Summary
Endoplasmic reticulum (ER) stress triggers tissue-specific responses that culminate in either cellular adaptation or apoptosis, but the genetic networks distinguishing these responses are not well understood. Here we demonstrate that ER stress induced in the developing zebrafish causes rapid apoptosis in the brain, spinal cord, tail epidermis, lens and epiphysis. Focusing on the tail epidermis, we uncover an apoptotic response that depends on Puma, but not on p53 or Chop. Puma is transcriptionally activated during this ER stress response in a p53-independent manner, and is an essential mediator of epidermal apoptosis. We demonstrate that the p63 transcription factor is upregulated to initiate this apoptotic pathway and directly activates puma transcription in response to ER stress. We also show that a mutation of human Connexin 31, which causes erythrokeratoderma variabilis, induces ER stress and p63-dependent epidermal apoptosis in the zebrafish embryo, thus implicating this pathway in the pathogenesis of inherited disease.
Keywords: ER stress, zebrafish, apoptosis, p63, puma, chop, erythrokeratodermia variabilis
Introduction
In all eukaryotic cells, the endoplasmic reticulum (ER) serves the critical cellular functions of protein translation, calcium storage, and folding and processing of membrane and secreted proteins (Rao et al., 2004). When the load of proteins to be folded in the ER exceeds the capacity of cellular chaperones to aid in the folding process, a response known as the ER stress pathway, or unfolded protein response (UPR; reviewed in Malhotra and Kaufman, 2007; Ron and Walter, 2007), is triggered. Shortly after the onset of ER stress, cells activate three proximal pathways, mediated by IRE1, ATF6, and PERK, to upregulate the transcription and translation of ER chaperones, amino acid biosynthesis enzymes, and the ERAD (endoplasmic reticulum-associated degradation) proteins. Successful engagement of these pathways can lower the unfolded protein load and return the cell to homeostasis. However, severe or prolonged stress can lead to an apoptotic response (Fribley et al., 2009), and the cellular outcome is central to a variety of human diseases including cancer, inherited skin disorders, diabetes, and neurodegenerative disorders (Lin et al., 2008 and Tatersall et al, 2009). The tissue-specific decisions responsible for apoptotic outcomes in diseased and normal cells are still poorly understood, warranting further research into how distinct vertebrate tissues integrate molecular signaling downstream of the proximal ER stress pathways to determine whether cells initiate programmed cell death or overcome the stress response and survive.
While the decisive events that induce cellular apoptosis after ER stress are not fully understood, the signaling pathways that sense and respond to ER stress have been extensively studied. Seminal work to elucidate these proximal ER stress pathways in yeast led to the discovery that the trans-ER membrane-bound IRE1 (inositol requiring enzyme-1) protein contains both kinase and endoribonuclease activities, which are triggered after stress in the ER (Cox et al., 1993; Mori et al., 1993). When the load of unfolded proteins in the ER lumen becomes excessive, the protein-folding chaperone BiP (Hspa5) detaches from IRE1 (Bertolotti et al., 2000; Okamura et al., 2000), resulting in trans-autophosphorylation of the IRE1 homodimer, which activates the endoribonuclease activity of IRE1 and the subsequent unconventional splicing of HAC1 mRNA (Sidrauski and Walter, 1997). This unconventional splicing alters the reading frame for translation of the HAC1 protein, resulting in the synthesis of a highly active transcription factor that can activate a wide array of downstream target genes, including those encoding protein chaperones and others encoding the ERAD components (Kopito, 1997; Travers et al., 2000). This work established the IRE1-HAC1 pathway as the most ancient and most conserved proximal ER stress pathway in eukaryotes.
Two additional ER stress pathways have been delineated in multicellular organisms (Ron and Walter, 2007). The ATF6 protein, a trans-ER membrane molecule, is directed to the Golgi apparatus upon activation via BiP uncoupling, where it undergoes cleavage by Site-1 and Site-2 proteases to become an active transcription factor (Haze et al., 1999). Like XBP1 (the target of mammalian IRE1), ATF6 can activate the transcription of a myriad of recovery genes to allow the stressed cell to repair its ER burden and survive. It also induces the transcription of XBP1, providing a cross-talk mechanism between the ATF6-and IRE1-mediated ER stress pathways (Yoshida et al., 2001). The third ER stress pathway directly triggers global translational attenuation to lessen the rate of unfolded proteins accumulating in the ER. When released by BiP, the trans-ER protein PKR-like endoplasmic reticulum kinase (PERK), directly phosphorylates eukaryotic translation initiation factor 2α (eIF2α) (Harding et al., 1999). This phosphorylation on serine 51 greatly diminishes the ability of eIF2α to initiate the cap-dependent translation of cellular proteins, thus reducing the overall amount of proteins in the ER. Remarkably, however, mRNA encoding the transcription factor ATF4 is preferentially translated due to the presence of small upstream ORFs (uORFs), resulting in a paradoxical upregulation of translation of this gene in response to phosphorylation of eIF2α (Harding et al., 2000). ATF4 then enters the nucleus as an active transcription factor that initiates the transcription of downstream target genes, including the pro-apoptotic transcription factor Chop. While no Chop homologue exists in worms, mammalian Chop contributes to some forms of ER stress-induced apoptosis and can kill cells when overexpressed (Matsumoto et al., 1996; Maytin et al., 2001; Zinszner et al., 1998).
Although work in yeast, worms, mice, and human cell culture has provided valuable insights into the proximal mechanisms that cells and tissues use to cope with ER stress, many of the downstream molecular events that determine stress-induced outcomes remain elusive. Here, we take advantage of the rapid chemical uptake properties of the zebrafish embryo to analyze the timing and spatial activation of ER stress, together with its apoptotic consequences, after the whole-animal application of thapsigargin or brefeldin A, two well-characterized ER stress-inducing compounds. In developing embryos treated with either drug, we demonstrate that cells within the lens, epiphysis, and tail epidermis undergo ER stress-induced apoptosis within 4 hours in a p53-independent manner. Further, we show that this rapid apoptotic response requires the BH3-only gene puma. Using morpholino knockdown, we demonstrate that the transcription factor p63 is required for puma expression, and further that p63 directly binds the puma promoter in vivo following an increase in p63 levels. Finally, we demonstrate that p63 mediates an in vivo apoptotic response to mutant Connexin 31, a protein produced as a result of a dominantly inherited mutation in humans that induces an ER stress response in the epidermis of the skin, contributing to the disease erythrokeratoderma variabilis (EKV).
Results
ER stressors induce p53-independent apoptosis in the brain, spinal cord, lens, epiphysis, and tail epidermis of early zebrafish embryos
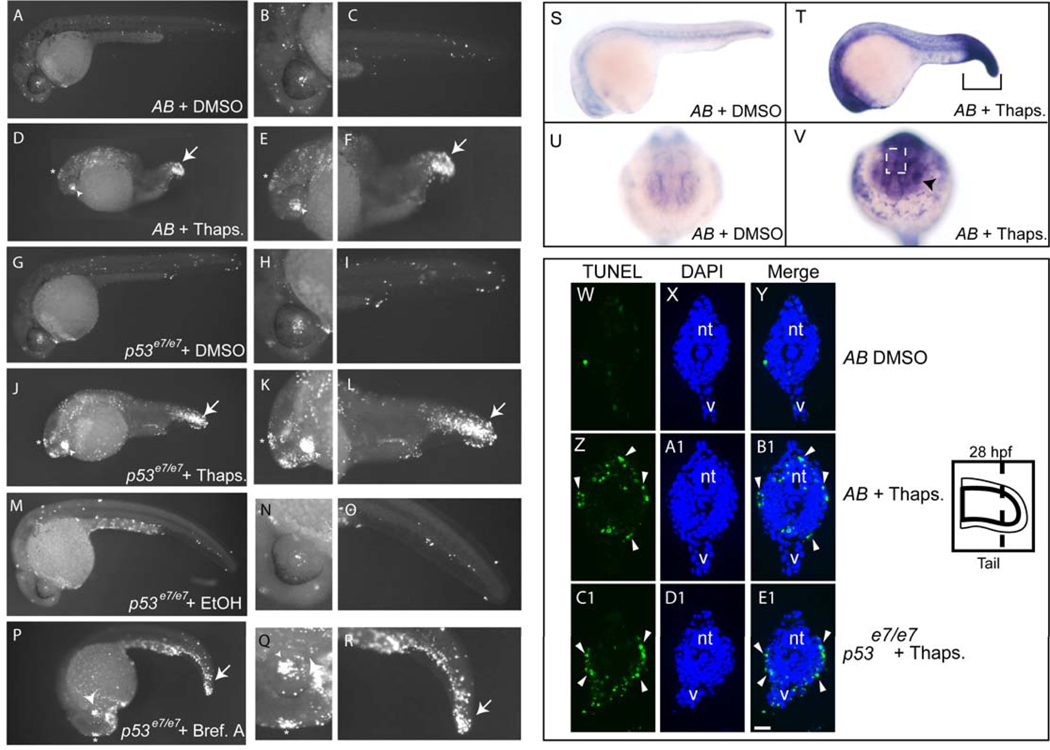
To test whether zebrafish undergo ER stress-induced apoptosis in vivo, we first treated 24-hour post-fertilization (hpf) embryos with thapsigargin, a potent ER calcium pump inhibitor that induces ER stress in mammalian cells (Pahl and Baeuerle, 1995). This treatment interval was chosen because most major developmental patterning is complete at this stage, allowing the assessment of tissue-specific cell death (Pyati et al., 2007). After 4 hours of continuous treatment with thapsigargin, we observed focal increases in apoptotic cell death as assayed by whole-mount TUNEL staining (Figure 1A–F). Increased TUNEL positivity was evident in the brain, spinal cord, lenses of the eyes, epiphysis (which ultimately forms the pineal gland), and tail epidermis (Figure 1). TUNEL staining was most pronounced in the latter three tissues, with less positivity in the brain and spinal cord. These data show that specific tissues within developing embryos are sensitive to thapsigargin-induced ER stress and rapidly undergo apoptosis.
