PubMed] [Google Scholar]
- Denecke J, Goldman MH, Demolder J, Seurinck J, Botterman J. The tobacco luminal binding protein is encoded by a multigene family. Plant Cell. 1991;3:1025–1035. doi: 10.1105/tpc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt ND, Hong B, Sussman MR, Harper JF. Targeting of two Arabidopsis H+-ATPase isoforms to the plasma membrane. Plant Physiol. 1996;112:833–844. doi: 10.1104/pp.112.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie L, Priddle J, Hawes C, Evans DE. A calcium pump at the higher plant nuclear envelope? FEBS Lett. 1998;429:44–48. doi: 10.1016/s0014-5793(98)00564-x. [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Evans DE, Williams LE. P-type calcium ATPases in higher plants: biochemical, molecular and functional properties. Biochim Biophys Acta. 1998;1376:1–25. doi: 10.1016/s0304-4157(97)00009-9. [DOI] [PubMed] [Google Scholar]
- Ferrol N, Bennett AB. A single gene may encode differentially localized Ca2+-ATPases in tomato. Plant Cell. 1996;8:1159–1169. doi: 10.1105/tpc.8.7.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foletti D, Guerini D, Carafoli E. Subcellular targeting of the endoplasmic reticulum and plasma membrane Ca2+ pumps: a study using recombinant chimeras. FASEB J. 1995;9:670–680. doi: 10.1096/fasebj.9.8.7768360. [DOI] [PubMed] [Google Scholar]
- Frisch DA, Harris-Haller LW, Yokubaitis NT, Thomas TL, Hardin SH, Hall TC. Complete sequence of the binary vector bin 19. Plant Mol Biol. 1995;27:405–409. doi: 10.1007/BF00020193. [DOI] [PubMed] [Google Scholar]
- Gavin O, Pilet PE, Chanson A. Tonoplast localization of a calmodulin-stimulated Ca2+-pump from maize roots. Plant Sci. 1993;92:143–150. [Google Scholar]
- Gens JS, Reuzeau C, Doolittle KW, McNally JG, Pickard BG. Covisualization by computational optical-sectioning microscopy of integrin and associated proteins at the cell membrane of living onion protoplasts. Protoplasma. 1996;194:215–230. doi: 10.1007/BF01882029. [DOI] [PubMed] [Google Scholar]
- Gill DL, Waldron RT, Rys-Sikora KE, Ufret-Vincenty CA, Graber MN, Favre CJ, Alfonso A. Calcium pools, calcium entry, and cell growth. Biosci Rep. 1996;16:139–157. doi: 10.1007/BF01206203. [DOI] [PubMed] [Google Scholar]
- Golovina VA, Blaustein MP. Spatially and functionally distinct Ca2+ stores in sarcoplasmic and endoplasmic reticulum. Science. 1997;275:1643–1648. doi: 10.1126/science.275.5306.1643. [DOI] [PubMed] [Google Scholar]
- Guerini D, Schroder S, Foletti D, Carafoli E. Isolation and characterization of a stable Chinese hamster ovary cell line overexpressing the plasma membrane Ca2+-ATPase. J Biol Chem. 1995;270:14. doi: 10.1074/jbc.270.24.14643. ,643–14,650. [DOI] [PubMed] [Google Scholar]
- Haas IG. BiP (GRP78), an essential hsp70 resident protein in the endoplasmic reticulum. Experientia. 1994;50:1012–1020. doi: 10.1007/BF01923455. [DOI] [PubMed] [Google Scholar]
- Harper JF, Hong B, Hwang I, Guo HQ, Stoddard R, Huang JF, Palmgren MG, Sze H. A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain. J Biol Chem. 1998;273:1099–1106. doi: 10.1074/jbc.273.2.1099. [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi KD, Zhen RG, Cunningham KW, Rea PA, Fink GR. CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc Natl Acad Sci USA. 1996;93:8782–8786. doi: 10.1073/pnas.93.16.8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofte H, Chrispeels MJ. Protein sorting to the vacuolar membrane. Plant Cell. 1992;4:995–1004. doi: 10.1105/tpc.4.8.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Berkelman T, Franklin AE, Hoffman NE. Characterization of a gene encoding a Ca2+-ATPase-like protein in the plastid envelope. Proc Natl Acad Sci USA. 1993;90:10. doi: 10.1073/pnas.90.21.10066. ,066–10,070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Ratterman DM, Sze H. Distinction between endoplasmic reticulum-type and plasma membrane-type Ca2+ pumps. Plant Physiol. 