Abstract
RNA-mediated, posttranscriptional gene silencing has been determined as the molecular mechanism underlying transgenic virus resistance in many plant virus-dicot host plant systems. In this paper we show that transgenic virus resistance in sugarcane (Saccharum spp. hybrid) is based on posttranscriptional gene silencing. The resistance is derived from an untranslatable form of the sorghum mosaic potyvirus strain SCH coat protein (CP) gene. Transgenic sugarcane plants challenged with sorghum mosaic potyvirus strain SCH had phenotypes that ranged from fully susceptible to completely resistant, and a recovery phenotype was also observed. Clones derived from the same transformation event or obtained after vegetative propagation could display different levels of virus resistance, suggesting the involvement of a quantitative component in the resistance response. Most resistant plants displayed low or undetectable steady-state CP transgene mRNA levels, although nuclear transcription rates were high. Increased DNA methylation was observed in the transcribed region of the CP transgenes in most of these plants. Collectively, these characteristics indicate that an RNA-mediated, homology-dependent mechanism is at the base of the virus resistance. This work extends posttranscriptional gene silencing and homology-dependent virus resistance, so far observed only in dicots, to an agronomically important, polyploid monocot.
Sugarcane (Saccharum spp. hybrid) ranks among the world's top 10 food crops and annually provides 60% to 70% of the sugar produced worldwide (Sugar and Sweetener Situation and Outlook Yearbook, 1997). Modern commercial sugarcane cultivars are interspecific hybrids derived from crosses of noble sugarcane, i.e. Saccharum officinarum L. (2n = 70–122). Crosses are most often made with Saccharum spontaneum L. (2n = 36–128), sometimes with Saccharum barberi (2n = 60–140) or Saccharum sinense (2n = 104–128), and rarely with Saccharum robustum (2n = 66–170) (Irvine, 1999). Sugarcane cultivars have ploidy levels that range from 5× to 14× (× = 5, 6, 8, 10, 12, or 14) and chromosomal mosaicism has been reported (Burner and Legendre, 1994, and refs. therein). The genetic complexity and low fertility of sugarcane render traditional breeding laborious and make it a prime candidate for improvement through genetic engineering.
Transgenic sugarcane plants have been obtained via particle gun bombardment of embryogenic callus (Bower and Birch, 1992; Gallo-Meagher and Irvine, 1996) and via electroporation of cells derived from embryogenic callus (Arencibia et al., 1995). Unlike many other members of the Poaceae in which regeneration is restricted to certain genotypes, most sugarcane cultivars tested to date have yielded regenerable calli. Therefore, introducing specific genetic improvements, such as virus resistance, directly into elite sugarcane varieties is a realistic goal.
SCMV has a monopartite, positive-strand RNA genome (Shukla et al., 1994). Recent taxonomic studies have shown that the SCMV complex comprises four or five different potyviruses, including strains of SrMV, SCMV, Johnson grass mosaic virus, and maize dwarf mosaic virus (Yang and Mirkov, 1997, and refs. therein). Members of the SCMV complex can cause mosaic symptoms and yield loss in susceptible members of sugarcane, maize, sorghum (Sorghum bicolor), and other poaceous plants. The worldwide epidemic spread of sugarcane mosaic in the 1920s caused a near collapse of the sugar industry in Louisiana, Argentina, and Brazil and was the main factor in replacing the highly susceptible noble cultivars with interspecific hybrid derivatives that were tolerant to SCMV. However, strains of SCMV and SrMV continue to cause losses in the sugarcane industry.
Several strategies have been used to engineer virus resistance in plants (for review, see Baulcombe, 1996). In CP and movement protein-mediated protection, a transgene-derived homolog of a viral protein is expressed in plants, which interferes with or prevents various stages of the viral life cycle, resulting in attenuated disease symptoms or resistance.
It is now well established that transgenes in plants can suppress expression of homologous endogenous genes, transgenes, or viral RNAs (for reviews, see Depicker and Van Montagu, 1997; Stam et al., 1997). Homology-dependent gene silencing can occur at the level of transcription or by a posttranscriptional process. Transcriptional silencing is often associated with increased methylation and inactivation of the promoter sequences of the affected genes. In PTGS, an unidentified cellular process is responsible for the specific degradation of the homologous RNA molecules. The expression level, number, and configuration of the integrated transgenes, as well as other less understood developmental and environmental factors, can all influence the occurrence of PTGS. There is now convincing evidence that PTGS is involved in many cases of RNA-mediated virus resistance; this type of resistance has been termed HDR (Mueller et al., 1995).
Recent experiments have shown that a signaling molecule is involved in the systemic spread of gene silencing and resulting virus resistance (Palauqui et al., 1997; Voinnet and Baulcombe, 1997). Gene silencing and virus resistance can be induced by virus infections of nontransgenic plants (Covey et al., 1997; Ratcliff et al., 1997; Al-Kaff et al., 1998). These results and others led to the hypothesis that gene silencing is a natural plant defense mechanism (for review of transgene silencing as a manifestation of cellular defense responses, see Matzke and Matzke, 1998). The recent demonstration that the expression of the P1/HC-Pro sequence of the tobacco etch potyviral genomic RNA can suppress PTGS provides direct evidence that PTGS functions as a host defense response and shows that viruses have evolved a mechanism to counter it (Anandalakshmi et al., 1998; Kasschau and Carrington, 1998).
PTGS and HDR have been conclusively demonstrated for a number of dicot plants with a variety of viruses (for review, see Baulcombe, 1996). Typically, this type of resistance is highly strain specific and two classes of resistance response are often encountered: a pre-established immune phenotype and a delayed recovery phenotype. Resistance is associated with reduced steady-state transgene mRNA levels, while the transgenes remain actively transcribed in the nucleus. Recent data indicate that transgene transcription is required for HDR (Sijen et al., 1996; English et al., 1997; Waterhouse et al., 1998). In a few studies, shorter than full-length transgene-derived RNA transcripts have been detected that may represent degradation products of the silencing mechanism (Goodwin et al., 1996; Tanzer et al., 1997). Finally, some cases of HDR are correlated with methylation in the transcribed region of the silenced transgenes (English et al., 1996; Sijen et al., 1996).
We have transformed sugarcane with an untranslatable form of the SrMV strain SCH CP gene using particle gun bombardment. Phenotypical and molecular data show that in virus-resistant sugarcane plants resistance relies on an RNA-mediated, homology-dependent mechanism. Our studies extend PTGS and its association with virus resistance to a monocot and reinforce the hypothesis of an ancestral PTGS pathway as a defense mechanism in plants.
MATERIALS AND METHODS
Constructs
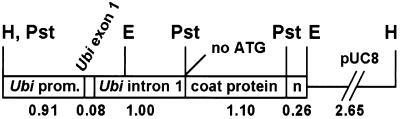
Plasmid pAHC20 (Christensen and Quail, 1996) contains the bar (bialaphos-resistance gene)-coding region under the control of the maize ubiquitin promoter, first exon, and first intron, followed by the nos (nopaline synthase gene) terminator and will be referred to as Ubi-bar in this paper. To construct Ubi-npt, a 963-bp BglII/ApaLI blunt-ended fragment from pCRII (Invitrogen, Carlsbad, CA) that contains the npt-coding sequence was inserted into the blunt-ended SalI sites of Ubi-bar to replace the bar-coding region. For construction of Ubi-hut, which contains the SrMV-SCH untranslatable CP ORF (hut), a 1.1-kb PCR product containing the SrMV-SCH CP-coding sequence and 77 nucleotides of the 3′UTR, was cloned into pCRII using the TA cloning kit (Invitrogen) yielding pSCH-1. Then, the CP ORF and 3′UTR sequences were isolated from pSCH-1 as a 1.1-kb blunt-ended EcoRI fragment and inserted into the blunt-ended SalI sites of Ubi-bar to replace the bar-coding region, yielding Ubi-hut (Fig. 1).