Figure 1. ER stressors trigger bip upregulation and apoptosis in the lens, epiphysis, and tail epidermis of 24 hpf zebrafish embryos.
24-hpf embryos from an AB incross or a stage-matched p53 homozygous mutant incross were treated with a DMSO vehicle or 5 µM thapsigargin for 4 hrs, fixed, and assayed for apoptosis by TUNEL labeling. (A–C) Scattered cells in AB DMSO-treated control embryos (n=29) showed TUNEL positivity. (D–F) Thapsigargin-treated AB embryos had increased TUNEL positivity that was concentrated in the epiphysis, lens, and tail (93% of embryos with increased apoptosis, n=30). (G–I) p53 homozygous mutant embryos treated with DMSO (n=31) showed comparable levels of TUNEL positivity to AB DMSO-treated embryos. (J–L) p53 homozygous mutant embryos treated with thapsigargin showed increased apoptosis in the same regions as in AB embryos (100% of embryos with increased apoptosis, n=31). Like thapsigargin treatment, Brefeldin A treatment (5 ug/ml) caused increased apoptosis in the lens, epiphysis, and tail (P–R) compared to vehicle-treated controls (M–O) (See also Supplemental Figure 1). Compared to DMSO-treated controls (S,U), bip expression was upregulated in embryos treated for 3 hours with thapsigargin (T, V). In cryosections, DMSO controls (W–Y) had minimal apoptosis, while thapsigargin treatment of both AB embryos (Z-B1) and p53 mutant embryos (C1–E1) caused extensive enriched apoptosis in the tail epidermis (white arrowheads). In A–R, Zoomed images of head and tail are shown at the right of corresponding whole-embryo images. Asterisks indicate epiphysis, arrowheads the lens, and arrows the tail in thapsigargin-treated embryos.
In mammals, ER stress-induced apoptosis can be triggered in either a p53-dependent or -independent manner, depending on the context (Galehdar et al.; Puthalakath et al., 2007; Reimertz et al., 2003). To test whether p53 is required for ER stress-induced apoptosis in the zebrafish, we treated embryos that carry a homozygous inactivating mutation within the DNA-binding domain of p53 (p53 e7/e7; Berghmans et al., 2005; Sidi et al., 2008) with thapsigargin. As in wild-type embryos, we observed a marked increase in apoptotic cells by TUNEL staining (Fig. 1G–L) in p53 mutants. To confirm that the observed tissue-specific apoptosis was due to ER stress, we treated embryos with brefeldin A, which disrupts ER to Golgi trafficking of membrane and secreted proteins (Samali et al., 2010). Similar to thapsigargin, brefeldin A triggered a p53-independent apoptotic response that was most prominent in the lens, epiphysis, and tail epidermis after 4 hours of treatment (Fig. 1M–R). Since thapsigargin treatment caused a more robust cell death response than did brefeldin A after 4 hours (compare Figure 1 J–L and P–R), we chose to use thapsigargin for most subsequent experiments, reserving brefeldin A to confirm key results. Taken together, these data show that ER stress-inducing drugs trigger apoptosis in zebrafish embryos independently of p53.
To determine the spatial extent of ER stress induced by thapsigargin, we examined expression of the ER stress recovery gene bip by whole-mount in situ hybridization (Fig. 1S–V). Similar to reports for mammalian cells, we observed a robust post-treatment increase of bip that was especially prominent in the tail epidermis, lens, and epiphysis, where we had observed apoptosis in our whole-mount TUNEL assays. Interestingly, we also observed increased bip expression in many parts of the head as well as in the hatching gland, where we saw lower levels of apoptosis. Similar staining was observed following brefeldin A treatment, confirming the selective sensitivity of distinct embryonic tissues to ER stressors (Supp. Fig. 1). Since the most robust apoptotic response to ER stress occurred in the tail epidermis (Fig. 1 F, L, R), we cryosectioned thapsigargin-treated embryos followed by TUNEL staining, to determine the precise tissue layer(s) that underwent cell death in this region. As shown in Figure 1 W-E1, the primary location of apoptotic cells in the tails of both AB and p53 mutant embryos was the epidermal cell layer (marked by arrowheads). Thus, ER stress triggers rapid apoptosis in the developing tail that is primarily localized to the epidermis, which later becomes the skin of the zebrafish.
ER stress activates proximal UPR pathways within 1 hour in zebrafish embryos
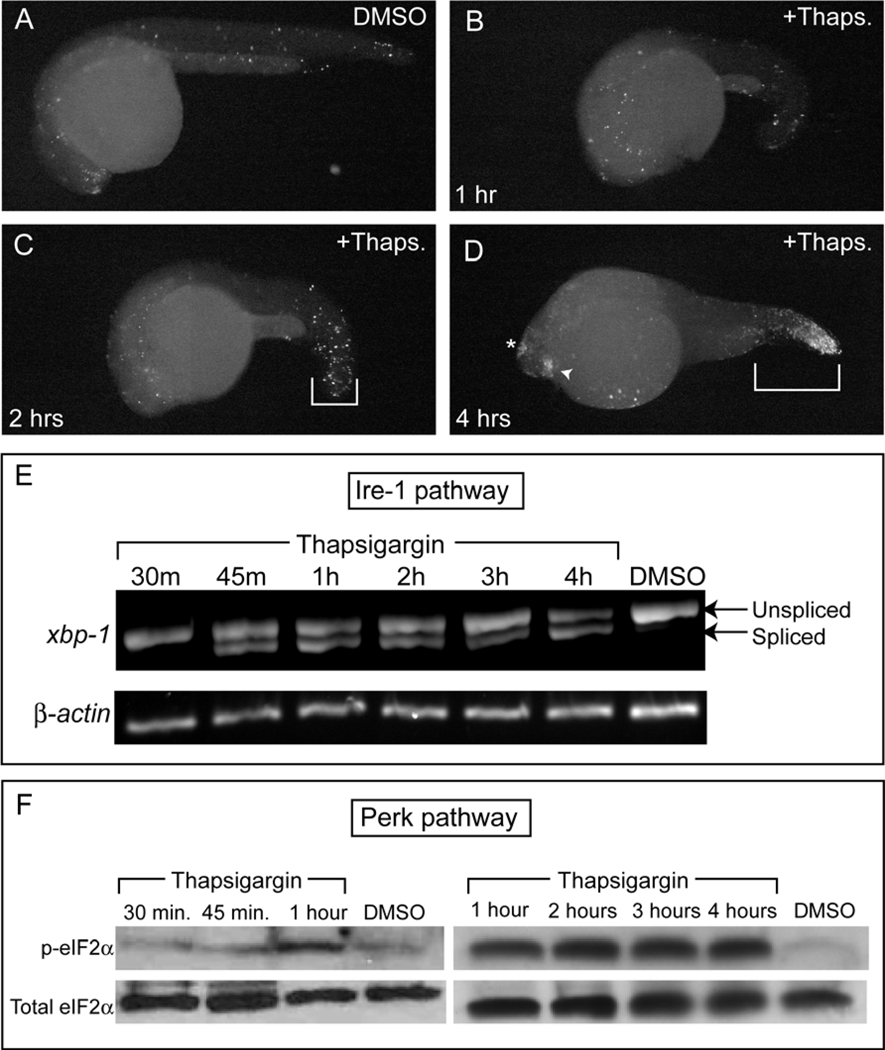
To asses the kinetics of ER stress-induced apoptosis in zebrafish embryos, we examined thapsigargin-treated embryos by TUNEL staining every hour for 4 hours. By 2 hours, we observed an increase in cell death, mainly in the tail, followed by a gradual increase in dying cells within the tail, lens, and epiphysis, until peak levels were reached at 4 hours post-treatment (Fig. 2A–D).
Figure 2. Thapsigargin treatment rapidly activates conserved vertebrate ER stress pathways to induce apoptosis.
TUNEL staining was performed in fixed embryos (A) after 4 hrs of DMSO treatment or after (B) 1 h, (C) 2 h, or (D) 4 h of thapsigargin treatment. (C) Increased apoptosis was first apparent in the tail by 2 h of thapisgargin treatment, but robust apoptosis in the lens, epiphysis, and tail was evident only after (D) 4 h of thapsigargin treatment compared to DMSO controls at the same stage. All images are representative of embryos examined at each timepoint (n=10 in all treatments except D, where n=9). (E) By RT-PCR, xbp-1 splicing in the Ire-1 pathway was first apparent after 45 mins of thapsigargin treatment, and was maintained over 4 hrs. Note the lower band present from 45 mins on, which corresponds to the non-conventional splice-form downstream of Ire-1 activation. (F) Anti-phospho eIF2α immunoblots showed that eIF2α was robustly phosphorylated in the Perk pathway from 1 hr through the end of the thapsigargin time-course. Total eIF2α was immunoblotted as a loading control after stripping of the original blot.
