Abstract
The human cytochrome P450 (CYP450) isozymes are the most important enzymes in the body to metabolize many endogenous and exogenous substances including environmental toxins and therapeutic drugs. Any unnecessary interactions between a small molecule and CYP450 isozymes may raise a potential to disarm the integrity of the protection. Accurately predicting the potential interactions between a small molecule and CYP450 isozymes is highly desirable for assessing the metabolic stability and toxicity of the molecule. The National Institutes of Health Chemical Genomics Center (NCGC) has screened a collection of over seventeen thousand compounds against the five major isozymes of CYP450 (1A2, 2C9, 2C19, 2D6 and 3A4) in a quantitative high throughput screening (qHTS) format. In this study, we developed support vector classification (SVC) models for these five isozymes using a set of customized generic atom types. The CYP450 datasets were randomly split into equal-sized training and test sets. The optimized SVC models exhibited high predictive power against the test sets for all five CYP450 isozymes with accuracies of 0.93, 0.89, 0.89, 0.85 and 0.87 for 1A2, 2C9, 2C19, 2D6 and 3A4, respectively, as measured by the area under the receiver operating characteristic (ROC) curves. The important atom types and features extracted from the five models are consistent with the structural preferences for different CYP450 substrates reported in the literature. We also identified novel features with significant discerning power to separate CYP450 actives from inactives. These models can be useful in prioritizing compounds in a drug discovery pipeline, or recognizing the toxic potential of environmental chemicals.
Introduction
The human body is constantly exposed to thousands of different molecules via different routes, such as swallowed, inhaled, injected, or absorbed through skin. In liver, many of these molecules will be transformed or metabolized by cytochrome P450 (CYP450) isozymes. The human CYP450 family contains 57 isozymes,1 which are predominantly involved in the phase I metabolism of xenobiotics, functioning as chemical processing machines.2 Inhibition or activation of the activities of CYP450 isozymes may cause undesirable drug-drug or food-drug interactions, as exemplified by the “grapefruit juice effect”.3 Accumulation of drug molecules resulting from suppression of activities of CYP450 enzymes increases the risk of adverse effects of the drug.4 Monitoring the interactions of drugs and environmental chemicals with CYP450 enzymes is critical in maintaining the integrity of this system. Using reliable predictive models as an alternative to laboratory testing provides the advantages of low cost, high speed and throughput. In addition, virtual compounds and compounds to be synthesized can also be predicted for their potential CYP450 liability.
Out of the 57 human CYP450 isozymes, the five most important isoforms, 1A2, 2C9, 2C19, 2D6 and 3A4, account for metabolizing 90% of the known drugs.4, 5 Crystal structures have been solved for four of the five aforementioned isozymes,6–9 while the only isozyme with no crystal structure available yet, CYP2C19, shares 91% sequence identity with CYP2C9.9 Although these recently solved X-ray crystal structures of CYP450 isozymes have helped to shed light into the steric and electronic features of the substrate binding sites, ligand specificity and ligand induced structural changes largely remain unknown.10 Indeed, analyses of crystal structures indicate large variations in the active site cavity volumes induced upon ligand binding,10 suggesting that the ligand binding sites of the CYP450 isozymes are adaptive and plastic.11 The conformational plasticity of CYP450 isozymes, as reflected by their capability of accommodating structurally diverse substrates and inhibitors, has prevented conclusive predictions from structure-based approaches, such as molecular docking and pharmacophore mapping. Alternatively, quantitative structure-activity relationships (QSAR), especially machine learning techniques, have been widely applied to assess the interactions between small molecules and CYP450 isozymes (Ref. 12 and references thereafter).12
Previously, QSAR models were largely based on small training sets of tens to hundreds of compounds. 12 As a result, the relatively small data size and limited structural diversity restricted the applicability of these models to larger data sets. High throughput screening (HTS) techniques have enabled in vitro screening of thousands to hundreds of thousands of compounds against different CYP450 isozymes,13 whereas the high false positive and false negative rates common to traditional single concentration HTS data made it less suitable to serve as training sets in machine learning. Another long-standing obstacle towards construction of a balanced training set is lack of inactive compounds, because the metabolic profiles of the “CYP450 clean” compounds, i.e. non-substrates and non-inhibitors, tend not to be discussed in literature. Recently, the National Institutes of Health (NIH) Chemical Genomics Center (NCGC) screened over 17,000 compounds against the five major CYP450 isozymes using the quantitative high throughput screening (qHTS) technique,14 where each compound was tested at 7–15 different concentrations.15 The high quality qHTS data has proved useful to overcome the hurdle toward the construction of robust and reliable CYP450 models.