Figure 1.
Schematic representation of the Ubi-hut plasmid (6.0 kb) linearized at the unique HindIII site showing the maize ubiquitin promoter (Ubi prom.), exon 1 and intron 1 plus the SrMV-SCH-untranslatable CP-coding region with 77 nucleotides of the 234 3′UTR sequence, and nos termination sequence (n) cloned into pUC8. Restriction sites: E, EcoRI; H, HindIII; Pst, PstI. Numerical values indicate approximate kilobase pairs.
The following phagemid clones were constructed for in vitro RNA synthesis and production of single-stranded DNA: (a) a 498-bp SphI fragment from the sugarcane Rubisco small subunit genomic clone scrbsc-1 (nucleotide position 1313–1811, see Tang and Sun, 1993) was subcloned in a 5′ to 3′ and a 3′ to 5′ orientation into the SphI site of pGem11Zf(+), yielding pIVING366-1 and pIVING366-2, respectively; (b) a 1.1-kb EcoRI fragment encompassing the SrMV-SCH CP-coding region and 77 nucleotides of the 3′UTR was isolated from pSCH-1 and subcloned in a 5′ to 3′ and a 3′ to 5′ orientation into the EcoRI site of pGem7Zf(−), yielding pIVING362-3 and pIVING362-4, respectively; (c) a 0.7-kb PstI fragment encompassing the 3′ part of the npt-coding region was isolated from Ubi-npt, rendered blunt with T4 DNA polymerase, and subcloned in a 5′ to 3′ and a 3′ to 5′ orientation into the SmaI site of pGem7Zf(−), yielding pIVING362-5 and pIVING362-6, respectively; and (d) a 0.6-kb PstI fragment containing the bar-coding region was isolated from Ubi-bar, made blunt with T4 DNA polymerase and subcloned in a 5′ to 3′ and a 3′ to 5′ orientation into the SmaI site of pGem7Zf(−), yielding pIVING362-7 and pIVING369-1, respectively. These plasmids were transformed into DH5αF′IQ cells (GIBCO-BRL). Unless stated otherwise, all recombinant DNA and bacterial manipulations were performed using standard techniques (Sambrook et al., 1989).
Sugarcane Transformation
Embryogenic callus cultures were established from young leaf bases and immature flowers of the commercial sugarcane (Saccharum spp. hybrid) varieties CP65-357 and CP72-1210. Transformation of callus by particle gun bombardment and regeneration of shoots were done as described previously (Gallo-Meagher and Irvine, 1996); 7- to 40-week-old calli were bombarded with an equimolar mixture of either Ubi-bar and Ubi-hut or Ubi-npt and Ubi-hut (4 μg DNA/480 μg particles). Two days after bombardment, calli were transferred to CI-3 medium containing either 1 mg/L bialaphos or 15 mg/L geneticin. Four weeks later, calli were transferred to 2K5N medium containing 2 mg/L kinetin, 5 mg/L NAA, and 1 mg/L bialaphos or 15 mg/L geneticin to promote shoot regeneration and inhibit development of nontransgenic tissue. Tissues were subcultured on this medium every 2 to 3 weeks for approximately 12 weeks. At this time, shoots were placed into Magenta boxes containing RG-2 with 4 mg/L indole butyric acid and either 3 mg/L bialaphos or 45 mg/L geneticin to induce roots. After about 8 weeks, shoots (5–15 cm) with well-developed roots were transferred to peat pots containing potting soil (Metromix, Scotts, Hope, AR) and placed in an environmental growth chamber at 30°C under 15 h of fluorescent and incandescent light. After 2 to 4 weeks, plants (15–30 cm high) were transferred to 15-cm-diameter pots and placed in the greenhouse.
Virus Isolates, Inoculation, and Resistance Tests
The virus strains SCMV-D, SrMV-SCM, SrMV-SCI, and a Texas isolate of SrMV-SCH were previously described (Yang and Mirkov, 1997). Virus strains were propagated on sorghum (Sorghum bicolor cv Rio). Sugarcane plants at the four- to eight-leaf stage were inoculated with sap extract (20 mL of a 0.1 m Na2SO3/0.1 m phosphate buffer, pH 7.0, containing 0.2 g of diatomaceous earth [Sigma] g−1 tissue) from virus-infected sorghum leaves. The plants were inoculated at least twice, at 2- to 3-week intervals. The infection rate on nontransgenic controls was more than 95%. Symptoms were scored visually and, in some cases, the virus level in leaves was quantified by ELISA, according to standard procedures (Converse and Martin, 1990), using SrMV-SCH alkaline-conjugated IgG at a 1:1000 dilution or by RNA gel-blot analysis.
DNA Gel-Blot Analysis
Total DNA was extracted from sugarcane leaves as described by Tai and Tanksley (1990) with slight modifications. Ten micrograms of digested DNA samples was electrophoresed on a 0.8% (w/v) agarose gel and transferred to membranes (Hybond N+, Amersham) by downward alkaline blotting (Koetsier et al., 1993). The membrane in Figure 2 was hybridized to a fluorescein-labeled DNA probe covering the complete SrMV-SCH CP-coding region. Labeling, hybridization, washing, and detection were performed according to the manufacturer's instructions (Gene Images, Amersham). The membrane in Figure 6 was hybridized to a 32P-labeled DNA probe covering part of the SrMV-SCH CP-coding region. Labeling was done using a nick translation kit (GIBCO-BRL); hybridization and washing were performed according to previously published procedures (Sambrook et al., 1989).
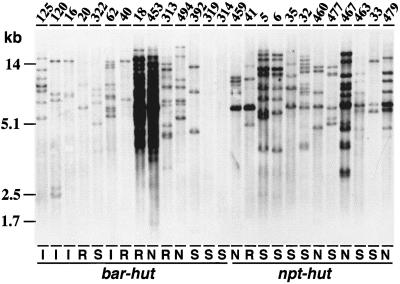
Figure 2.
DNA gel-blot analysis of CP transgene integration patterns and copy number in sugarcane transgenics. DNA gel blot of total DNA (10 μg) digested with HindIII and hybridized with a fluorescein-labeled 1.1-kb EcoRI fragment containing the SrMV-SCH CP and 3′UTR. Numbers above the lanes refer to individual sugarcane transgenics. I, R, and S denote immune, recovered, and susceptible plants, respectively; N refers to plants that were not inoculated and with an undetermined phenotype. Transgenics were selected on bialaphos (bar-hut) or on geneticin (npt-hut), as indicated. DNA size markers are shown on the left.
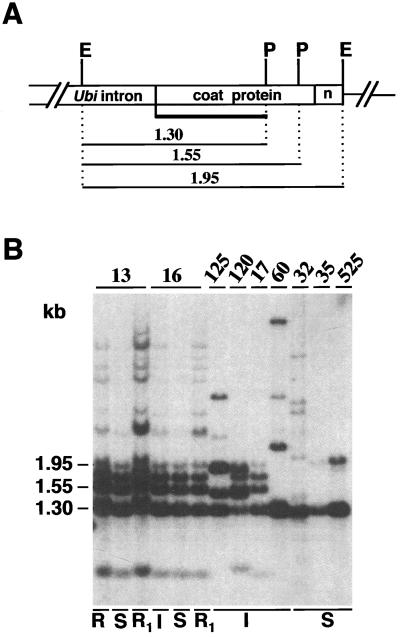
Figure 6.