In mammalian cells, ER stressors activate proximal ER stress pathways through three major trans-ER membrane proteins: IRE1, ATF6, and PERK (Malhotra and Kaufman, 2007; Ron and Walter, 2007). In the zebrafish embryo, we interrogated the kinetics of the Ire-1 and Perk pathways, measuring over time the endogenous ER-membrane associated “unconventional” splicing of xbp-1 (as a direct readout of IRE1 activation; Calfon et al., 2002) and the phosphorylation of eIF2α (as a direct readout of PERK activation; Harding et al., 1999), using cell lysates prepared from pooled thapsigargin-treated embryos. Analysis of xbp-1 splicing indicated that the Ire-1 pathway was activated after only 45 minutes of thapsigargin treatment (Fig. 2E) and was sustained over 4 hours. Measurements of endogenous levels of phosphorylated eIF2α indicated that Perk was activated after 1 hour of treatment and was continuously activated for 4 hours (Fig. 2F). Thus, thapsigargin-treated zebrafish embryos rapidly activate proximal ER stress pathways, as indicated by the prompt induction of xbp-1 splicing by Ire-1 at 45 minutes post- treatment and by phosphorylation of eIF2α after an additional 15 minutes.
Chop is dispensable for the rapid ER stress-induced apoptotic pathway but is essential for late ER stress-induced apoptosis
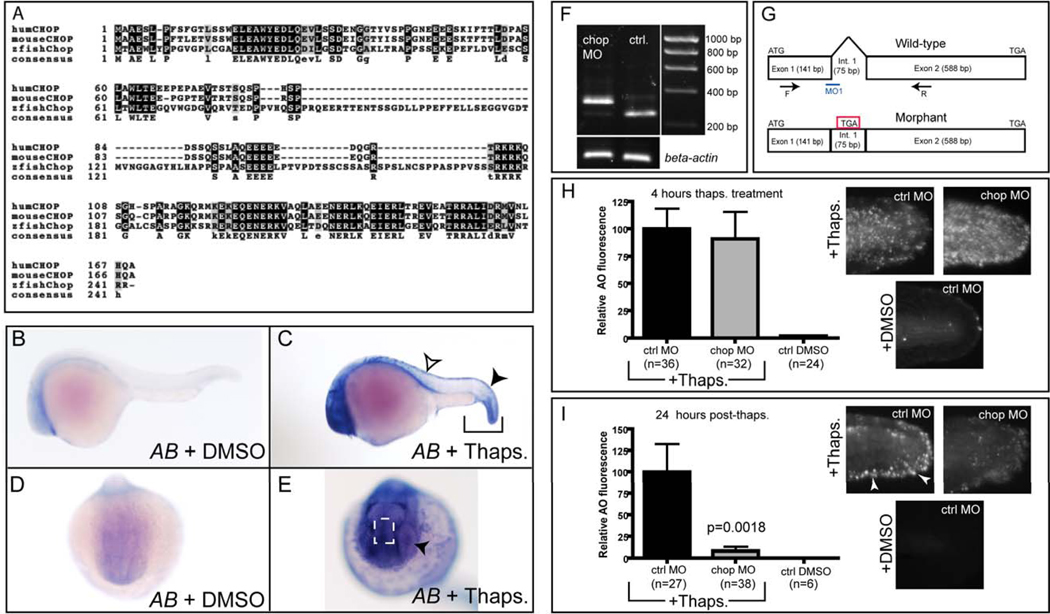
In mammalian systems, the pro-apoptotic transcription factor Chop is necessary to elicit peak levels of cell death following ER stress induction in some contexts (Zinszner et al., 1998). We thus sought to determine whether the zebrafish genome harbors a chop orthologue and whether Chop plays a critical pro-apoptotic role in the zebrafish epidermis as in some mammalian tissues. By performing a BLAST search in the Ensembl genome server using the human CHOP sequence, we identified a single highly similar sequence located on chromosome 5 of the zebrafish genome. This putative orthologue showed a close syntenic relationship to mouse and human CHOP, with coding regions for similar genes in direct proximity (www.ensembl.org). Based on our cloned and verified cDNA fragment for the full-length to Chop coding region, the predicted zebrafish Chop protein, though divergent in length from human and mouse isoforms, shares particularly high sequence identity in the C-terminal DNA-binding domain (brackets in Fig. 3A). Thus, we conclude that the zebrafish genome contains a single zebrafish chop orthologue that encodes a protein with a highly conserved DNA binding-domain sequence.
Figure 3. Zebrafish Chop is dispensable for rapid ER stress-induced apoptosis, but is required for 24 hr apoptosis in the caudal fin fold.
(A) Alignment of zebrafish Chop with human and mouse orthologues. Consensus sequence is listed below the aligned sequences. (B, D) DMSO-treated embryos have minimal chop expression, while embryos treated with thapsigargin for 3 hours (C) have high levels of chop expression in the tail (brackets), including the tail epidermis (black arrowhead). Expression was also observed in the dorsal trunk epidermis (open arrowhead in C). (E) An anterior view reveals additional chop expression in the epiphysis region (dashed white box) and the lens (black arrowhead). Images in B–E represent 100% of embryos examined; n≥14 embryos per condition. (F) 1% agarose gel showing alternate splice product generated 28 hours after chop morpholino (MO) injection compared to uninjected controls (Ctrl.); both groups of embryos were treated with thapsigargin at 24 hpf. (G) Depiction of splice-blocking event in chop morphant mRNA compared to wild-type, as determined by sequencing. (H) chop morpholino (5 ng) was injected into p53 homozygous mutant embryos, which were then grown to 24 hpf, treated with thapsigargin for 4 hrs, and assayed for cell death by AO staining. Cropped photos were quantitated for total fluorescence intensity with Volocity software, and values were normalized to controls. (I) chop morpholino-injected embryos were grown to 24 hpf, treated with thapsigargin for 4 hrs, washed into egg medium, left for 24 hrs, and then stained with AO and assayed as in (H). All data are represented as means +/− SEM.
Next, we wished to determine whether zebrafish chop, like human CHOP, is transcriptionally activated following ER stress. In 24-hpf embryos treated with thapsigargin for 3 hours, we observed robust induction of this pro-apoptotic factor at the end of treatment (Fig. 3 B–E). Similar to bip (Fig. 1), the expression of chop was tissue-restricted and enriched in the tail (brackets in Fig. 3C), lens (black arrowhead in Fig. 3E) and epiphysis region (box in Fig. 3E). Chop expression was also induced in the dorsal epidermis (white arrowhead in Fig. 3C), where we had observed minimal apoptosis in our TUNEL assays (Fig. 1). Importantly, brefeldin A triggered chop expression in similar tissues and with similar kinetics (Supp. Fig. 1), confirming that zebrafish chop is generally ER stress-responsive and not solely responsive to thapsigargin.
To test whether zebrafish Chop is required in vivo for the ER stress-induced apoptotic pathway in tail epidermis, we injected a chop-specific splice-blocking morpholino (Robu et al., 2007) into p53 homozygous mutant embryos (to eliminate any nonspecific morpholino toxicity that could confound our results (Figure 3 F , G). Next, to quantify the level of apoptosis induction, we treated morpholino-injected embryos with thapsigargin for 4 hours starting at 24 hpf and performed AO staining in live embryos to mark dead and dying cells. Remarkably, despite nearly complete knockdown of Chop (Figure 3F), we still observed marked ER stress-induced cell death after 4 hours of thapsigargin treatment (Figure 3H), indicating that this apoptotic pathway was rapidly triggered independently of both Chop and p53.
Multiple studies in mammalian cell culture have shown that Chop deficiency attenuates ER stress-induced apoptosis over a prolonged period (Puthalakath et al., 2007; Zinszner et al., 1998). Thus, we asked whether a delayed ER stress-induced cell death response occurs that is distinct from the rapid apoptotic response shown in Fig. 1, and whether it depends on Chop. We injected either a control morpholino or the chop morpholino into p53 mutant embryos, treated 24 hpf embryos with thapsigargin for 4 hours, and then washed out the drug and assayed for cell death by AO staining 24 hours later. Since AO staining provides a transient image of dying cells during development (Abrams et al., 1993), we reasoned that this vital dye would mark only cells that were dying or had died within 24 hours post- treatment. Indeed, control morphants displayed cell death at 24 hours post- treatment that was distinct from the cell death we observed in the 4-hour apoptotic assay (Figure 3I). This cell death was enriched in the caudal fin fold (white arrowheads in Fig. 3I), in cells that normally develop into the tail fins (Webb and Kimelman, 2005), with significantly decreased cell death in the lateral region of the tail. In contrast to control morpholino-injected embryos, chop morpholino-injected embryos showed very few dying cells in the tail at this stage (Figure 3I). These data are consistent with the expression pattern of chop in the caudal fin fold epidermis following ER stress (black arrowhead in Fig. 3C), suggesting that developing caudal fin cells die at 24 hours post- treatment in a Chop-dependent manner that does not require p53. Importantly, we could block all ER stress-induced cell death by overexpression of mRNA encoding Bcl-2 (Supp. Fig. 2), confirming that intrinsic mitochondrial apoptosis is induced by ER stress in this system. We conclude that zebrafish embryos activate both a rapid Chop-independent apoptotic pathway within 4 hours of ER stress induction, and a separate Chop-dependent apoptotic pathway 24 hours later. Interestingly, 5-dpf thymocytes were also sensitive to ER stress, indicating that these pathways can still be triggered beyond early development (Fig. S3).