Materials and methods
Data sets
The 17,143-compound data set was downloaded from PubChem (PubChem AID: 1851). The compounds were tested at multiple concentrations against five recombinant CYP450 isozymes (1A2, 2C9, 2C19, 2D6, and 3A4).14 These five isozymes were assayed with a bioluminescent-based detection technique where the activity of firefly luciferase is coupled to the metabolism of pro-luciferin CYP substrates.16 The luciferase-based P450-Glo™ Screening Systems were obtained from Promega (Madison, WI) for CYP 1A2 (V9770), CYP 2C9 (V9790), CYP2 C19 (V9880), CYP 2D6 (V9890), and CYP 3A4 Luciferin-PPXE (V9910) and were adapted for 1,536-well microplates and an automated protocol. The control compounds furafylline for 1A2 (F124), sulfaphenazole for CYP 2C9 (S0758), ketoconazole for CYP 2C19 (K1003), quinidine for 2D6 (Q3625), and ketoconazole for 3A4 (K1003) were purchased from Sigma Aldrich (St. Louis, MO). Recombinant P450 enzymes were obtained from baculovirus constructs expressed in insect cells (BD/Gentest). These enzymatic assays detect both inhibitors and activators of the P450 isozymes. It is important to note that in addition to inhibitors, substrates may also decrease the bioluminescent signal in these assays as both types of compounds reduce the amount of free enzyme available to catalyze the conversion of pro-luciferin substrates. Therefore, inhibition in the present dataset may be due to either inhibitors or substrates.
The qHTS assay was performed in 1,536-well plates and a concentration-response curve was generated for every compound with concentrations ranging from 0.24 nM to 40 μM. Analysis of compound concentration–response data was performed as previously described.15 Briefly, raw plate reads for each titration point were first normalized relative to the positive control compound (−100%) and DMSO-only wells (0%) and then corrected by applying a NCGC in house pattern correction algorithm using compound-free control plates (i.e., DMSO-only plates) at the beginning and end of the compound plate stack. Concentration–response titration points for each compound were fitted to a four-parameter Hill equation, yielding concentrations of half-maximal activity (AC50) and maximal response (efficacy) values. Compounds were designated as Class 1–4 according to the type of concentration–response curve observed.15 Curve classes are heuristic measures of data confidence, classifying concentration–responses on the basis of efficacy, the number of data points observed above background activity, and the quality of fit. Compounds with class 1.1, 1.2, 2.1 were defined as active. Compounds with class 4 curves were defined as inactive and compounds with other curve classes were considered inconclusive and excluded from the modeling exercises. Inconclusive compounds were excluded from the data set. The remaining compounds were processed through a Pipeline Pilot17 protocol to remove salts, redundant and heavy metal containing compounds. The preprocessed data set for each of the five CYP450 isozymes was randomly split into a training set and a test set of equal size. The active percentages of the training and test sets are roughly equal for each CYP450 isozyme, as shown in Table 1.
Table 1.
Summary of training and test sets of five CYP450 isozymes.