DNA gel-blot analysis of differential methylation patterns in total DNA from resistant and susceptible leaf tissue. A, Diagram showing the position of the EcoRI (E) and PvuII (P) sites within the CP transgene. The solid line represents the 0.75-kb probe used for hybridization of the blot shown in B. The 1.30-kb fragment indicates complete digestion (no methylation) of the PvuII and EcoRI sites. The 1.55- and 1.95-kb fragments indicate partial digestion (partial methylation) of the PvuII site(s). B, DNA gel blot of total DNA (10 μg) digested with EcoRI and PvuII and hybridized with the probe shown in A. Numbers refer to sugarcane transgenics: 125, 120, 17, and 60 are immune (lane I); 32, 35 and 525 are susceptible (lane S). For plants 13 and 16: lanes S and R1 indicate symptomatic and resistant tissue harvested from the same plant before and after recovery; lanes R and I indicate resistant tissue from a different 13 and 16 clone that did not show symptoms upon re-inoculation. Hybridizing bands that correspond to the fragments shown in A are indicated on the left.
RNA Gel-Blot Analysis
Total RNA was extracted from sugarcane leaves as previously described (Jones et al., 1985). RNA samples (15 μg of total RNA or the indicated amounts of poly(A+) RNA) were electrophoresed on a 1.5% (w/v) agarose, 0.45 m formaldehyde, and 20 mm Hepes gel, pH 7.4, transferred to Hybond N+ membranes by downward alkaline blotting (Ingelbrecht et al., 1998), and hybridized with a fluorescein-labeled DNA probe covering the complete SrMV-SCH CP- coding region. Labeling, hybridization, washing, and detection were performed according to the manufacturer's instructions (Gene Images, Amersham). Poly(A+) RNA was isolated using the Oligotex mRNA Midi kit (Qiagen, Chatsworth, CA).
Isolation of Nuclei and Nuclear Run-Off Transcription Assays
Nuclei were isolated from fresh leaf tissue of field-grown sugarcane and the transcription assay and the conditions for prehybridization were as previously described (Ingelbrecht and de Carvalho, 1992). Denatured, double-stranded DNA fragments (approximately 0.25 μg per slot) or single-stranded phagemid-derived DNA (1 μg per slot) or single-stranded in vitro-generated RNA (0.5 μg per slot) were slot blotted onto a nitrocellulose membrane using a microfiltration apparatus (Bio-Rad), according to the manufacturer's instructions. Single-stranded DNA was prepared from phagemids using the helper phage M13KO7, according to standard procedures (Sambrook et al., 1989). In vitro transcription reactions were performed using an in vitro transcription system (Promega) with the following linearized plasmids: pIVING366-1 and -2 digested with NotI for antisense and sense scrbcs-1 sequences respectively; pIVING362-3 and -4 digested with XhoI for antisense and sense CP RNA, respectively; the plasmid SCUBI561 containing a sugarcane polyubiquitin cDNA clone (Albert et al., 1995) was linearized with BamHI and HindIII to produce antisense and sense ubiquitin RNA, respectively.
RESULTS
Transgene Constructs and Plant Transformation
The establishment and maintenance of a collection of seven SCMV strains that currently cause disease in sugarcane throughout the world was previously described (Yang and Mirkov, 1997). This collection includes the SCMV strains A, B, D, and E and the SrMV strains SCH, SCI, and SCM. For each strain, 2 kb of the 3′ terminus, encompassing the complete CP ORF, was sequenced. The nucleotide sequence identity of the CP genes within the strains of each group is more than 95%, whereas the nucleotide sequence identity between the two groups averages approximately 75%.
To engineer resistance in sugarcane, the CP-coding region and 77 nucleotides of the 234 3′UTR nucleotides of SrMV-SCH were fused to the maize ubiquitin-1 promoter with the first exon and intron and the 3′-terminator sequence of the nopaline synthase gene, yielding the plasmid Ubi-hut, illustrated in Figure 1. The Ubi-1 promoter region was previously shown to drive high levels of gene expression in sugarcane (Gallo-Meagher and Irvine, 1993). In potyvirus-infected plants the CP gene product is produced by proteolytic cleavage of a polyprotein, and the CP ORF does not have a translation initiation codon at its 5′-proximal end. In the CP ORF the first start codon is located 17 nucleotides downstream of the CP 5′-proximal end, in a different reading frame from the CP ORF. The first start codon in-frame with the CP ORF is located approximately 313 nucleotides further downstream. Upon translation, this would yield a CP gene product that is truncated by about one-third (111 amino acids) of its normal size. A truncated CP gene product was never detected by ELISA or in western blots (data not shown).
The efficiency of cobombardment is very high in sugarcane; therefore, we used this strategy for generating transgenic plants expressing the Ubi-hut CP gene. Embryogenic callus derived from the commercial sugarcane varieties CP65-357 and CP72-1210 was bombarded with Ubi-hut in combination with Ubi-bar or Ubi-npt and selected on bialaphos- or geneticin-containing medium, respectively (see Methods). Approximately 220 plants were regenerated and transferred to the greenhouse for virus inoculations.
Transgene Integration Patterns
To assess the number of unique transformation events among the 220 plants, DNA gel-blot analysis was performed. Plant DNA was digested with HindIII and hybridized with the SrMV-SCH CP-coding region. In total, 29 different CP hybridization profiles were identified and the transgenics were grouped accordingly in groups numbered 1 through 29. More than one-half of the 220 plants belonged to only three groups and most of these redundant plants were discarded for practical reasons. The CP hybridization pattern of transgenic plants representing 26 groups appears in Figure 2. HindIII-digested DNA from at least one plant of each group was then hybridized to bar and/or npt gene sequences. Because HindIII cuts only once within the plasmid sequences, the copy number of the CP transgenes and the selectable marker genes can be estimated by scoring the number of bands on the DNA gel blots. All bands were scored once, irrespective of size and intensity of hybridization. The estimated gene copy numbers for the 29 groups are summarized in Table I; the plants shown in Figure 2 are listed in this table according to their group number.
Table I.
Summary of transgene copy number in transgenic sugarcane, as determined by hybridization of HindIII-digested DNA to the CP, bar, and npt probes
| Group No. | Copy No.
|
Plant No. | ||
|---|---|---|---|---|
| CP | bar | npt | ||
| bar-hut | ||||
| 1 | 13 | 4 | – | 313 |
| 2 | 7 | 4 | – | 125 |
| 3 | 5 | 5 | – | 120 |
| 4 | 3 | 6 | – | 16 |
| 5 | 2 | 1 | – | 20 |
| 6 | 5 | 2 | – | 322 |
| 7 | 10 | 5 | – | 62 |
| 8 | 6 | 2–5 | – | NRd |
| 9 | 2 | 5 | – | 40 |
| 10 | ≥15 | 5 | – | 18 |
| 11 | ≥15 | 13 | – | 453 |
| 12 | 8 | 4 | – | 494 |
| 13 | 3 | 1–3 | – | 392 |
| 14 | 1 | 1 | – | 319 |
| 15 | 0 | 1 | – | 314 |
| npt-hut | ||||
| 16 | 5 | – | ≥15 | 459 |
| 17 | 4 | – | 6 | 41 |
| 18a | 12 | – | 13 | 5 |
| 19a | 10 | – | 13 | 6 |
| 20 | 4 | – | 2–4 | 35 |
| 21 | 13 | – | ≥15 | 32 |
| 22 | 5 | – | 10 | 460 |
| 23 | 6 | – | 13 | 477 |
| 24b | 13 | – | ≥15 | 467 |
| 25b | 14 | – | ≥15 | NR |
| 26c | 7 | – | 13 | 463 |
| 27c | 5 | – | 8 | NR |
| 28 | 3 | – | 5 | 33 |
| 29 | 8 | – | 13 | 479 |
a,b,c Groups with related transgene integration patterns.