1997;113:535–548. doi: 10.1104/pp.113.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. Retrieval of transmembrane proteins to the endoplasmic reticulum. J Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Hofte H, Chrispeels MJ. An intrinsic tonoplast protein of protein storage vacuoles in seeds is structurally related to a bacterial solute transporter (GIpF) Plant Cell. 1990;2:525–532. doi: 10.1105/tpc.2.6.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin NJ, Settle VJ. The sarcoplasmic reticulum Ca2+-ATPase, SERCA1a, contains endoplasmic reticulum targeting information. Biochem Biophys Res Commun. 1992;186:219–227. doi: 10.1016/s0006-291x(05)80796-x. [DOI] [PubMed] [Google Scholar]
- Kuo TH, Liu BF, Yu Y, Wuytack F, Raeymaekers L, Tsang W. Co-ordinated regulation of the plasma membrane calcium pump and the sarco(endo)plasmic reticular calcium pump gene expression by Ca2+ Cell Calcium. 1997;21:399–408. doi: 10.1016/s0143-4160(97)90051-8. [DOI] [PubMed] [Google Scholar]
- Lanini L, Bachs O, Carafoli E. The calcium pump of the liver nuclear membrane is identical to that of endoplasmic reticulum. J Biol Chem. 1992;267:11. ,548–11,552. [PubMed] [Google Scholar]
- Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- Liang F, Cunningham KW, Harper JF, Sze H. ECA1 complements yeast mutants defective in Ca2+ pumps and encodes an endoplasmic reticulum-type Ca2+-ATPase in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:8579–8584. doi: 10.1073/pnas.94.16.8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Malmstrom S, Askerlund P, Palmgren MG. A calmodulin-stimulated Ca2+-ATPase from plant vacuolar membranes with a putative regulatory domain at its N-terminus. FEBS Lett. 1997;400:324–328. doi: 10.1016/s0014-5793(96)01448-2. [DOI] [PubMed] [Google Scholar]
- McClean IW, Nakane PK. Periodate-lysine-paraformaldehyde fixative: a new fixative of immunoelectron microscopy. J Histochem Cytochem. 1974;22:1077. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- Meyer P, Saedler H. Homology-dependent gene silencing in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:23–48. doi: 10.1146/annurev.arplant.47.1.23. [DOI] [PubMed] [Google Scholar]
- Nagahashi J, Kane AP. Triton-stimulated nucleoside diphosphatase activity: subcellular localization in corn root homogenates. Protoplasma. 1982;112:167–173. [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rudolph HK, Antebi A, Fink GR, Buckley CM, Dorman TE, LeVitre J, Davidow LS, Mao JI, Moir DT. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+-ATPase family. Cell. 1989;58:133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- Santella L, Carafoli E. Calcium signaling in the cell nucleus. FASEB J. 1997;11:1091–1109. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Serrano R. Purification of the proton pumping ATPase from plant plasma membranes. Biochem Biophys Res Commun. 1984;121:735–740. doi: 10.1016/0006-291x(84)90243-2. [DOI] [PubMed] [Google Scholar]
- Sheen J. Ca2+-dependent protein kinases and stress signal transduction in plants. Science. 1996;274:1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- Topfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH. A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 1987;15:5890. doi: 10.1093/nar/15.14.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley FM, Pelham HR. The KKXX signal mediates retrieval of membrane proteins from the Golgi to the ER in yeast. Eur J Cell Biol. 1994;64:211–216. [PubMed] [Google Scholar]
- Wimmers LE, Ewing NN, Bennett AB. Higher plant Ca2+-ATPase: primary structure and regulation of mRNA abundance by salt. Proc Natl Acad Sci USA. 1992;89:9205–9209. doi: 10.1073/pnas.89.19.9205. [DOI] [PMC free article] [PubMed] [Google Scholar]