| P450 Isoforms | |||||
|---|---|---|---|---|---|
| 1A2 | 2C19 | 2C9 | 2D6 | 3A4 | |
| Training Set | 7208 | 6038 | 6627 | 7788 | 6800 |
| POS/NEG | 2874/4334 | 2701/3337 | 2521/4106 | 1061/6727 | 2334/4466 |
| Postive % | 39.87% | 44.73% | 38.04% | 13.62% | 34.32% |
| Test Set | 7128 | 5923 | 6530 | 7761 | 6738 |
| Positive % | 39.51% | 44.25% | 37.73% | 13.88% | 32.34% |
Molecular descriptors
Atom types were employed as molecular descriptors in this study. The original atom type casting tree was designed to reflect the chemical environment of each atom type, according to whether the atom is aromatic, whether the atom is in a ring, whether the atom is next to a functional group, etc.18 This original tree, largely based on a medicinal chemist’s intuition, was subject to a recursive optimization cycles in terms of where to further split the tree, where to stop splitting, and where to combine the branches, in order to make the best prediction of logP values in the Starlist data set containing over 11,000 structurally diverse compounds.18 The optimized tree output 218 atom types, featuring 88 different carbon types, 7 hydrogen types, 55 nitrogen types, 31 oxygen types, 8 halide types, 23 sulfur types, and 6 phosphorus types.18 Together with 26 correction factors to catch a number of whole molecule features, the original set contained 254 molecular descriptors. In this study, the following correction factors were added, to represent the molecular globularity, molecular rigidity, lipophilicity, and group functionality: 1) Polar surface area (PSA), 2) Fraction of sp2 hybrid atoms, 3) Number of macro-rings (ring size > 6), 4) Fraction of ring atoms, 5) Number of hydrogen bond acceptors (HBA), 6) Number of hydrogen bond donors (HBD), 7) Number of naphthalines, 8) Number of sugar rings, 9) Presence/absence of a steroid scaffold, and 10) Number of branching atoms. Therefore, a series of 264 numerical values comprise the final set of the molecular descriptors.
Support vector machine (SVM)
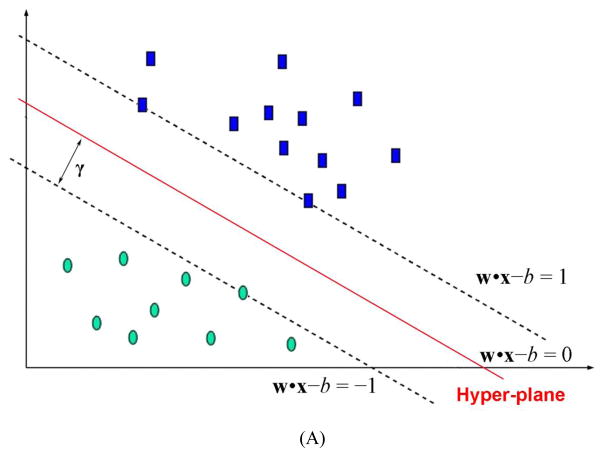
SVM is a supervised machine learning method, capable of deciphering subtle patterns in noisy and complex datasets.19, 20 SVM is one of the most popular kernel methods, enabling a smooth introduction of nonlinearity thus allowing application of linear algorithm to solve nonlinear problems. Like many other classification methods, a separation hyper-plane is to be determined in SVM to maximize the separation over a training set. What makes SVM different from other classification methods is that the algorithm is designed to find the balance point between maximizing separation of data points in a training set and minimizing generalization errors.21 Since it has been proved that minimizing generalization errors is equivalent to maximizing the margin between the separating hyper-planes,22 (Fig. 1A) the SVM classification problem can be solved by solving the constrained optimization problem:
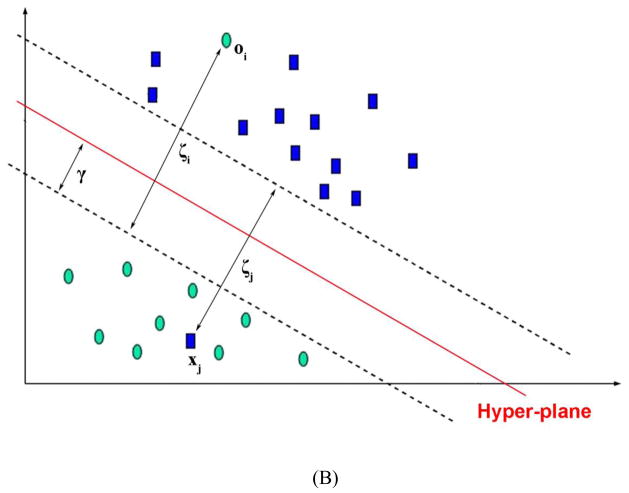
Figure 1.