NR, Not represented in the DNA gel blot of Figure 2.
As shown in Table I, the CP transgene copy number varies between 0 and more than 15, with about one-half of the plants having 4 to 9 copies. All plants selected on geneticin had both npt and CP sequences as shown in Figure 2. Insertion of only the bar selectable marker gene occurred once; plant 314 had no CP sequences but contained a single bar insertion. This indicates a high degree of cotransformation, in agreement with previous results on biolistic cotransformation of sugarcane and other monocots such as rice and barley (Wan and Lemaux, 1994; Bower et al., 1996; Qu et al., 1996). Note that plants 5 and 6 have a similar, yet clearly distinct, CP hybridization profile. Comparable observations were made for plants in groups 24 and 25 and also in groups 26 and 27 (data not shown). Plants with highly related transgene integration patterns most likely originated from the same transformation event and might represent chimeras for the CP transgene and/or the selectable marker gene.
Virus-Inoculated Transgenic Plants Display an Immune, Recovery, or Susceptible Phenotype That Is Not Strictly Correlated with Genotype
Transgenic plants were repeatedly inoculated with SrMV-SCH at the four- to eight-leaf stage, as described in Methods. The plants were grown in the greenhouse for 8 to 10 months and then transplanted to the field where they were exposed to natural infection. The incidence of infection of nontransgenic controls in the field was approximately 30%.
Symptom development was periodically monitored by visual inspection and, in selected cases, also by ELISA or RNA gel-blot analysis. Three different responses were observed: an immune, a susceptible, and a recovery phenotype. Immune plants never developed symptoms even though some were inoculated up to six times. Susceptible and recovery plants developed mosaic symptoms 2 to 5 weeks after inoculation on the newly developing leaves. In susceptible plants these symptoms persisted and were most obvious on the younger leaf tissue. The recovery phenomenon was manifested by a gradual reduction in mosaic symptoms in successive leaves until leaves emerged that were completely symptomless and virus free. Resistant tissue from a recovery plant was phenotypically similar to immune tissue. In mature plants these two phenotypes could no longer be distinguished because of senescence of the older, symptomatic leaves in the recovered plants. The characteristic mosaic pattern of an inoculated, susceptible leaf is compared to an immune, symptomless leaf in Figure 3.
Figure 3.
Phenotypes of SrMV-SCH-inoculated transgenic sugarcane plants. Leaves were taken from the same position on each of the plants. S, Susceptible leaf with typical mosaic symptoms; I, immune leaf without symptoms.
The time required for full recovery was variable. For example, recovery occurred relatively fast for plants in group 4; i.e. virus-free leaves appeared approximately 2 months after initial symptom development. However, symptoms persisted for approximately 1 year in plants 18 (group 10) and 313 (group 1) before recovery. Table II summarizes the phenotypes of individual transgenics from 25 different groups. Note that plants with the same transgene integration pattern did not necessarily display the same phenotype, as illustrated in Table II. Groups 2, 4, and 7 contain immune and recovery plants, whereas groups 1, 10, 17, and 20 include recovery and susceptible individuals. Group 6 includes immune, recovery, and susceptible individuals. Resistant plants tend to have an intermediate CP gene copy number, ranging from 4 to 10, whereas the number of CP insertions in most susceptible plants is either lower or higher.
Table II.
Phenotypes of individual transgenic sugarcane plants
| Group No. | Plant No.
|
CP Copy No. | ||
|---|---|---|---|---|
| Immune | Recovery | Susceptible | ||
| 3 | 120, 124 | 5 | ||
| 2 | 125 | 100, 103, 107, 119, 123 | 7 | |
| 4 | 11, 16, 17 | 12, 13, 15, 23 | 3 | |
| 7 | 60, 61, 62 | 3, 4, 38, 39, 57 58, 159 | 10 | |
| 5 | 20 | 2 | ||
| 9 | 40 | 2 | ||
| 22 | 368 | 5 | ||
| 29 | 31 | 8 | ||
| 1 | 313 | 315 | 13 | |
| 10 | 18 | 22 | ≥15 | |
| 17 | 41, 46, 52, 56 | 48, 95 | 4 | |
| 20 | 30 | 25, 35, 36 | 4 | |
| 14 | 319 | 1 | ||
| 15 | 314 | 0 | ||
| 21 | 32 | 13 | ||
| 11 | 543 | ≥15 | ||
| 13 | 392 | 3 | ||
| 18 | 5, 24 | 12 | ||
| 19 | 6 | 10 | ||
| 23 | 359, 376, 469, 475, 476, 477 | 6 | ||
| 24 | 377 | 13 | ||
| 25 | 356 | 14 | ||
| 26 | 463 | 7 | ||
| 28 | 33 | 3 | ||
| 6 | 323 | 399, 424 | 322, 400, 525 | 5 |
Virus Resistance Requires a High Degree of Sequence Similarity
To determine whether virus resistance was strain specific, SrMV-SCH-resistant shoots were reinoculated with SrMV-SCH, with the closely related strains SrMV-SCI and -SCM, or with the more distantly related strain SCMV-D. Initial experiments had indicated that the resistance phenotype of group 4 transgenics, derived from either a recovered or immune plant, was maintained in shoots obtained after vegetative propagation. Stalks from different transformants of this group were cut into two- to four-bud sections and shoots were produced from these cuttings. Twenty-five shoots were sap inoculated with each of these virus strains.
As summarized in Table III, most shoots challenged with the more distantly related strain SCMV-D (22 of 25) developed symptoms. Symptom development was far less frequent after inoculation with the more closely related strains SrMV-SCI or -SCM (7 and 8 of 25, respectively). In SrMV-SCH challenge inoculations, 3 of 25 plants developed symptoms, indicating that the original resistance broke down at low frequency. However, these plants had completely recovered 3 months after inoculation, as demonstrated by RNA gel-blot analysis (data not shown). Symptomatic and recovered tissue was harvested from two of these plants and used for the study of DNA methylation patterns (see below).
Table III.
Response of SrMV-SCH-resistant shoots to challenge inoculations with SrMV and SCMV virus strains
| Inoculated Virus Strain | CP Sequence Identitya | Plants with Symptoms
|
|
|---|---|---|---|
| Transgenicb | Nontransgenic control | ||
| % | no. | ||
| SrMV-SCH | 100 | 3/25 | 8/10 |
| SrMV-SCM | 95 | 8/25 | 5/5 |
| SrMV-SCI | 95 | 7/25 | 3/3 |
| SCMV-D | 75 | 22/25 | 5/5 |
Nucleotide sequence identity between the CP ORF of the transgene and the inoculated virus strain.
SrMV-SCH-resistant transformants from group 4 were vegetatively propagated by cuttings and shoots were sap inoculated with different SrMV and SCMV strains at the three- to five-leaf stage. Symptoms were visually scored 2 months after infection.
