Abstract
Background
An Italian interlaboratory study was run in 2010 to assess the performance of Blood Transfusion Services in detecting the genome of West Nile virus (WNV) in plasma.
Materials and methods
Each laboratory received a panel of samples containing four samples negative for WNV and six positive samples with a nominal viral concentration close to or below the 95% detection limit of two commercially available nucleic acid amplification tests (NAT) for WNV, the PROCLEIX® WNV kit and the Cobas® TaqScreen West Nile Virus kit.
Results
Ten laboratories took part in the study. All correctly identified the positive samples with a viral concentration above the 95% detection limit. No pre- or post-analytical errors were observed.
Conclusions
The interlaboratory study run in 2010 allowed participants to assess the performance of the NAT methods applied in their seasonal routine screening of blood donations.
Keywords: WNV-RNA, interlaboratory study, NAT, quality
Introduction
West Nile virus (WNV), an arbovirus belonging to the Flaviviridae family, is transmitted by the bite of an infected mosquito. The virus infects many species of birds, horses and other mammals, including humans, as incidental hosts in which infections are mainly (80%) asymptomatic1. Symptomatic WNV infections range from a mild febrile syndrome termed West Nile fever to meningo-encephalitis, and possibly death. The risk of this disease increases with age and appears to be significantly higher in immunocompromised individuals, especially in organ transplant recipients.
Over the past 10 years the diffusion of WNV has been reported in several European countries indicating that the virus is among the emerging threats for both human and veterinary public health in Europe2. WNV infection has become an important topic in relation to blood, tissue and organ safety following several reports of contaminated blood supplies and organs during the large WNV outbreak in the United States3–4. In Italy, after the reports of the first two cases of human WNV neuroinvasive infection and taking into consideration data from epidemiological surveillance on horses and wild birds, in September 2008 the following precautionary measures were adopted for blood safety5: deferral for 28 days of donors who have spent at least one night in affected areas and introduction of WNV RNA screening by nucleic acid amplification testing (NAT) of all blood donations from donors living in the areas of Bologna and Ferrara. On the basis of human and animal surveillance activities that confirmed an intense circulation of WNV in the area of the Po river and the risk estimates obtained applying a mathematical model6, testing of donations for WNV RNA was implemented for all donations from donors resident in the interested areas during the summer of 2009 and the summer of 2010 (from July 15 to November 15). Two CE-marked commercial assays, the Cobas® TaqScreen West Nile Virus kit (Roche Molecular Systems, Branchburg, NJ, USA) and the PROCLEIX® WNV kit (Gen-Probe Inc., San Diego, CA, USA) were used for routine donor screening which was done on mini-pools of six donations and on single donations, respectively.
In 2010, the National Blood Centre and the National Centre for Research and Evaluation of Immunobiological Products (CRIVIB) of the Italian Institute of Health (ISS) organised an interlaboratory study of diagnostic methods for the detection of WNV-RNA by NAT aimed at providing all participant testing laboratories a valuable way to monitor the quality of their analytical performance and competence of their operators.
Materials and methods
Participants
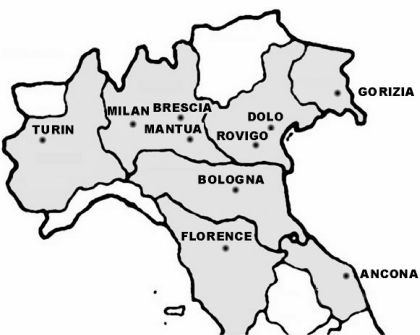
The interlaboratory study involved seven blood banks in the Regions of Veneto, Emilia Romagna and Lombardy which performed WNV RNA screening routinely and four additional blood banks that were ready to start NAT testing in the case of extension of the epidemiological area of WNV circulation (Tuscany, Marche and Friuli Venezia Giulia and Piedmont) (Figure 1).
Figure 1.
Geographical distribution of the blood bank laboratories participating in the WNV interlaboratory study.
All participating laboratories were asked to complete an agreement form and a questionnaire regarding the NAT methods applied. A code was assigned to each laboratory: the complete list is reported in appendix 1.
Nucleic acid amplification techniques
All participants used commercial NAT assays for the detection of WNV RNA in plasma. These assays were based on transcription-mediated amplification (PROCLEIX® WNV kit; Gen-Probe Inc., San Diego, CA, USA) (TMA) or on real-time polymerase chain reaction (Cobas® TaqScreen West Nile Virus kit; Roche Molecular System, Branchburg, NJ, USA) (CTS).