Illustration of (A) hyper-planes with maximal margin. (B) Soft margin tolerating a number of misclassified data points associated with a penalty. The support vectors are composed of the data points adjacent to the hyper-planes and those misclassified.
Maximizing the margin
The constrained optimization problem is solvable for linearly separable data sets upon mapping into a high dimensional feature space, as illustrated in Fig. 1A, by using Lagrange multipliers. The problem might become unsolvable when noisy or complex data sets are involved. Cortes and Vapnik introduced the concept of soft margin to tackle with the non-separable training data, by allowing misclassified data points.23 The method introduces slack variables, ξi, which measure the degree of misclassification of the data point i.(Fig. 1B) Non-zero ξi is penalized by a cost parameter, C, in the objective function, and the optimization becomes a tradeoff between a large margin and a small error penalty.
In this study we used LIBSVM, a software implementation of SVM developed by Chang and Lin.24 The kernel used is the Gaussian Radial Basis Function (RBF):
The tunable parameters, C and γ, are optimized using an exhausted searching method. A python driven grid-based method was applied to maximize the prediction accuracy in a 7-fold cross validation (CV) of the training data. The ROC curve was employed to evaluate the predictive power of the model against the equal-sized test set.
Results and Discussion
CYP1A2
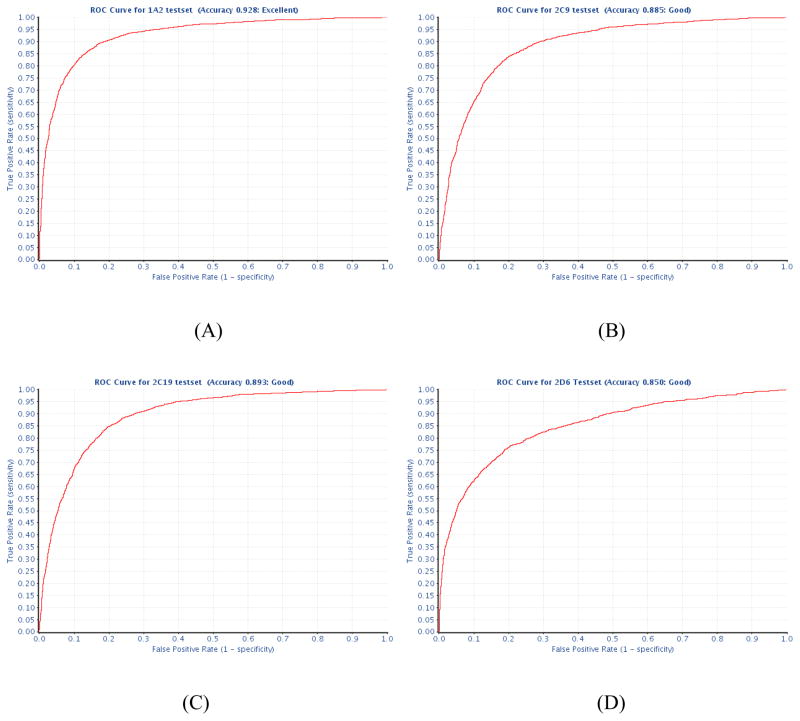
The training set for CYP1A2 contains 7208 compounds with 2874 (39.87%) active and the test set with 2816 (39.51%) actives of 7128 compounds. It turned out that the combination of C = 1.2 and γ = 1.0 gave the best CV accuracy of 87.5%. The area under the curve (AUC) of the ROC plot for CYP1A2 was 0.93, (Fig. 2A) indicating an excellent predictive power of the model. The top 10% compounds ranked by the calculated probability of being active are 95.9% active (683/712), while the bottom 10% compounds are only 1.26% active (9/712). The first half of the rank ordered compounds contains 91.9% actives of the whole data set.