Puma is transcriptionally activated following ER stress and is necessary for rapid epidermal apoptosis
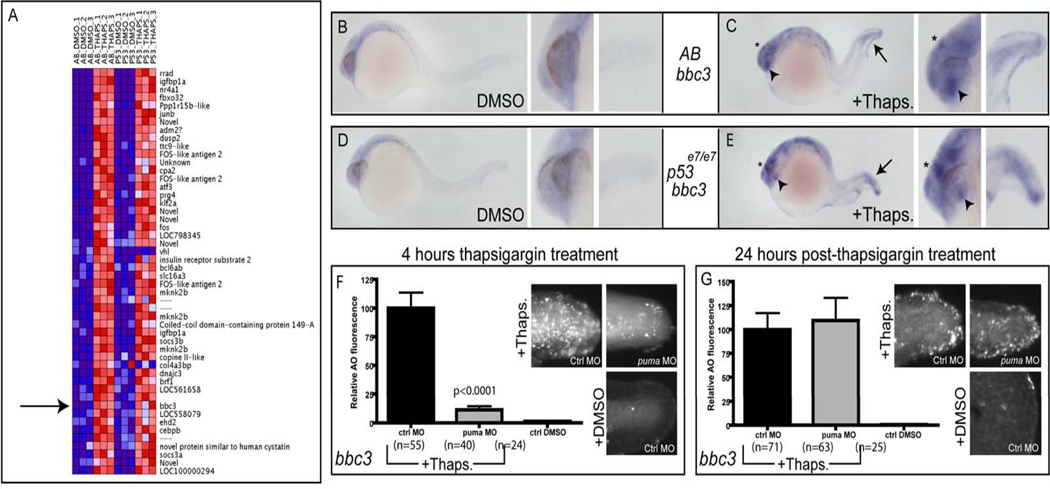
To discover which genes are transcriptionally activated and critical for the Chop-independent cell death program, we performed cDNA microarray analysis using dissected tails from pooled thapsigargin-treated embryos in both the AB and p53 mutant backgrounds. By using dissected tail tissue only, we reasoned that we could decrease the influence of developmental gene expression elsewhere in the organism and maximize the fold induction of ER stress-responsive genes in the tail epidermis. Among the list of the 50 most upregulated genes after thapsigargin treatment, there were numerous known mammalian ER stress-induced genes, including igfbp1a, junb, dusp2, atf3, fos, and dnajc3. These results indicate that thapsigargin induces a rapid ER stress response in the zebrafish epidermis similar to that in mammals. Interestingly, when we analyzed the 50 most highly ER stress-activated genes, we discovered that the pro-apoptotic BH3-only gene puma was among this group (Fig. 4A), showing an approximate 6-fold upregulation in thapsigargin-treated tails compared to DMSO controls. Since Bcl-2 overexpression could block the early apoptotic pathway (Supp. Fig 2), we considered puma to be a strong candidate for the pro-apoptotic BH3-only gene that was activated to kill tail epidermal cells. Indeed in thapsigargin-treated embryos, puma was induced in many parts of the brain as well as the lens, epiphysis, and tail, where rapid ER stress-induced apoptosis was most robust (Fig. 4B, C). Furthermore, as observed in the microarrays, the transcriptional activation of puma was p53-independent (Figure 4D,E), supporting a role for this factor in ER stress-induced apoptosis. Importantly, we observed a similar increase in puma transcription following brefeldin A treatment, supporting the hypothesis that puma activation is part of the general response to ER stress induction, and not specific to thapsigargin treatment (Supp. Fig. 3).
Figure 4. puma expression is increased in a p53-independent manner following thapsigargin treatment.
(A) Microrray analysis of dissected tail tissue revealed increased puma expression following thapsigargin treatment in both AB and p53 mutant embryos. (B,C) Compared to DMSO-treated controls (B), thapsigargin-treated embryos (C) had increased puma expression in the tail (arrow), epiphysis (asterisk), and lens (arrowhead). This increased expression was also observed in thapsigargin-treated p53 mutant embryos (E) compared to DMSO-treated controls (D). (F) Knockdown of puma attenuated 4-hour ER stress-induced apoptosis, but not (G) 28 hour ER stress-induced apoptosis (See also Supplemental Figures 2 and 3).
To determine whether Puma is critical for epidermal ER stress-induced apoptosis in vivo, we injected a puma splice-blocking morpholino (Sidi et al., 2008) into p53 mutant embryos and treated the embryos with thapsigargin for 4 hours (Fig. 4F, G). Puma knockdown strongly blocked the 4-hour apoptotic response in the lateral tail epidermis (Fig. 4F), showing that puma transcriptional induction is the apoptotic “trigger” downstream of ER stress. This result was confirmed with brefeldin A (Supp. Fig. 3), indicating that puma is required for the apoptotic response to multiple ER stressors. Despite its requirement for the 4-hour Chop-independent apoptotic response, Puma was not required for the 24-hour Chop-dependent apoptotic response (Fig. 4G), further demarcating these two pathways. Thus, ER stress activates puma expression within 3 hours in the tail epidermis in a Chop and p53-independent manner, while upregulation of puma is critical for the subsequent 4-hour apoptotic response in vivo.
p63 is required for the activation of puma and apoptotic death that occurs 4 hours after ER stress in the epidermis
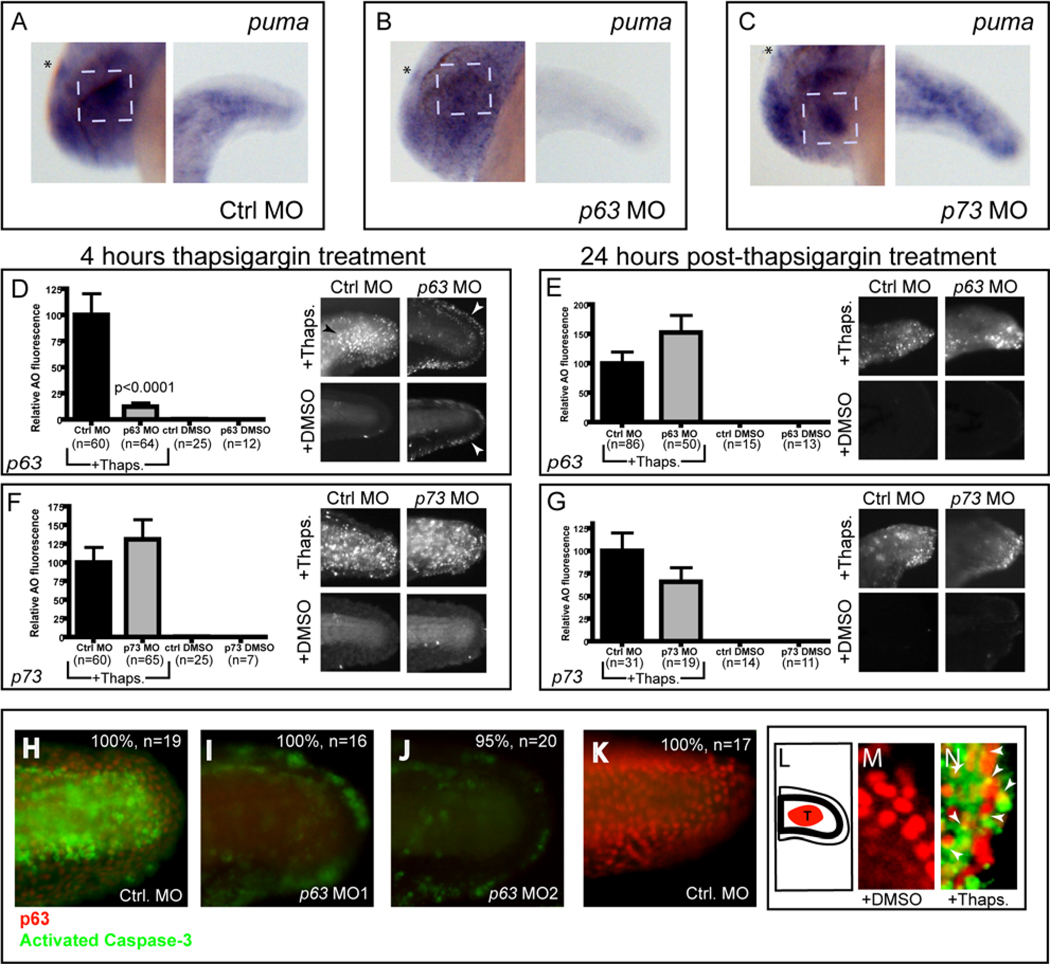
Because p63 and p73 partially overlap with p53 in their transcriptional targets (Harms et al., 2004), we could not be certain about the stringency of the requirement for p63 in puma activation. Thus, using published morpholinos against both p63 (Sidi et al., 2008) and p73 (Rentzsch et al., 2003) (Supp. Fig. 4). We compared the effects of these knockdowns on puma expression in homozygous p53 mutant fish after induction of ER stress. As shown in Figure 5, p63 knockdown, but not p73 knockdown, markedly reduced puma activation after 3 hours of thapsigargin treatment in the lens, epiphysis, and tail epidermis (Fig. 5B) compared with findings in controls (Fig. 5A) or p73 morphants (Fig. 5C). This reduction was especially pronounced in the tail epidermis, where puma expression was nearly absent in p63 morphants.