Both assays were validated by the test kits’ manufacturers for analytical sensitivity, expressed as the 95% detection limit, using dilution series of reference material: 10 copies/mL for TMA and 40 copies/mL for CTS (as reported in the respective Product Inserts).
Among the 11 laboratories that participated in this study, five used the TMA assay, five used the CTS assay and one used both TMA and CTS assays.
Negative and positive samples
The negative samples were prepared using a plasma pool made up of 25 donations that had tested negative for hepatitis C virus, hepatitis B virus, human immunodeficiency virus and WNV by serological and NAT tests. The positive samples were obtained by spiking this negative plasma pool with two Italian reference preparations of WNV RNA, both of which were heat inactivated:
- WNV-RNA Ref. Prep. batch ISS 0109 obtained by diluting a secondary standard of WNV (supplied by Roche) in negative plasma. The starting material has the same lineage (lineage 1) as the Health Canada Reference Reagent (HC-SC WNV Nat Ref 001/03) and its assigned titre is directly traceable to the Canadian Reference Reagent7. The ISS 0109 batch has a final concentration of 1000 copies/mL of WNV RNA.
- WNV-RNA Ref. Prep. batch ISS 0710 was obtained by diluting an “Italian strain” of WNV RNA, lineage 18 (kindly provided by V. Sambri, Bologna) in negative plasma. The preparation has a concentration of about 135 copies/mL of WNV RNA when calibrated against the secondary standard of WNV (see below) or about 274 copies/mL as determined by quantitative polymerase chain reaction analysis (WNV LightMix, TimBiol).
At the time this interlaboratory study was performed, a definitive viral concentration had not yet been assigned to this reference preparation, so the results obtained from participants regarding this material were not used for the determination of the outcome (see study design).
West Nile virus panels
Two panels of five vials each were designed. Panel A consisted of two negative plasma samples and three WNV-positive plasma samples made from half-log dilutions of WNV RNA Ref. Prep. batch ISS0109 in negative plasma (1:3.16, 1:10 and 1:31.6 corresponding to 316, 100 and 31.6 copies/mL, respectively). Panel B also consisted of two negative plasma samples and three WNV positive plasma samples, in this case made from half-log dilutions of WNV RNA Ref. Prep. batch ISS0710 in negative plasma (1:6.3, 1:20 and 1:63 corresponding to 21–43, 14-7 and 4-2 copies/mL respectively).
Negative and positive samples were distributed in frozen aliquots of 3.0 mL. Before distribution to participants, the panels were tested by an external laboratory using the TMA assay in order to confirm the positivity/negativity of the samples.
Study design
Participants were asked to test panels A and B in two different runs using their routine NAT assay for WNV RNA, without further dilution of the samples and testing each sample singly. It was requested that results be reported in a qualitative way as either positive i.e. WNV RNA detected or negative i.e. WNV RNA was not detected.
Data sheets and a method form were provided to participants so that all the relevant information could be recorded.
On completion of the study, participants had to fax the data sheet to the ISS. Within 24–48 hours, participants were informed via e-mail about the correctness of their results.
In order to evaluate the outcome (favourable or not favourable) of each individual laboratory the following proficiency criteria were adopted. First, the two samples of panel A (codes 2 and 4) containing 316 and 100 copies/mL of WNV RNA respectively, had to be correctly detected as positive because the viral concentrations were well within the 95% detection limit of both commercial assays. Second, no false-positive results were allowed for the four negative samples of panels A and B (codes 1, 3, 8 and 10).
The other positive samples in both panels with viral concentrations at or below the 95% detection limit of either or both methods (samples 5, 6, 7 and 9) were not used to determine the outcome of the participating laboratories. In fact participants could miss the target virus as a consequence of its random distribution in the plasma.
A “general” assessment of the performance of each laboratory was made by assigning the following scores: +0.5 for correctly identified negative samples and positive samples at 316 and 100 copies/mL; +1.0 for positive samples at 31 and 22–46 copies/mL; +2 for positive samples at 7–14 copies/mL; and +3 for positive samples at 2–4 copies/mL.
In the case of an incorrectly identified sample, a score of −3 was assigned to negative samples and positive samples at 316 and 100 copies/mL. For the remaining positive samples with lower viral loads, the score assigned was “0” (zero) when not correctly identified.
In the case of a discrepancy between the value reported by the participant and the expected result, the ISS worked closely with the reporting laboratory in order to find the root cause of the error and to determine whether it was an error in the pre-analytical phase (e.g. exchange of samples), an error in the post-analytical phase (e.g. interpretation of results, transcription error) or an unspecified analytical error.