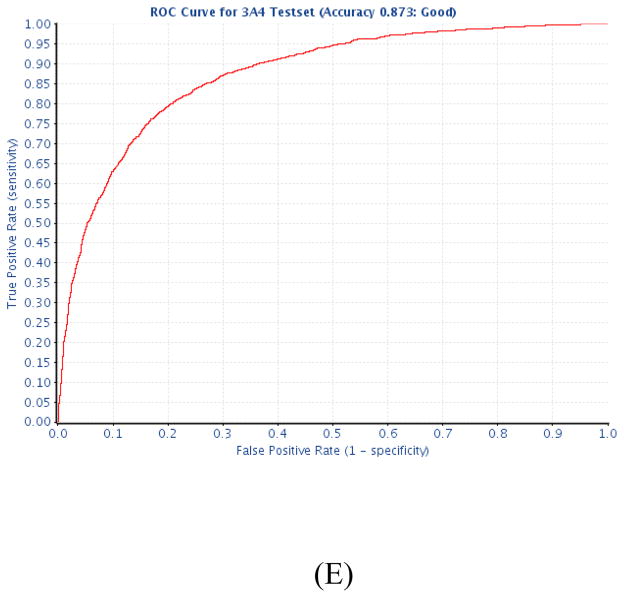
Figure 2.
Test ROC curves for (A) CYP1A2, (B) CYP2C9, (C) CYP2C19, (D) CYP2D6, and (E) CYP3A4.
The crystal structure of human CYP1A2 revealed a rather compact, flat, and closed active site.7 A survey of the known CYP1A2 substrate structures also indicated that the enzyme favored lipophilic, neutral, and planar polyaromatic or polyheteroaromatic small molecules.4, 25 The substrate structural features deduced from both the crystal structure of CYP1A2 and its substrates are in good agreement with the determinant features derived from the CYP1A2 model.
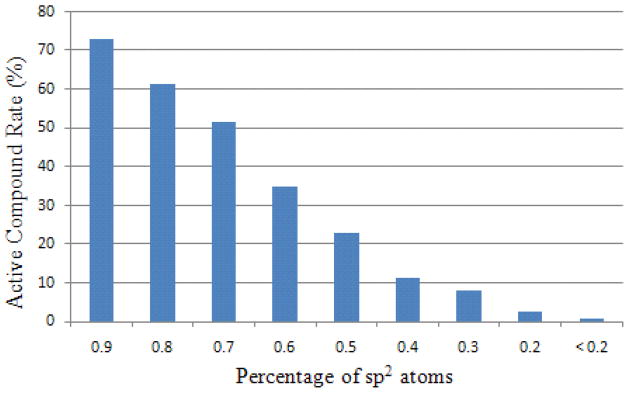
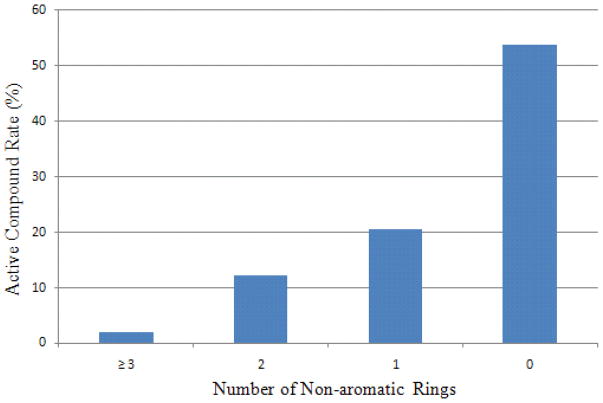
Although SVM itself cannot determine the importance of each structural feature, algorithms have been developed to couple feature selection strategies with SVM.26, 27 The top ranked features for CYP1A2, as measured by an F-score, included the fraction of sp2 hybrid atoms in a molecule, the count of aliphatic hydrogen atoms, the number of non-aromatic rings, the count of bridge carbon atoms connecting to one heteroatom in a fused ring system, and the count of hydrogen bond acceptors (HBA). As shown in Fig. 3, for example, the portion of active compounds gradually decreased from 72.8% for those with over 90% sp2 atoms to less than 1% for those with 20% or less sp2 atoms, compared with the average active rate of 39.87%. Compounds with and without bridge carbon atoms have a 61.1% and 27.9%, respectively, chance of being active, indicating that the CYP1A2 active site favors fused ring systems. A clear increasing trend of active rate was also observed for molecules with a decreasing number of non-aromatic rings (Fig. 4), which could not be derived from any structure-based approaches or QSAR models based on smaller training data sets. Introducing aliphatic rings to a molecule is a medicinal chemist’s common strategy to improve the hydrophilicity and solubility of the molecule.28
Figure 3.