Most Resistant Plants Contain Reduced CP Transgene Steady-State mRNA Levels
To determine CP transgene and viral RNA levels in immune and susceptible plants, total RNA was extracted from leaves of selected 1-year-old, field-grown plants. When probed for CP sequences, the symptomatic plants 35, 463, and 525 showed a smear of RNA extending from approximately 10 kb, i.e. the size of the viral genomic RNA, to a few hundred bases, as shown in Figure 4A. This smear of viral RNA, which had been previously observed in potyvirus-infected material (Vance, 1991), prevented detection of the transgenic CP mRNA in the inoculated susceptible plants. As expected, no viral RNA was detected in the immune plants 17, 60, 120, and 323. The level of steady-state CP mRNA in plants 17, 60, and 120 is either below the detection limit or lower than in plant 323, which had a CP mRNA level comparable to that of the noninoculated susceptible plant 463 (Fig. 4B).
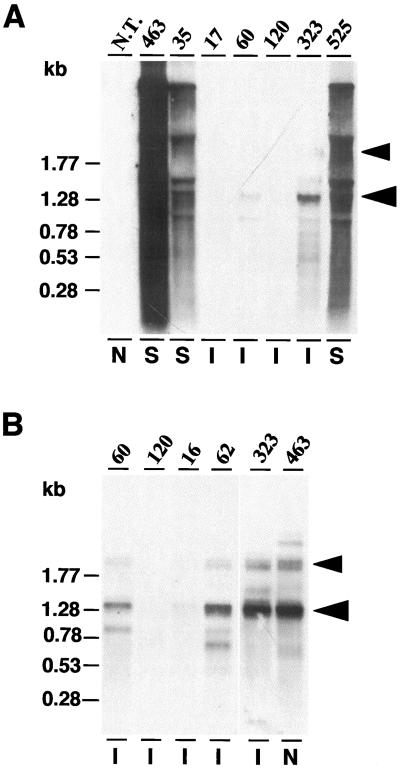
Figure 4.
RNA gel-blot analyses of CP transgenic RNA and viral RNA levels in selected immune and susceptible transgenic sugarcane plants and a nontransgenic control. A, RNA gel blot of total RNA (15 μg per lane) hybridized with a fluorescein-labeled 1.1-kb PstI fragment containing the SrMV-SCH CP ORF and 3′UTR. Numbers on top indicate sugarcane transgenics: N.T. is a nontransgenic, noninoculated (N) control; 463, 35, and 525 are susceptible (S) plants; 17, 60, 120, and 323 are immune (I) plants. RNA size markers are indicated on the left. The larger arrowhead on the right indicates the 1.3-kb CP mRNA; the smaller arrowhead indicates transcripts that cross-hybridize with the ubiquitin intron 1 sequence. B, RNA gel blot of poly(A+) RNA from immune (I) plants 60, 120, 16, 62, and 323 and a noninoculated (N) susceptible control, 463; 0.9 μg of poly(A+) RNA was loaded for plants 60 and 120; 2.5 μg for plants 16 and 62; and 0.8 μg for plants 323 and 463. RNA sizes are indicated on the left. Probes and hybridization were as in A.
The RNA gel blots in Figure 4 show multiple CP RNA transcripts. The 1.3-kb mRNA, indicated with the larger arrowhead, corresponds to the predicted mRNA product of the Ubi-hut construct. The doublet indicated with the smaller arrowhead was observed in all plants with detectable levels of the 1.3-kb CP mRNA, including the noninoculated, susceptible plant 463 (Fig. 4B). These two bands were polyadenylated (Fig. 4B) and cross-hybridized with the ubiquitin intron 1 sequence (data not shown), suggesting that they represent incompletely processed transcripts. Some less-than-full-length, polyadenylated transcripts were specific for the immune plants 60 and 62, as shown in Figure 4B.
CP Transgenes in Resistant and Susceptible Plants Are Actively Transcribed
To compare transcriptional activity of the transgenes in susceptible and resistant plants, nuclei were isolated from the same leaf material that was used for the RNA gel-blot analyses in Figure 4. In vitro run-off transcription reactions were performed, and labeled nRNA was hybridized to slot-blotted DNA containing CP, bar, and npt gene sequences. Transcription along the 2.65-kb vector sequences downstream of the chimeric genes was also measured. The endogenous polyubiquitin genes served as an internal control.
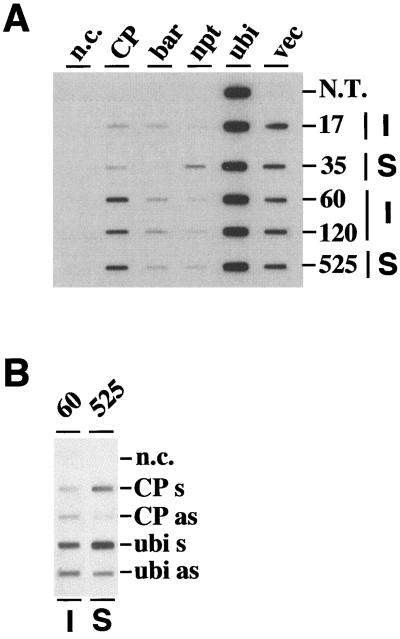
As illustrated in Figure 5A, the CP transgenes were transcribed in all transgenic plants but not in the nontransgenic control. The bar gene was transcribed in plants 17, 60, 120, and 525 and the npt gene was transcribed in plant 35, as expected. Overall, the transcription rates of the CP transgenes correlated well with their estimated copy number (Table I). The immune phenotype was not correlated with a higher CP transcription rate. Together, Figures 4 and 5A show that the CP transgenes in the immune plants 17, 60 and 120 were actively transcribed, whereas their steady-state CP mRNA level was low or below the detection limit. This suggests that posttranscriptional degradation of the CP transcripts occurs, in agreement with a gene-silencing-related, virus-resistance mechanism.
Figure 5.
Analyses of transcription rates for selected immune and susceptible transgenic sugarcane plants and a nontransgenic control. Nuclei were isolated from leaves of 1-year-old, field-grown plants and used in nuclear run-off assays. A, Autoradiogram showing 32P-labeled nRNAs hybridized to DNA fragments containing gene-specific sequences: CP, 1.1-kb coding sequence of the SrMV-SCH CP; bar, 0.58-kb coding sequence of the bar gene; npt, 0.76-kb 3′ part of the npt-coding sequence; ubi, 0.74-kb 5′ part of the SCUBI561 sugarcane polyubiquitin cDNA; vec, the 2.65-kb pUC8 sequences present downstream of the chimeric CP and marker genes; n.c., negative control (no DNA). Numbers on the right refer to plants used as a source of nuclei: 17, 60, and 120 are immune (I); 35 and 525 are susceptible (S) plants; N.T. is a nontransgenic, noninoculated control. B, Autoradiogram showing 32P-labeled nRNAs of the immune plant 60 and the susceptible plant 525 hybridized to in vitro-synthesized single-stranded RNA. Abbreviations are as in A. Sense (s) and antisense (as) refers to the polarity of the nascent RNA.
Initial run-off assays using phagemid-derived single-stranded DNA showed strong hybridization signals with phagemids that did not carry an insert. This “background” hybridization was consistently observed in transgenic plants but not in a nontransgenic control (data not shown). There is sequence similarity between pUC8 and the phagemid, which suggested that the pUC8 vector sequences were actively transcribed. Figure 5A confirms that the 2.65-kb vector sequences present downstream of the transgenes were indeed transcribed at similar rates in all transgenic plants.
In Figure 5B, in vitro-transcribed, single-stranded RNA was bound to the membrane. As shown, both sense and antisense strands of the CP transgenes were transcribed to an equal extent in the immune plant 60. Antisense transcription of the CP transgenes also occurred in the susceptible plant 525 but at a much lower rate. Surprisingly, antisense transcription was also evident for the endogenous ubiquitin genes.