Figure 5. p63, but not p73, is required for ER stress-induced puma expression and 4 hr apoptosis.
(A, B) Compared to thapsigargin-treated control morphants (100% with high puma expression, n=16) thapsigargin-treated p63 morphants had reduced puma expression in the epiphysis, lens, and tail (78% with reduced expression, n=18). (C) Thapsigargin-treated p73 morphants had similar puma expression (94%, n=18) compared to control morphants (Asterisk=epiphysis, dashed box=lens) (D) p63 and control morphants were assayed for apoptosis by AO staining at 4 hrs after thapsigargin or DMSO treatment, or (E) washed out and assayed by AO staining 24 hrs later (See also Supplemental Figure 4 and Supplemental Figure 5). In (D), note the weak AO staining in the outer fin epidermis (white arrowheads) of p63 morphants, regardless of thapsigargin treatment, but the nearly complete block of cell death in the lateral epidermis compared to the same region in thapsigargin-treated control morphants (black arrowhead). (F,G) p73 morphants were processed the same as for p63 morphants. p73 morpholino injection does not block either (F) 4 hr thapsigargin-induced apoptosis or (G) 28 hr thapsigargin-induced apoptosis. (H–K) Embryos were injected with either control morpholino (H), p63 MO1 (I) or p63 MO2 (J), then treated with thapsigargin for 4 hours and assayed for p63 expression (red) and activated Caspase-3 (green) by immunofluorescence microscopy. Note the severe reduction in p63 and Caspase-3 expression in both p63 morphants (I,J) compared to control morphants (H). (K) Control embryos treated with DMSO showed extensive p63 expression with minimal Caspase-3 positivity. (L–N) Confocal analysis of co-immunofluorescence in embryos treated with DMSO (M) or thapsigargin (N) to analyze p63 expression (red) and activated Caspase-3 (green). Note the extensive co-localization of activated Caspase-3 with p63 in (N), marked with arrowheads. (L) Shows a schematic representation of the imaged region in (M) and (N). All graphical data are represented as means +/− SEM.
The above results suggest that knockdown of p63 would likely, given the importance of Puma in this response, block ER stress-induced apoptosis in the epidermis. To test this prediction, we treated p63 and control morpholino-injected p53 mutant embryos with thapsigargin at 24 hpf and performed AO staining 4 hours later. Since p63 and p73 loss-of-function have been reported to increase apoptosis that is unrelated to ER stress (Pietsch et al., 2008), we also examined DMSO control embryos for each of these morpholino injections. p63 morphants treated with thapsigargin had markedly decreased AO fluorescence compared to controls treated in the same manner (Figure 5D). Both thapsigargin-treated and DMSO-treated p63 morphants showed weak AO positivity in the outer fin epidermis at this stage (white arrowheads in Fig. 5D), supporting previously described roles for p63 in fin development (Bakkers et al., 2002; Lee and Kimelman, 2002). However, neither of these conditions with p63 knockdown displayed the robust AO positivity of control morphants treated with thapsigargin, particularly in the lateral epidermis of the tail (black arrowhead in Fig. 5D). Repeated AO staining after brefeldin A treatment yielded similar results (Supp. Fig. 5), confirming the involvement of p63 in epidermal apoptosis induced by ER stress. By contrast, p63 knockdown did not block the late Chop-dependent apoptotic pathway (Fig. 5E) while p73 knockdown failed to block either the early or late apoptotic response (Fig. 5F, G). Importantly, we observed similar results in AB embryos (Supp. Fig. 5), showing that p63 is not simply redundant with p53 in this apoptotic response, but, rather acts independently of p53 to induce apoptosis in epidermal cells following ER stress.
To confirm our p63 knockdown results with a second apoptotic assay, we performed dual immunofluorescence for activated Caspase-3 to mark apoptotic cells (in green) and for p63 (in red) to indicate the extent of p63 protein deficiency (Fig. 5H–K). Compared to the standard control morpholino, the p63 splice-blocking morpholino effectively reduced p63 protein levels and eliminated Caspase-3 activation in the lateral epidermis of thapsigargin-treated embryos, confirming that p63 is required for Caspase-3 activation following ER stress.
Unlike mammals, which have six different isoforms of p63 (Yang et al., 1998), the only isoform of p63 thus far detected in zebrafish is ΔNp63α (Bakkers et al., 2002; Lee and Kimelman, 2002). Nonetheless, since our p63 splice-blocking morpholino could affect all potential isoforms of p63 (Sidi et al., 2008), we repeated our apoptosis assay using a second morpholino designed to block only the translation of ΔNp63. As shown in Figure 5J, the ΔNp63-specific morpholino blocked 4-hour apoptosis similar to the pan-p63 morpholino, showing that the pro-apoptotic effects of p63 are elicited through ΔNp63.
Finally, to determine the spatial distribution of p63 in the tail epidermis relative to the dying cells observed in the 4-hour apoptotic response, we performed dual confocal immunofluorescence using the p63 antibody together with the activated-Caspase-3 antibody. By examining single optical sections obtained from confocal microscopy, we analyzed clusters of p63-positive cells within the tail epidermis of 28-hpf embryos (schematized by red region in Figure 5L). While DMSO-treated embryos showed minimal activation of Caspase-3 (Fig. 5M), embryos treated with thapsigargin for 4 hours showed clear Caspase-3 activation in the tail epidermis (Fig. 5N). In other systems, the activated form of Caspase-3 has been shown to translocate from the cytoplasm to the nucleus during apoptosis (Kamada et al., 2005); accordingly, we observed activated Caspase-3 staining that partially overlapped with p63-positive nuclear staining in approximately 50% of cells within the tail epidermis (see arrowheads in Fig. 5N), indicating that apoptosis was occurring in p63-expressing cells. Together, these data show that in the tail epidermis, p63 mediates a 4-hour ER stress-induced apoptotic response via transcriptional activation of puma.
p63 is activated by ER stress and directly binds to the puma promoter
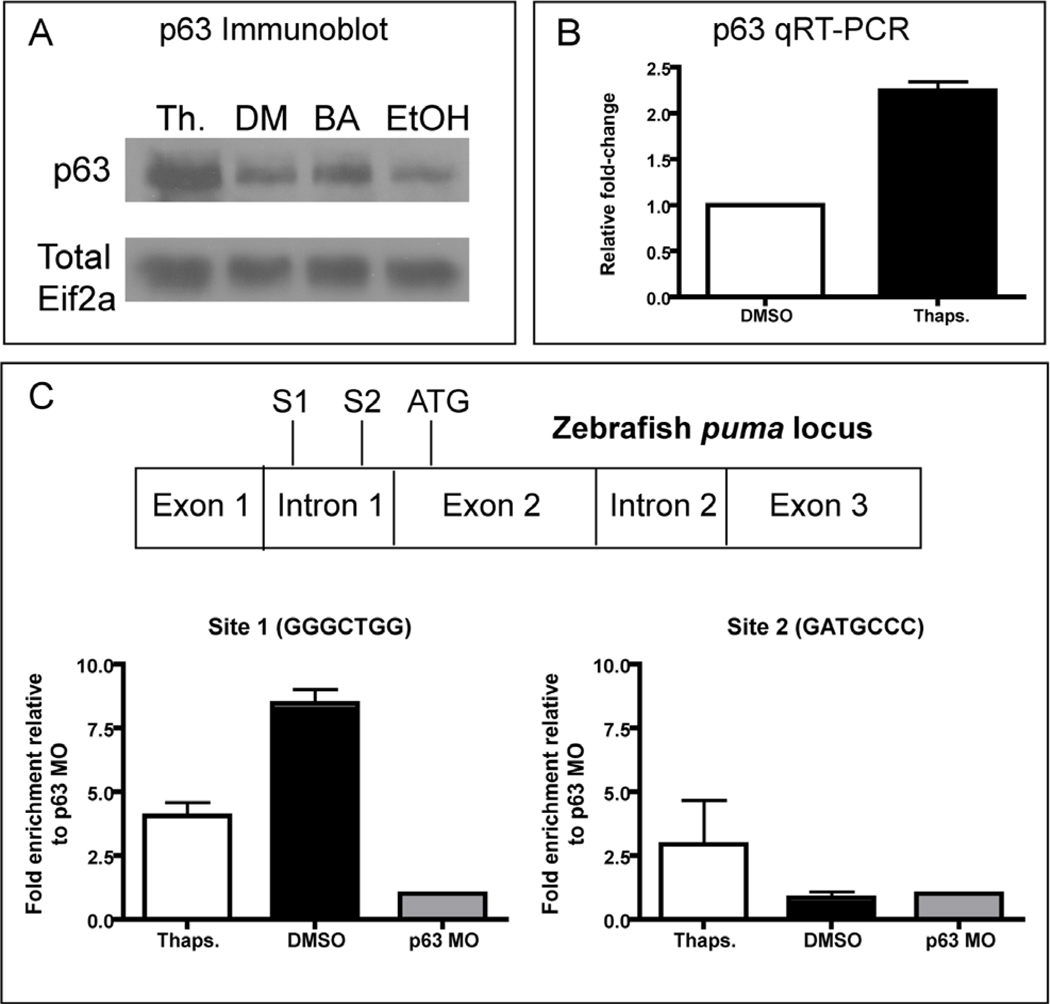
Given the link between p63 and the apoptotic response of epithelial cells to ER stress, we sought to elucidate the mechanism(s) underlying p63 activation and its role in the ER stress response. One major mechanism for p63 activation in other systems is an increase in protein levels, either through increased p63 transcription or stabilization of the p63 protein (Antonini et al., 2006; Rossi et al., 2006a; Rossi et al., 2006b). We initially examined whether p63 protein levels were increased by performing immunoblotting for p63 from pooled thapsigargin-treated p53 mutant embryos compared to DMSO controls. We noted an increase of p63 protein levels in thapsigargin-treated embryos, indicating that p63 levels rise in response to ER stress (Fig. 6A). To confirm that this effect was not specific to thapsigargin, we also compared brefeldin A-treated with ethanol-treated control lysates, observing an increase in p63 levels after treatment with brefeldin A (Fig. 6A) that was not as robust as for thapsigargin treatment, consistent with the somewhat lower levels of epidermal apoptosis triggered by this drug. To test whether p63 was increased transcriptionally, we performed qRT-PCR on pooled thapsigargin-treated embryos compared to DMSO controls (Fig 6B). This led to an approximate 2-fold increase in p63 transcript levels, consistent with our Western blotting results. Thus, when ER stress activates p63 transcription, the resultant in p63 protein levels likely contributes to the activation of puma and subsequent apoptosis in the epidermis.