Shipment
Samples were shipped on dry ice and participants were asked to check the integrity of the parcel, the presence of dry ice and the status of the samples immediately upon receipt. This information was faxed to the ISS using the acknowledgement of receipt sheet. Each participant also completed a responsibility sheet to acknowledge that the samples received were potentially infectious.
Results
The interlaboratory study started in August 2010 with the distribution of 12 panels A and 12 panels B to 11 participating laboratories (five used TMA, five used CTS and one used both TMA and CTS).
Applying the evaluation criteria reported in the study design, all 11 participating laboratories (100%) passed the minimum requirements (favourable) for successful participation (Table I).
Table I.
Overall results and scores assigned to each sample/participant.
| Panel A | Panel B | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID sample | 2 | 4 | 5 | 1 | 3 | 9 | 7 | 6 | 8 | 10 | |
| Copies/mL | 316 | 100 | 31 | Negative | Negative | 22–46 | 7–14 | 2–4 | Negative | Negative | |
| Score* | +0.5 | +0.5 | +1.0 | +0.5 | +0.5 | +1.0 | +2.0 | +3.0 | +0.5 | +0.5 | |
| Lab code | Method | Panel A | Panel B | Total Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | TMA | POS | POS | POS | neg | neg | POS | POS | neg | neg | neg | 7.0 |
| 15 | TMA | POS | POS | POS | neg | neg | POS | POS | POS | neg | neg | 10.0 |
| 27 | TMA | POS | POS | POS | neg | neg | POS | POS | neg | neg | neg | 7.0 |
| 72 | TMA | POS | POS | POS | neg | neg | POS | POS | POS | neg | neg | 10.0 |
| 98 | TMA | POS | POS | POS | neg | neg | POS | neg | POS | neg | neg | 8.0 |
| 130 | TMA | POS | POS | POS | neg | neg | POS | POS | POS | neg | neg | 10.0 |
| 20 | CTS | POS | POS | POS | neg | neg | POS | neg | neg | neg | neg | 5.0 |
| 44 | CTS | POS | POS | POS | neg | neg | POS | neg | POS | neg | neg | 8.0 |
| 46 | CTS | POS | POS | POS | neg | neg | POS | POS | neg | neg | neg | 7.0 |
| 51 | CTS | POS | POS | neg | neg | neg | Neg | neg | neg | neg | neg | 3.0 |
| 129 | CTS | POS | POS | POS | neg | neg | POS | POS | neg | neg | neg | 7.0 |
| 130 | CTS | POS | POS | POS | neg | neg | POS | neg | neg | neg | neg | 5.0 |
| Overall result positive/tested | 12/12 | 12/12 | 11/12 | 0/12 | 0/12 | 11/12 | 7/12 | 5/12 | 0/12 | 0/12 | ||
see the study design section
In detail, with regard to panel A, all participants correctly identified samples 2 and 4 with concentrations of about 316 and 100 copies/mL of WNV RNA, respectively. Sample 5, containing 31 copies/mL was identified correctly by all laboratories with the exception of laboratory 51 (CTS method). This result is expected considering the 95% detection limit of the CTS assay.
With regard to panel B, containing dilutions of an “Italian strain” of WNV, all laboratories except one (laboratory 51) correctly identified sample 9 with a concentration of about 22–46 copies/mL of WNV RNA. Sample 7, with a concentration of about 7–14 copies/mL of WNV RNA, was identified as positive in five out of the six laboratories using the TMA assay and in two out of the six laboratories using the CTS test. Finally, sample 6 with a concentration of about 2–4 copies/mL of WNV RNA was identified as positive in four out of the six laboratories using the TMA assay and in one out of the six laboratories using the CTS assay. From retrospective analysis, participants that failed to detect WNV-positive samples were not able to identify any deviation in the laboratory’s procedure. Considering the low level of viral target in these WNV-positive samples, the false negative results could be attributed to the 95% detection limit of the assays.
All laboratories correctly identified the negative samples (1, 3, 8 and 10).
With regards to the “performance” of laboratories, three laboratories using the TMA assay (15, 72 and 130) reported an excellent overall result obtaining the highest score, six laboratories, three TMA users and three CTS users (11, 27, 44, 46, 98 and 129), reported a good result with a total score between 7 and 8, while three laboratories that used the CTS test achieved a score between 3 and 5. These results, obtained on a limited number of tests, reflected the difference in analytical sensitivity between the two kits.