Specific Sites Located in the Transcribed Region of the CP Transgenes Are More Extensively Methylated in Most Resistant Plants
To examine whether virus resistance was correlated with increased methylation of the CP transgenes, we performed DNA gel-blot analyses using total DNA isolated from resistant and susceptible leaf tissue.
This analysis was first performed on isogenic leaf material with and without symptoms. We used two recovered plants obtained after vegetative propagation and reinoculation with SrMV-SCH for this purpose (see above): one was derived from transformant 13, which originally had a recovery phenotype, and one was from transformant 16, which originally was immune. These two plants were showing clear symptoms and accumulated viral CP, as determined by ELISAs 2 months after inoculation. Four weeks later, both plants had developed new, symptomless leaves that were free of viral RNA, as judged by RNA gel-blot analysis (data not shown). Total DNA was isolated from leaf tissue with and without symptoms from these two plants. As a control, DNA was also isolated from two clones that did not develop symptoms upon reinoculation; one clone was derived from plant 13 and one was from plant 16. This DNA was digested with EcoRI and PvuII, both of which are sensitive to cytosine methylation, and probed with a CP gene-specific fragment, illustrated in Figure 6A. Figure 6B indicates that the CP genes are more extensively methylated in recovered (R and R1) or immune (I) tissue than in leaves with symptoms (S), as judged by the presence of the higher-Mr bands in lanes R, I, and R1 versus the S lanes.
We then compared the methylation status of the same sites among independently transformed resistant and susceptible plants. As shown in Figure 6B, most immune plants displayed the 1.55- and/or the 1.95-kb bands in addition to the 1.3-kb band, indicating partial methylation of one or both the PvuII sites. By contrast, only the 1.3-kb band was observed in the sensitive plants and in one immune plant, suggesting that these sites are not detectably methylated in these plants. The origin of the intense 1.8-kb band in lanes 13, 16, 17, and 120 is unclear. It might represent a junction fragment between plant and plasmid DNA or rearranged CP gene sequences. Using the same plant material, we could not detect methylation of the 10 Sau3AI and 4 HpaII sites that were located in the 5′-ubiquitin promoter sequences (data not shown).
DISCUSSION
Resistance to SrMV-SCH was obtained in transgenic sugarcane plants expressing an untranslatable form of the SrMV-SCH CP gene. Several lines of evidence indicate that the underlying resistance mechanism in the investigated clones is related to PTGS.
In general, transgenic sugarcane plants challenged with SrMV-SCH fell into three phenotypic classes: (a) plants that were susceptible and showed symptoms throughout the 2-year monitoring period; (b) recovery plants, i.e. plants that initially showed symptoms but, during further growth, developed symptomless leaves that were virus free; and (c) immune plants that did not show symptoms and were virus free (Table II). The time required for recovery was variable; it was relatively fast in group-4 plants, which were completely virus free 2 to 3 months after initial symptom development, but not in plants 18 and 313, in which symptoms persisted for approximately 1 year. SrMV-SCH-resistant shoots were resistant to challenge inoculations with SrMV-SCH and the closely related SrMV-SCI and -SCM strains (95% nucleotide similarity) but not to inoculation with the more distantly related SCMV strain D (75% nucleotide similarity, Table III). The immune and recovery phenotype and the strain-specific resistance are well-documented characteristics of gene-silencing-related, virus-resistance phenomena in dicots (Lindbo et al., 1993; Smith et al., 1994; Mueller et al., 1995; Tenllado et al., 1995, 1996; Goodwin et al., 1996; Pang et al., 1996; Prins et al., 1996). The inoculation experiments also demonstrated that shoots obtained after vegetative propagation of immune or recovered plants remained resistant. The fact that sugarcane is a vegetatively propagated crop may therefore prove advantageous in maintaining valuable clones, because PTGS can be reversed during meiosis.
In total, 220 transgenic plants were regenerated; they could be classified into 29 different groups based on their CP and marker-transgene-integration patterns (Fig. 1; Table I). Transformants with the same CP transgene integration pattern did not necessarily display the same phenotype upon inoculation. In fact, five groups had both recovered and susceptible individuals (Table II), and one of these groups (group 6) even included an immune individual. It has been previously reported that isogenic transgenic lines can display different levels of resistance upon virus inoculation in dicots (Smith et al., 1994; Sijen et al., 1996). This differential response occurred more frequently when the length of the homologous sequence between the transgene and the inoculated virus was reduced (Sijen et al., 1996). This result is comparable to what we observed in our inoculation studies using different virus strains. When SrMV-SCH resistant plants were infected with a virus strain with decreasing levels of sequence homology, from 100% to 95% to 75%, an increasing number of plants developed symptoms, from 3 to 7 or 8 to 22 (of 25), indicating a positive correlation between the level of resistance and the sequence homology between the CP ORF of the infecting virus and the transgene. This result further suggests that the transition from a susceptible to a resistant phenotype is gradual and continuous. As a whole, we can conclude that the resistance mechanism in sugarcane holds quantitative aspects and can operate with variable efficiency in plants with certain genetic backgrounds.
On RNA gel blots, the susceptible plants 35 and 525 showed a smear of viral RNA that obscured the transgene-derived CP mRNA (Fig. 4). No viral RNA could be detected in the immune plants 17, 60, and 120, and the steady-state transgene CP mRNA level in these plants was undetectable or reduced, compared with the noninoculated susceptible control 463, which is in agreement with a PTGS-related, virus-resistance mechanism (Lindbo et al., 1993; Smith et al., 1994; Mueller et al., 1995; Pang et al., 1996; Sijen et al., 1996). The steady-state CP mRNA level in the immune plant 323 was higher than in the other immune plants and similar to that in the noninoculated susceptible plant 463. Perhaps the resistance mechanism in this plant is not related to PTGS. On the other hand, Tenllado et al. (1995) and Marano and Baulcombe (1998) described putative examples of HDR where resistant plants retained high levels of the homologous transgenic mRNA. RNA gel-blot analyses revealed a number of transcripts in addition to the expected 1.3-kb CP mRNA. Two polyadenylated transcripts of approximately 2.4 kb were observed in all plants with detectable levels of the 1.3-kb CP mRNA. These bands cross-hybridize with the Ubi-1 intron sequence, suggesting that they represent incompletely processed CP transcripts. Because these bands also occur in the noninoculated susceptible control 463, they are not related to transgene silencing. Some less than full-length, polyadenylated bands were detected in the immune plants 60 and 62 but not in the noninoculated, susceptible control 463 (Fig. 4B). Truncated transcripts have previously been shown in other cases of HDR (Goodwin et al., 1996; Tanzer et al., 1997) and may represent intermediates of the RNA-degradation mechanism.
The immune plants 17, 60, and 120 having low or undetectable CP steady-state mRNA levels display active transcription of the CP transgenes in the nucleus, as shown in nuclear run-off assays (Fig. 5). This result suggests that the CP mRNA level in these plants is down-regulated by a posttranscriptional mechanism, as expected for HDR (Lindbo et al., 1993; Mueller et al., 1995; Prins et al., 1996; Sijen et al., 1996). There was no obvious correlation between transcription rates of the CP transgenes and an immune or susceptible phenotype. In the immune plant 60, both sense and antisense transcription of the CP transgenes occurred at similar rates. Antisense transcription was also detected for the CP genes of the susceptible plant 525 but at a much lower rate than sense transcription. Most surprisingly, we also detected antisense transcription for the endogenous ubiquitin genes. Although highly unexpected, this finding might not be unprecedented, because a previous report (Christensen and Quail, 1989) described antisense transcription for the maize ubiquitin genes and possibly also for maize rDNA. Whether antisense transcription of these host genes should be considered background, as suggested by these authors, or indicates a true event is not clear at present. Finally, the run-off assays clearly showed that the pUC8 vector sequences located downstream of the CP and/or marker genes were actively transcribed in all transgenic plants. Fortuitous transcription of vector sequences might result from transcriptional readthrough originating from the transgenic Ubi-1 promoter or from endogenous plant promoters that flank the integrated transgenes. The presence of truncated or rearranged plasmid sequences, often observed in biolistic transformation, possibly enhanced this unexpected transcription.