Figure 6. p63 is transcriptionally increased and directly binds the puma promoter.
(A) Western blot to analyze p63 protein levels following 4 hours of thapsigargin (Th.) treatment, DMSO (DM) treatment, Brefeldin A (BA) treatment, or Ethanol (EtOH) treatment. Note the increased levels of p63 following both thapsigargin and brefeldin A treatment relative to their respective vehicle-only controls. Total Eif2-alpha antibody was used to control for loading. (B) p63 qRT-PCR revealed an increase in p63 transcript levels following 3 hours of thapsigargin treatment relative to controls (the sum of 3 experiments is represented). (C) Chromatin Immunoprecipitation to determine p63 binding to the puma promoter. Data are representative of experiments from two sets of biological samples, and are normalized to the p63 morpholino-injected negative control sample (p63 MO).
To determine whether p63 binds directly to the puma promoter following ER stress to activate gene transcription, we performed chromatin immunoprecipitation (ChIP) from thapsigargin and DMSO-treated control embryos using an anti-p63 antibody (Fig. 5C). The zebrafish puma locus contains two conserved p53 response elements in the first intron (Ensembl.org), similar to mouse and human puma (Wu et al., 2005), leading us to ask whether p63 binds to these response elements in vivo during thapsigargin-induced ER stress. As a negative control for this experiment, we used p63 morphant embryos, which express much lower levels of p63 protein (see Fig. 5H–K). After measuring the fold enrichment in immunoprecipitated chromatin compared to input chromatin by qPCR and normalizing the result to the fold-enrichment in our p63 morphant sample, we found that p63 constitutively binds Site 1 (GGGCTGG) within the puma first intron (Fig. 6C). The fold-enrichment was higher in DMSO samples than in thapsigargin-treated samples (~8-fold vs. ~4-fold), indicating that p63 binding to this site cannot explain the ER-stress induced increase in puma expression. Importantly, we observed enrichment of the levels of p63 binding to Site 2 (GATGCCC) in the thapsigargin-treated sample (~2.5-fold), but not in the DMSO control sample (Fig. 5C). Thus, our ChIP results show that Site 1 in the puma first intron is constitutively bound by p63, while Site 2 is bound only upon thapsigargin treatment, and thus is likely involved in the p63-mediated transcriptional upregulation of puma during the ER stress response.
Connexin 31 mutants trigger ER stress-induced apoptosis through p63
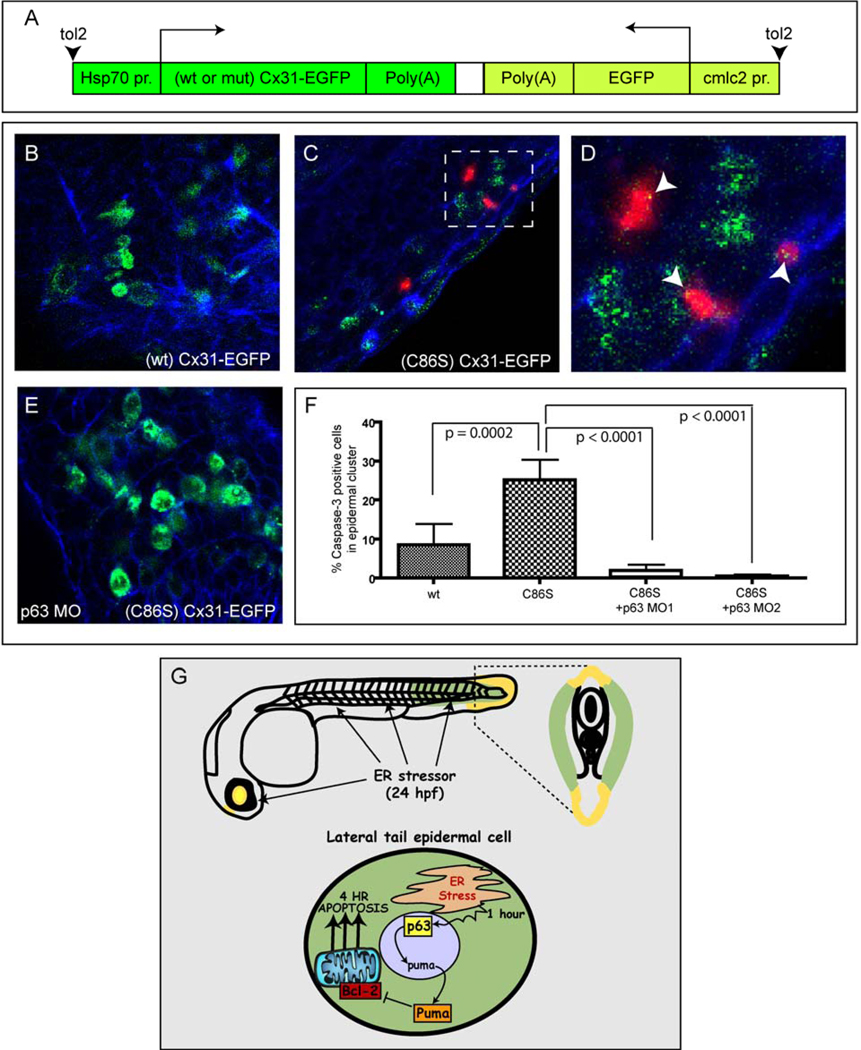
To investigate the clinical relevance of the epidermal p63-ER stress pathway, we generated a zebrafish model of human erythrokeratoderma variabilis (EKV). This hereditary disease of the skin epidermis, characterized by hyperkeratosis and erythema, is caused by mutations in the hemichannel protein Connexin 31 (Cx31) (Hunzeker et al., 2008). Recently, the Cx31 mutation C86S, a dominantly inherited mutation leading to the human disease, was shown to trigger apoptosis in cultured epidermal cells through defective intracellular trafficking and ER stress, rather than through impaired hemichannel function (Tattersall et al., 2009). To determine whether we could model this response in vivo in the zebrafish and whether p63 is required for the apoptotic response, we generated heat-shock inducible constructs containing human Cx31 mutant and wild-type cDNAs fused to EGFP and flanked by tol2 transposable element sites (Fig 7A). Since coinjection of tol2 mRNA yields high levels of plasmid integration and low levels of mosaicism (Kawakami, 2007), we injected the wild-type or C86S mutant Cx31 constructs with tol2 into fertilized eggs of homozygous p53 mutant embryos at the one-cell stage. We then heat-shocked the embryos at 24 hpf to induce expression of the Cx31-EGFP coding sequences within each of the two constructs. As shown in Fig. S6A–D, heat-shock for 30 minutes triggered nearly ubiquitous expression of the Cx31-EGFP constructs throughout the embryos within 2 hours. We next assessed the phenotypes resulting from expression of the Cx31-EGFP constructs, observing a severe phenotype in (C86S)Cx31-EGFP embryos, characterized by disruption of the skin epidermis in 85% of the embryos (n=20) by 3 hours post heat-shock, suggesting induction of an apoptotic response. This phenotype resembles the one obtained in vitro that is thought to contribute to human EKV disease (Tattersall et al., 2009).
Figure 7. p63 is required for apoptosis induced by a mutant Connexin 31 construct associated with human EKV disease.
(A) Overview of the Cx31 DNA constructs used in this study. (B–F) p53 mutant embryos were injected with wt or (C86S) mutant Cx31-EGFP constructs, then heat shocked at 24 hpf to induce Cx31-EGFP expression, fixed at 2 hrs later, and processed for anti-EGFP (green), anti-activated Caspase-3 (red) and phalloidin staining (blue) (See also Supplementary Figure 6). (B) (wt) Cx31-EGFP caused minimal apoptosis. (C) (C86S) Cx31-EGFP caused a higher level of epidermal apoptosis than (wt) Cx31-EGFP. (D) Zoomed region from (C). (E) Co-injection of p63 MO1 blocked the apoptosis induced by (C86S) Cx31-EGFP. (F) Quantification of apoptosis expressed as % Caspase-3 positive cells in an EGFP positive epidermal cluster. At least 8 embryos per condition were used for quantitation. (G) A model for the mechanism of ER stress-induced apoptosis in the developing epidermis.