Discussion
Participation in interlaboratories studies, in addition to being a requirement for the accreditation and certification of laboratories, is certainly a very good way for a laboratory to verify the correctness of its analytical results and to compare its performance with that of other laboratories. In fact, through the evaluation of the results obtained in such studies, a laboratory can evaluate the effectiveness of its quality system, estimate the ability of analytical procedures (in terms of methods, equipment and reagents) to generate results that meet the parameters and the technical specifications of the assay, and, last but not least, detect any possible weakness in the pre- and post-analytical steps of the process.
This article presents the results obtained during an Italian interlaboratory study organised by the National Blood Centre and CRIVIB in 2010 for blood transfusion centres that screen blood donations for WNV RNA. Two WNV samples, both of lineage 1, were used: a preparation with a well-defined viral concentration expressed in copies/mL and traceable to the Health Canada reagent and a sample of WNV isolated from an Italian patient which had not been completely characterised at the time of the present study. For this reason, the favourable or not favourable outcome assigned to each participant was based only on the samples with viral concentrations of 316 and 100 copies/mL of WNV and on the negative samples.
Based on the design of the study, all participants conducted the interlaboratory study with favourable results, correctly identifying negative and positive samples. The observed differences in performance between the two NAT assays in terms of the probability of detecting low level WNV RNA samples reflect their 95% detection limits stated by the manufactures. However, given the limited number of samples used in this study - only two WNV strains, both of lineage 1 - the results of this study do not provide enough evidence for Health Authorities to issue specific indications concerning the choice of NAT methods. Additional studies including different well-characterised WNV strains, possibly of both lineages 1 and 2, are needed to get more in-depth data about the sensitivity of current WNV NAT assays and their ability to detect low-level viraemic donations. The establishment of well-characterised and universally accepted reference materials is essential for this purpose.
Appendix 1. Participants of the interlaboratory study
- G. Salvoni, AOU “Ospedali Riuniti” Torrette di Ancona
- W. Marani, Ospedale Maggiore C.A. Pizzardi, Bologna
- A. Pierro AOU, S. Orsola-Malpighi. Bologna
- M. Statuto, AO Spedali Civili, Brescia
- M. Stecchina, Ospedale San Giovanni di Dio, Gorizia
- G. Gesu/D. Fanti AO Niguarda Ca’ Granda, Milano
- C. Peduzzi, AOU Careggi, Firenze
- P. Barin, Ospedale Civile, Dolo
- P. Ghiazza, AO. OIRMS- Sant’Anna, Torino
- P. Vecchi/M. De Maria, AO Policlinico, Modena
- R. Potenza/S. Bellini, Ospedale Civile, Rovigo
References
- 1.Petersen LR, Marfin AA, Gubler DJ. West Nile virus. JAMA. 2003;290:524–8. doi: 10.1001/jama.290.4.524. [DOI] [PubMed] [Google Scholar]
- 2.Sabirovic M, Roberts H, Lopez M, et al. International disease monitoring, October to December 2009. Veterinary Record. 2010;166:160–2. doi: 10.1136/vr.c569. [DOI] [PubMed] [Google Scholar]
- 3.Pealer LN, Marfin AA, Petersen LR, et al. Transmission of West Nile virus through blood transfusion in the United States in 2002. N Engl J Med. 2003;349:1236–45. doi: 10.1056/NEJMoa030969. [DOI] [PubMed] [Google Scholar]
- 4.Iwamoto M, Jernigan DB, Guasch A, et al. Transmission of West Nile virus from an organ donor to four transplant recipients. N Engl J Med. 2003;348:2196–203. doi: 10.1056/NEJMoa022987. [DOI] [PubMed] [Google Scholar]
- 5.Grazzini G, Liumbruno GM, Pupella S, et al. West Nile virus in Italy: a further threat to blood safety, a further challenge to the blood system. Blood Transf. 2008;6:235–7. doi: 10.2450/2008.0065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biggerstaff BJ, Petersen LR. A modeling framework for evaluation and comparison of trigger strategies for switching from minipool to individual-donation testing for West Nile virus. Transfusion. 2009;49:1151–9. doi: 10.1111/j.1537-2995.2009.02112.x. [DOI] [PubMed] [Google Scholar]
- 7.Saldanha J, Shead S, Heath A, et al. Collaborative study to evaluate a working reagent for West Nile virus RNA detection by nucleic acid testing. Transfusion. 2005;45:97–102. doi: 10.1111/j.1537-2995.2005.04151.x. [DOI] [PubMed] [Google Scholar]
- 8.Rossini G, Carletti F, Bordi L, et al. Phylogenetic analysis of West Nile virus isolates, Italy, 2008–2009. Emerg Infect Dis [serial on the Internet] 2011 May; doi: 10.3201/eid1705.101569. [DOI] [PMC free article] [PubMed] [Google Scholar]