A correlation between methylation of transgenes in the transcribed region and resistance has been found in several examples of HDR in dicots (Smith et al., 1994; English et al., 1996; Sijen et al., 1996) as well as in our study. Using leaf material with and without symptoms isolated from the same plant before and after recovery, we could demonstrate that specific sites located in the coding region of the CP transgenes were more extensively methylated in resistant tissue (Fig. 6). Methylation of the CP transgenes was most pronounced in tissue that had just recovered, suggesting that DNA methylation occurs primarily during the initial establishment of resistance. When comparing independently transformed susceptible and resistant plants, increased methylation of these sites was found in most but not all resistant plants. We could not detect clear methylation of sites located in the Ubi-1 promoter in resistant or susceptible tissue.
Various models have been proposed to explain PTGS and how it is involved in virus resistance, including the RNA threshold model (for review, see Dougherty and Parks, 1995) and the ectopic pairing model (for review, see Baulcombe and English, 1996). In essence, these models propose that enhanced turnover of transgene and viral RNA is mediated by cRNA molecules that are synthesized by a plant-encoded, RNA-dependent RNA polymerase. The trigger for turnover is unknown, but Dougherty and Parks (1995) proposed that this cRNA is synthesized in response to the presence of overexpressed RNA molecules. According to the ectopic pairing model, specific DNA-DNA interactions result in the formation of aberrant RNA, which is copied into cRNA and leads to the degradation of homologous RNA molecules. Recently, Waterhouse et al. (1998) proposed an interesting variant of these models. These authors suggested that PTGS is induced by double-stranded RNA, which serves as a template for the production of cRNA by the RNA-dependent RNA polymerase. Our run-off data do not support a model that is based strictly on exceeding a critical level of RNA because the transcription rate of the CP transgenes in immune plants is not always higher than in susceptible transgenics. As mentioned, in the immune plant 60, the CP transgenes were transcribed in both directions at similar rates. This could potentially lead to the formation of double-stranded RNA, consistent with the model of Waterhouse et al. (1998). However, we could not detect CP antisense transcripts in total RNA preparations from this plant, as judged by RNA gel-blot analysis (data not shown).
In a recent review of epigenetic transgene silencing, Matzke and Matzke (1998) pointed out that the genomes of agriculturally important cereal crops are often substantially more complex than that of Arabidopsis or of the Nicotiana species, which are generally used in these studies. These authors then raised the issue whether or not the variations in base composition between dicots and monocots could be responsible for different types of gene silencing. Sugarcane has a particularly complex genome, because it is an allopolyploid with variable chromosome numbers and ploidy levels. Despite these differences, we have also recognized the hallmarks of PTGS and HDR in dicot plants in our study. Our data provide evidence that an ancestral pathway for posttranscriptional gene regulation and RNA surveillance is conserved between monocots and dicots. This is consistent with reports of related gene-silencing mechanisms in fungi (for review, see Cogoni and Macino, 1997), Paramecium (Ruiz et al., 1998), and Caenorhabditis elegans (Fire et al., 1998). Still, we are only beginning to understand these phenomena; studies at the molecular level may reveal differences between PTGS and HDR in monocots versus dicots. The recent identification of a viral suppressor of PTGS in tobacco etch virus offers approaches to directly address these questions. From a more applied perspective, our data demonstrate the feasibility of this approach to engineering virus resistance in this agronomically important group of plants.
ACKNOWLEDGMENTS
We would like to thank Josefina Bustamante, Mercedes Campos, Teresa De La Garza, and Eduardo Hernandez for plant cell tissue culture and plant maintenance. We are appreciative to Dr. F. Moonan for helpful discussions throughout this work and to Dr. Z.N. Yang for help with constructing the plasmids Ubi-npt and Ubi-hut. We are grateful to Dr. H. Albert and Dr. W. Tang for providing the plasmids SCUBI561 and scrbcs-1.
Abbreviations:
- CP
coat protein
- cRNA
complementary RNA
- HDR
homology-dependent resistance
- ORF
open reading frame
- PTGS
posttranscriptional gene silencing
- SCMV
sugarcane mosaic virus
- SrMV
sorghum mosaic virus
- 3′UTR
3′-untranslated region
Footnotes
This work was supported by the Texas Higher Education Coordinating Board Advanced Technology Program (grant nos. 999902-029 and 999902-188) and a grant from the International Consortium for Sugarcane Biotechnology.
LITERATURE CITED
- Albert HH, Carr JB, Moore PM. Nucleotide sequence of a sugarcane polyubiquitin cDNA (accession no. L41658) (PGR 95-045) Plant Physiol. 1995;109:337. [Google Scholar]
- Al-Kaff NS, Covey SN, Kreike MM, Page AM, Pinder R, Dale PJ. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science. 1998;279:2113–2115. doi: 10.1126/science.279.5359.2113. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB. A viral suppressor of gene silencing in plants. Proc Natl Acad Sci USA. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arencibia A, Molina P, de la Riva G, Selman-Housein G. Production of transgenic sugarcane (Saccharum officinarumL.) plants by intact cell electroporation. Plant Cell Rep. 1995;14:305–309. doi: 10.1007/BF00232033. [DOI] [PubMed] [Google Scholar]
- Baulcombe DC. Mechanisms of pathogen-derived resistance to viruses in transgenic plants. Plant Cell. 1996;8:1833–1844. doi: 10.1105/tpc.8.10.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe DC, English JJ. Ectopic pairing of homologous DNA and post-transcriptional gene silencing in transgenic plants. Curr Opin Biotechnol. 1996;7:173–180. [Google Scholar]
- Bower R, Birch RG. Transgenic sugarcane plants via microprojectile bombardment. Plant J. 1992;2:409–416. [Google Scholar]
- Bower R, Elliott AR, Potier BAM, Birch RG. High-efficiency, microprojectile-mediated co-transformation of sugarcane, using visible or selectable markers. Mol Breed. 1996;2:239–249. [Google Scholar]
- Burner DM, Legendre BL. Cytogenetic and fertility characteristics of elite sugarcane clones. Sugar Cane. 1994;1:6–10. [Google Scholar]
- Christensen AH, Quail PH. Sequence analysis and transcriptional regulation by heat shock of polyubiquitin transcripts from maize. Plant Mol Biol. 1989;12:619–632. doi: 10.1007/BF00044153. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Conservation of transgene-induced post-transcriptional gene silencing in plants and fungi. Trends Plant Sci. 1997;11:438–443. [Google Scholar]
- Converse RH, Martin RR (1990) ELISA methods for plant viruses. In R Hamptom, E Ball, S De Boer, eds, Serological Methods for Detection and Identification of Viral and Bacterial Plant Pathogens. APS Press, St. Paul, MN, pp 179–196
- Covey SN, Al-Kaff NS, Lángara A, Turner DS. Plants combat infection by gene silencing. Nature. 1997;385:781–782. [Google Scholar]