Next, we reduced the level of expression by limiting the time of heat shock to 10 minutes, and injected specific cells into the embryos at the 2- to 4-cell stage to produce mosaic expression. This allowed us to observe the fates of subsets of skin epidermal cells that expressed the Cx31-EGFP constructs (Fig. 7A). To measure the level of apoptosis induced by (C86S)Cx31-EGFP expression, we injected this construct or the control (WT)Cx31-EGFP construct and subjected the embryos to heat shock for 10 min at 24 hpf. Using dual immunofluorescence to detect EGFP (green) and activated Caspase-3 (red), and phalloidin (blue) to mark the epidermal cell boundaries (Fig. 7 B–E), we found that ~25% of the (C86S)Cx31-EGFP-positive cells underwent apoptosis, compared to ~10% of the (WT)Cx31-EGFP positive cells (p= 0.0002) (Fig. 7F). As shown by a representative image in Figure 7E, knockdown of p63 transcripts blocked the epidermal apoptosis caused by the (C86S)Cx31-EGFP mutant construct. Quantitatively, that was a nearly complete loss of the apoptotic cells with each of the p63 MOs (Fig. 7F; p< 0.0001). These results support the role of ER stress- induced apoptotic cell death in the pathogenesis of human EKV and indicate that this response occurs through the p63-mediated, p53-independent apoptotic axis identified in this study.
Discussion
We have shown that ER stress pathways are activated within 1 hour of thapsigargin treatment in the tail epidermis of 24-hpf zebrafish embryos. This highly conserved ER stress response triggers 4-hour apoptosis through direct activation of puma by p63 in the epidermal cell layer, independently of Chop, p53, and p73. This pathway is activated by the mutant (CS86)Cx31 protein in human EKV disease, likely because mutant Cx31 protein activates the unfolded protein response and induces ER stress in epidermal cells as in mammals.
High levels of ER stress in the developing epidermis trigger rapid p53-independent apoptosis
Development of the mammalian epidermis involves ER stress as part of the physiological barrier formation process en route to skin maturation (Celli et al., 2010). Additionally, we and others have observed that bip and xbp, two markers of ER stress induction, are constitutively expressed in the developing zebrafish epidermis (data not shown). Based on these findings, we propose that the epidermis continuously induces low-level ER stress during early development, owing to the massive amounts of membrane and secreted proteins that are produced to form the epidermal basement membrane (Webb et al., 2007). Another tissue in the developing zebrafish, the hatching gland, also expresses these ER stress markers, and a previous study showed that Xbp loss causes massive ER dysfunction indicative of ER stress (Bennett et al., 2007). We propose that the epidermis, like the lens, epiphysis, and hatching gland, is highly sensitized to ER stress induction, and that this tissue undergoes robust apoptosis after excessive ER stress. Thus, when zebrafish embryos are treated at 24 hpf with pharmacological stressors such as thapsigargin and brefeldin A or genetic stressors such as (C86S)Cx31-EGFP, the epidermis is quickly overloaded with ER stress, leading to the death of epidermal cells via induction of puma by the transcription factor p63. Furthermore, this activation is independent of p53, which mediates DNA damage-induced apoptosis in the developing zebrafish spinal cord (Berghmans et al., 2005; Langheinrich et al., 2002; Sidi et al., 2008).
Thapsigargin is a robust and rapid inducer of ER stress in the early embryo
Our tissue-restricted microarray data allow us to implicate puma as the critical factor mediating ER stress-induced epidermal apoptosis. They also underscore the activity of thapsigargin in inducing a robust ER stress response within 3 hours in the tail epidermis. Although the calcium-mediated axis-kinking phenotype caused by thapsigargin is often severe, a previous study of the accordion mutant, which has the axis-kinking phenotype, did not describe any overt apoptosis during development (Hirata et al., 2004). This mutant lacks the function of a major muscle-specific calcium ATPase pump, resembling the pharmacological effects of thapsigargin treatment in the muscle. In addition, our results were confirmed with a second drug, brefeldin A, that inhibits transport of proteins from the ER to Golgi through a mechanism independent of calcium flux. Thus, thapsigargin and brefeldin A each appear to induce a specific ER stress response in the epidermis via p63 and puma within the 4-hour window of our analysis.
p63 directly activates puma to trigger ER stress-induced epidermal apoptosis
By knocking down the transcripts encoding the p53 homologs p63 and p73, we uncovered a role for p63 in the epidermal ER stress response. After its ER stress-mediated activation, p63 directly binds the promoter of the BH3-only cell death inducer puma and activates its transcription. Using chromatin immunoprecipitation from whole zebrafish embryos, we found that p63 binds constitutively to one of its response elements within puma intron 1. Following ER stress induction, there was increased binding of p63 to a second response element within the same intron, suggesting that this site is responsible for the marked increase in puma expression after ER stress. Based on these results and our in situ hybridization data showing that ER stress-induced puma expression in the epidermis is markedly diminished after p63 knockdown, we propose that after ER stress, p63 binds to site 2 within the puma first intron and triggers the expression of this BH3-only gene. Subsequently, cells within the tail epidermis undergo apoptosis, which can be blocked by Bcl-2 overexpression.
p63 is required for apoptosis induced by the human Connexin 31 mutation C86S
The p63-mediated ER stress pathway we have identified appears to have a role in human disease. The C86S germline mutation in the GJB3 gene, which encodes the Cx31 protein, causes a skin disease called EKV, which is characterized by erythematous patches along with hyperkeratosis. In mammalian keratinocytes, (C86S) Cx31 but not wild-type Cx31, was recently shown to induce cell death through ER stress rather than through disruption of hemichannels at the cell membrane (Tattersall et al., 2009). In addition to the pharmacological induction of ER stress, we show that human Cx31-EGFP with a C86S mutation induces apoptosis in the developing tail epidermis, similar to that observed after treatment with thapsigargin or brefeldin A. By using two different p63 morpholinos, we show further that the (C86S)Cx31-EGFP construct causes apoptosis in a p63-dependent manner. This implicates the p63-dependent apoptotic axis in the pathogenesis of EKV, where it presumably contributes to the erythema and hyperproliferation of the skin epidermis characteristic of this disease.
Experimental Procedures
Thapsigargin treatment
Thapsigargin (10 mM, Sigma St. Louis) in DMSO was diluted 1:20 in E3 egg water, and 10 µl was then added to 1 ml of egg water in a single well of a 12-well tissue culture plate (BD Falcon, Canaan, CT) to make a 5 µM final concentration. 20–25 embryos, no older than 24 hpf at the start of the experiment, were placed in each well. For rapid apoptosis induction, embryos were left for 4 hrs in the dark at 28.5°C and processed for either Acridine Orange staining, TUNEL staining, or anti-activated Caspase-3 immunohistochemistry. For the induction of late apoptosis, embryos were left in thapsigargin for 4 hrs in the dark at 28.5°, washed 3 times with E3 egg medium, and replaced at 28.5° for 24 hrs. Cell death was then assayed by AO staining.
Apoptosis quantitation with Volocity software
To quantify AO or TUNEL positivity in the tail, we mounted stained embryos laterally in 0.8% low melting point agarose within 60×15mm petri dishes (Falcon). Subsequently, stained embryos were visualized by fluorescent microscopy with a Nikon SMZ1500 zoom stereomicroscope (Nikon Instruments Inc., Melville, NY) using a 488 nm filter for the green fluorescent signal; photographs were taken with a Nikon Digital Sight DS-2MBWc black and white camera with NIS Elements software. All images were acquired at the same magnification, exposure, and gain. Subsequently, acquired images were rotated and cropped to a 3” × 3” square extending from the tip of the tail anteriorly using Adobe Photoshop software. Only images in focus were used for Volocity analysis, in which fluorescence intensity was calculated by first setting the threshold intensity above background in a representative control morpholino-injected embryo treated with thapsigargin. This was done for each experiment, since the threshold varied between experiments. Next, the total fluorescence intensity at or above the threshold value was calculated for each image. Means were calculated for each treatment using Prism software, and Microsoft Excel software was used to convert all data points to a percentage of the control mean. Unless otherwise noted, three independent experiments were conducted for each morpholino injection and were summed for graphical illustration and statistical analysis in Prism. Unpaired student’s t-tests were performed in Prism to determine the P-value for each treatment compared to controls. All findings are reported as means +/− SEM.
Chromatin Immunoprecipitation
For each sample (Ctrl MO + DMSO, Ctrl MO + Thapsigargin, p63 MO1 + Thapsigargin), input chromatin was saved following sonication to determine enrichment. Chromatin immunoprecipitation was performed using Dynabeads as described elsewhere for whole zebrafish embryos: https://wiki.zfin.org/display/prot/Chromatin+Immunoprecipitation+528ChIP%29+Protocol+using+Dynabeads. 4A4 anti-human p63 antibody (Santa Cruz) was used for immunoprecipitation. The sequences for the primers used for subsequent qPCR of the puma promoter and additional information can be found in the Supplementary Methods.
Microarrays
Tails from 20 embryos in each experimental condition were dissected and processed using Trizol (Ambion) to extract RNA. Three biological samples on separate days were generated and submitted simultaneously to the DFCI Microarray Core Facility for processing. GeneChip Zebrafish Genome Array chips (Affymetrix) were used for hybridization, representing ~14900 transcripts. Subsequently, data processing was performed using dChip software (Cheng Li laboratory, DFCI) to generate a list of transcripts with p values <.05 and fold-change of >1.5 per sample for AB +Thapsigargin vs. AB + DMSO RNAs. This list of genes was then sorted in descending order of fold change and fold-change values for the same gene list were included from p53 mutant samples (Thapsigargin vs. DMSO-treated). The top 50 upregulated genes following thapsigargin treatment were used to generate a heatmap in GenePattern (Broad Institute).
Caspase-3/Connexin 31-EGFP immunofluorescence and confocal microscopy
Heat-shocked embryos were fixed for 1 hour in 4% Paraformaldehyde at room temperature, then washed into PBST and permeabilized in PBS + .1% Triton for 30 minutes at room temperature. Embryos were blocked in 2% FBS/PBST for 1 hour, and then incubated in primary antibody overnight at 4 degrees Celsius. The anti-activated Caspase-3 antibody was used at a dilution of 1:200, anti-EGFP at 1:500. For Caspase-3 detection, an Alexa 647 goat anti-rabbit secondary antibody was used, while for EGFP detection, an Alexa 488 goat anti-mouse secondary antibody was used. Subsequently, embryos were washed in PBST and stained in phalloidin for 1 hour prior to imaging. Imaging was performed using a Leica SP5 confocal microscope with a 20X water immersion lens. Counts were performed in clusters of EGFP positive and EGFP/Caspase 3 positive cells between the yolk extension and the end of the tail.
Highlights.
ER stress induces p53-independent apoptosis in the epidermis of zebrafish embryos
This rapid ER stress-induced apoptotic axis depends on Puma and p63, but not Chop
p63 directly activates puma expression in response to ER stress to trigger apoptosis
p63 is also required for ER stress induced by an inherited Connexin 31 mutation
Supplementary Material
Acknowledgments
We thank John Gilbert, Elspeth Payne, and Samuel Sidi for critical readings of the manuscript. Confocal microscopy images for this study were acquired in the Confocal and Light Microscopy Core Facility at the Dana-Farber Cancer Institute with the assistance of Lisa Cameron. UJP was supported by the Alex’s Lemonade Stand Foundation Young Investigator Award, UJP and SC by NIH 3R01CA119066-04S1, and ATL by NIH R01CA119066.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams JM, White K, Fessler LI, Steller H. Programmed cell death during Drosophila embryogenesis. Development (Cambridge, England) 1993;117:29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- Antonini D, Rossi B, Han R, Minichiello A, Di Palma T, Corrado M, Banfi S, Zannini M, Brissette JL, Missero C. An autoregulatory loop directs the tissue-specific expression of p63 through a long-range evolutionarily conserved enhancer. Mol Cell Biol. 2006;26:3308–3318. doi: 10.1128/MCB.26.8.3308-3318.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkers J, Hild M, Kramer C, Furutani-Seiki M, Hammerschmidt M. Zebrafish DeltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev Cell. 2002;2:617–627. doi: 10.1016/s1534-5807(02)00163-6. [DOI] [PubMed] [Google Scholar]
- Bennett JT, Joubin K, Cheng S, Aanstad P, Herwig R, Clark M, Lehrach H, Schier AF. Nodal signaling activates differentiation genes during zebrafish gastrulation. Developmental biology. 2007;304:525–540. doi: 10.1016/j.ydbio.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nature cell biology. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Celli A, Mackenzie DS, Crumrine DS, Tu CL, Hupe M, Bikle DD, Elias PM, Mauro TM. Endoplasmic reticulum Ca(2+) depletion activates XBP1 and controls terminal differentiation in keratinocytes and epidermis. Br J Dermatol. doi: 10.1111/j.1365-2133.2010.10046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collavin L, Lunardi A, Del Sal G. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ. 17:901–911. doi: 10.1038/cdd.2010.35. [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Fribley A, Zhang K, Kaufman RJ. Methods in molecular biology. Vol. 559. Clifton, NJ: 2009. Regulation of apoptosis by the unfolded protein response; pp. 191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galehdar Z, Swan P, Fuerth B, Callaghan SM, Park DS, Cregan SP. Neuronal Apoptosis Induced by Endoplasmic Reticulum Stress Is Regulated by ATF4-CHOP-Mediated Induction of the Bcl-2 Homology 3-Only Member PUMA. J Neurosci. 30:16938–16948. doi: 10.1523/JNEUROSCI.1598-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Molecular cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci. 2004;61:822–842. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Saint-Amant L, Waterbury J, Cui W, Zhou W, Li Q, Goldman D, Granato M, Kuwada JY. Development. Vol. 131. Cambridge, England: 2004. accordion, a zebrafish behavioral mutant, has a muscle relaxation defect due to a mutation in the ATPase Ca2+ pump SERCA1; pp. 5457–5468. [DOI] [PubMed] [Google Scholar]
- Hunzeker CM, Soldano AC, Levis WR. Erythrokeratoderma variabilis. Dermatol Online J. 2008;14:13. [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- Jette CA, Flanagan AM, Ryan J, Pyati UJ, Carbonneau S, Stewart RA, Langenau DM, Look AT, Letai A. BIM and other BCL-2 family proteins exhibit cross-species conservation of function between zebrafish and mammals. Cell Death Differ. 2008;15:1063–1072. doi: 10.1038/cdd.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada S, Kikkawa U, Tsujimoto Y, Hunter T. Nuclear translocation of caspase-3 is dependent on its proteolytic activation and recognition of a substrate-like protein(s) J Biol Chem. 2005;280:857–860. doi: 10.1074/jbc.C400538200. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8 Suppl 1:S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- Langheinrich U, Hennen E, Stott G, Vacun G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr Biol. 2002;12:2023–2028. doi: 10.1016/s0960-9822(02)01319-2. [DOI] [PubMed] [Google Scholar]
- Lee H, Kimelman D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev Cell. 2002;2:607–616. doi: 10.1016/s1534-5807(02)00166-1. [DOI] [PubMed] [Google Scholar]
- Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annual review of pathology. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Minami M, Takeda K, Sakao Y, Akira S. Ectopic expression of CHOP (GADD153) induces apoptosis in M1 myeloblastic leukemia cells. FEBS Lett. 1996;395:143–147. doi: 10.1016/0014-5793(96)01016-2. [DOI] [PubMed] [Google Scholar]
- Maytin EV, Ubeda M, Lin JC, Habener JF. Stress-inducible transcription factor CHOP/gadd153 induces apoptosis in mammalian cells via p38 kinase-dependent and -independent mechanisms. Exp Cell Res. 2001;267:193–204. doi: 10.1006/excr.2001.5248. [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochemical and biophysical research communications. 2000;279:445–450. doi: 10.1006/bbrc.2000.3987. [DOI] [PubMed] [Google Scholar]
- Pahl HL, Baeuerle PA. A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by transcription factor NF-kappa B. Embo J. 1995;14:2580–2588. doi: 10.1002/j.1460-2075.1995.tb07256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietsch EC, Sykes SM, McMahon SB, Murphy ME. The p53 family and programmed cell death. Oncogene. 2008;27:6507–6521. doi: 10.1038/onc.2008.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Pyati UJ, Look AT, Hammerschmidt M. Zebrafish as a powerful vertebrate model system for in vivo studies of cell death. Semin Cancer Biol. 2007;17:154–165. doi: 10.1016/j.semcancer.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11:372–380. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- Reimertz C, Kogel D, Rami A, Chittenden T, Prehn JH. Gene expression during ER stress-induced apoptosis in neurons: induction of the BH3-only protein Bbc3/PUMA and activation of the mitochondrial apoptosis pathway. J Cell Biol. 2003;162:587–597. doi: 10.1083/jcb.200305149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch F, Kramer C, Hammerschmidt M. Specific and conserved roles of TAp73 during zebrafish development. Gene. 2003;323:19–30. doi: 10.1016/j.gene.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA, Croce CM, Melino G. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci U S A. 2006a;103:12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, De Simone M, Pollice A, Santoro R, La Mantia G, Guerrini L, Calabro V. Itch/AIP4 associates with and promotes p63 protein degradation. Cell Cycle. 2006b;5:1816–1822. doi: 10.4161/cc.5.16.2861. [DOI] [PubMed] [Google Scholar]
- Samali A, Fitzgerald U, Deegan S, Gupta S. Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int J Cell Biol. 2010:830307. doi: 10.1155/2010/830307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, Pascual J, Imamura S, Kishi S, Amatruda JF, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Tattersall D, Scott CA, Gray C, Zicha D, Kelsell DP. EKV mutant connexin 31 associated cell death is mediated by ER stress. Hum Mol Genet. 2009;18:4734–4745. doi: 10.1093/hmg/ddp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Webb AE, Kimelman D. Methods in molecular biology. Vol. 289. Clifton, NJ: 2005. Analysis of early epidermal development in zebrafish; pp. 137–146. [DOI] [PubMed] [Google Scholar]
- Webb AE, Sanderford J, Frank D, Talbot WS, Driever W, Kimelman D. Laminin alpha5 is essential for the formation of the zebrafish fins. Developmental biology. 2007;311:369–382. doi: 10.1016/j.ydbio.2007.08.034. [DOI] [PubMed] [Google Scholar]
- Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Molecular cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes & development. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.