- Depicker A, Van Montagu M. Post-transcriptional gene silencing in plants. Curr Opin Cell Biol. 1997;9:373–382. doi: 10.1016/s0955-0674(97)80010-5. [DOI] [PubMed] [Google Scholar]
- Dougherty WG, Parks TD. Transgenes and gene suppression: telling us something new? Curr Opin Cell Biol. 1995;7:399–405. doi: 10.1016/0955-0674(95)80096-4. [DOI] [PubMed] [Google Scholar]
- English JJ, Davenport GF, Elmayan TE, Vaucheret H, Baulcombe DC. Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J. 1997;12:597–603. [Google Scholar]
- English JJ, Mueller E, Baulcombe DC. Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell. 1996;8:179–188. doi: 10.1105/tpc.8.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gallo-Meagher M, Irvine JE. Effects of tissue type and promoter strength on transient GUS expression in sugarcane following particle bombardment. Plant Cell Rep. 1993;12:666–670. doi: 10.1007/BF00233416. [DOI] [PubMed] [Google Scholar]
- Gallo-Meagher M, Irvine JE. Herbicide resistant transgenic sugarcane plants containing the bargene. Crop Sci. 1996;36:1367–1374. [Google Scholar]
- Goodwin J, Chapman K, Swaney S, Parks TD, Wernsman EA, Dougherty WG. Genetic and biochemical dissection of transgenic RNA-mediated virus resistance. Plant Cell. 1996;8:95–105. doi: 10.1105/tpc.8.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelbrecht I, de Carvalho F (1992) Isolation of nuclei and in vitro run-off transcription. In D Inzé, D Van Der Straeten, M Van Montagu, eds, Practical Course on Plant Molecular Biology. EMBO, Laboratorium voor Genetica, Gent, Belgium, pp 117–132
- Ingelbrecht IL, Mandelbaum CI, Mirkov TE. Highly sensitive northern hybridization using a rapid protocol for downward alkaline blotting of RNA. BioTechniques. 1998;25:420–425. doi: 10.2144/98253st03. [DOI] [PubMed] [Google Scholar]
- Irvine JE (1999) Saccharum species as horticultural classes. Theor Appl Genet (in press)
- Jones JDG, Dunsmuir P, Bedbrook J. High level expression of introduced chimeric genes in regenerated transformed plants. EMBO J. 1985;4:2411–2418. doi: 10.1002/j.1460-2075.1985.tb03949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- Koetsier PA, Schorr J, Doerfler W. A rapid optimized protocol for downward alkaline southern blotting of DNA. BioTechniques. 1993;15:260–262. [PubMed] [Google Scholar]
- Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG. Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell. 1993;5:1749–1759. doi: 10.1105/tpc.5.12.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marano MR, Baulcombe DC. Pathogen-derived resistance targeted against the negative-strand RNA of tobacco mosaic virus: RNA strand-specific gene silencing? Plant J. 1998;13:537–546. [Google Scholar]
- Matzke MA, Matzke AJM. Epigenetic silencing of plant transgenes as a consequence of diverse cellular defense responses. CMLS Cell Mol Life Sci. 1998;54:94–103. doi: 10.1007/s000180050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller E, Gilbert J, Davenport G, Brigneti G, Baulcombe DC. Homology-dependent resistance: transgenic virus resistance in plants related to homology-dependent gene silencing. Plant J. 1995;7:1001–1013. [Google Scholar]
- Palauqui J-C, Elmayan T, Pollien J-M, Vaucheret H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang SZ, Jan F-J, Carney K, Stout J, Tricoli DM, Quemada HD, Gonsalves D. Post-transcriptional transgene silencing and consequent tospovirus resistance in transgenic lettuce are affected by transgene dosage and plant development. Plant J. 1996;9:899–909. [Google Scholar]
- Prins M, Resende RD, Anker C, van Schepen A, de Haan P, Goldbach R. Microbe Interact. 1996;9:416–418. doi: 10.1094/mpmi-9-0416. [DOI] [PubMed] [Google Scholar]
- Qu R, De Kochko A, Zhang L, Marmey P, Li L, Tian W, Zhang S, Fauquet CM, Beachy RN. Analysis of a large number of independent transgenic rice plants produced by the biolistic method. In Vitro Cell Dev Biol Plant. 1996;32:233–240. [Google Scholar]
- Ratcliff F, Harrison BD, Baulcombe DC. A similarity between viral defense and gene silencing in plants. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- Ruiz F, Vayssié L, Klotz C, Sperling L, Madeddu L. Homology-dependent gene silencing in Paramecium. Mol Biol Cell. 1998;9:931–943. doi: 10.1091/mbc.9.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shukla DD, Ward CW, Brunt AA (1994) The sugarcane mosaic virus subgroup. In The Potyviridae. CAB International, Wallingford, UK, pp 360–371
- Sijen T, Wellink J, Hiriart J-B, van Kammen A. RNA-mediated virus resistance: role of repeated transgenes and delineation of targeted regions. Plant Cell. 1996;8:2277–2294. doi: 10.1105/tpc.8.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HA, Swaney SL, Parks TD, Wernsman EA, Dougherty WG. Transgenic plant virus resistance mediated by untranslatable sense RNAs: expression, regulation, and fate of nonessential RNAs. Plant Cell. 1994;6:1441–1453. doi: 10.1105/tpc.6.10.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M, Mol JNM, Kooter JM. The silence of genes in transgenic plants. Ann Bot. 1997;79:3–12. [Google Scholar]
- Sugar and Sweetener–Situation and Outlook Yearbook (1997) U.S. Department of Agriculture, Washington, DC
- Tai TH, Tanksley SD. A rapid and inexpensive method for isolation of total DNA from dehydrated plant tissue. Plant Mol Biol Rep. 1990;8:297–303. [Google Scholar]
- Tang W, Sun SSM. Sequence of a sugarcane ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit gene. Plant Mol Biol. 1993;21:949–951. doi: 10.1007/BF00027128. [DOI] [PubMed] [Google Scholar]
- Tanzer MM, Thompson WF, Law MD, Wernsman EA, Uknes S. Characterization of post-transcriptionally suppressed transgene expression that confers resistance to tobacco etch virus infection in tobacco. Plant Cell. 1997;9:1411–1423. doi: 10.1105/tpc.9.8.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenllado F, García-Luque I, Serra MT, Díaz-Ruíz JR. Nicotiana benthamianaplants transformed with the 54-kDa region of the pepper mild mottle tobamovirus replicase gene exhibit two types of resistance responses against viral infection. Virology. 1995;211:170–183. doi: 10.1006/viro.1995.1389. [DOI] [PubMed] [Google Scholar]
- Tenllado F, García-Luque I, Serra MT, Díaz-Ruíz JR. Resistance to pepper mild mottle tobamovirus conferred by the 54-kDa gene sequence in transgenic plants does not require expression of the wild-type 54-kDa protein. Virology. 1996;219:330–335. doi: 10.1006/viro.1996.0257. [DOI] [PubMed] [Google Scholar]
- Vance VB. Replication of potato virus X is altered in coinfections with potato virus Y. Virology. 1991;182:486–494. doi: 10.1016/0042-6822(91)90589-4. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC. Systemic signalling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- Wan Y, Lemaux PG. Generation of large numbers of independently transformed fertile barley plants. Plant Physiol. 1994;104:37–48. doi: 10.1104/pp.104.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang M-B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZN, Mirkov TE. Sequence and relationships of sugarcane mosaic and sorghum mosaic virus strains and development of RT-PCR-based RFLPs for strain discrimination. Phytopathology. 1997;87:932–939. doi: 10.1094/PHYTO.1997.87.9.932. [DOI] [PubMed] [Google Scholar]