The compound active rate decreases with decreasing percentages of sp2 hybrid atoms in CYP1A2 model.
Figure 4.
The relationship between the percentage of actives in the training set and the number of non-aromatic rings for CYP1A2.
CYP2C9
The training and test sets for CYP2C9 contain 6627 (38.0% active) and 6530 (37.7% active) compounds, respectively. The combination of C = 2.0 and γ = 1.0 yielded the best CV accuracy of 82.9% for the training set. The predictive accuracy, as measured by the AUC from the ROC plot, for CYP2C9 was 0.89 (Fig. 2B).
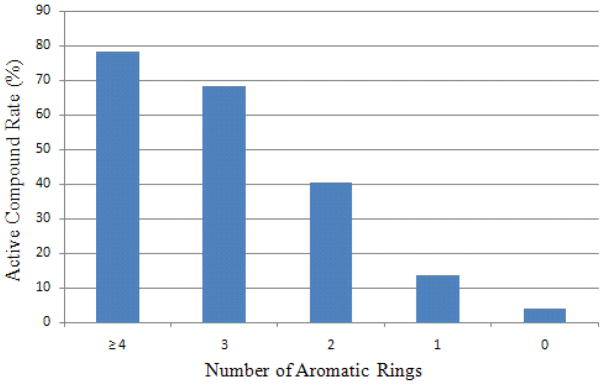
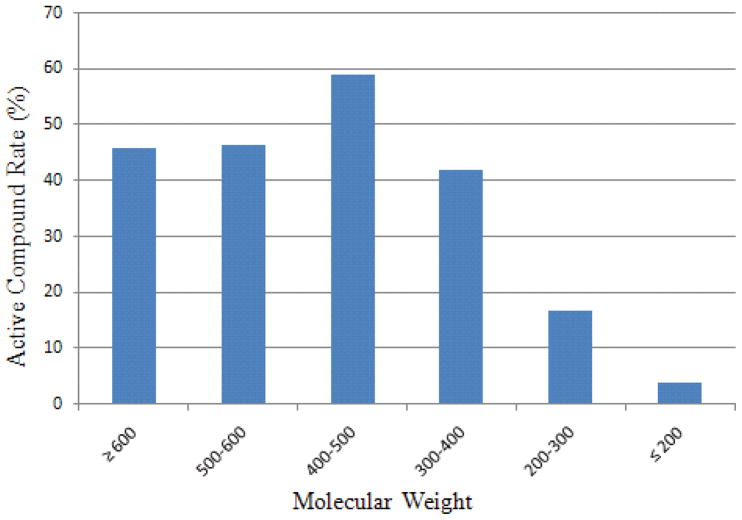
CYP2C9 has a significantly larger active site than CYP1A2, capable of accommodating multiple substrates and inhibitors.9, 10 CYP2C9 substrates are mostly negatively charged at physiological pH, while neutral compounds can also bind tightly to CYP2C9 due to its vast binding pocket.4, 25 Interestingly, the top ranked features selected by the model that confer the compound activity do not include any negative charge related atom types (the first negatively charged atom type ranked #16). Instead, the number of aromatic rings, molecular weight (MW), together with percentage of sp2 atoms are the first tier of features with the most discerning power. Compounds with four or more aromatic rings are 78.3% active, while compounds with no aromatic ring are only 4.1% active. The more aromatic rings in a compound, the more likely it is CYP2C9 active. (Fig. 5) There was no preference of CYP2C9 inhibition, when the MW of a compound was 300 or above. However, the active rate dropped sharply to 16.8% and 3.7% if a compound has a molecular weight between 200 and 300, or below 200. (Fig. 6)
Figure 5.
CYP2C9 training set active rate as a function of the number of aromatic rings.
Figure 6.
The impact of molecular weight on CYP2C9 compound active rate.
CYP2C19
The training and test sets of CYP2C19 contain the least compounds suitable for modeling among the five isozymes, but the data sets are well balanced with around 44% comprised of actives (Tab. 1). Coincidently, the optimized CV results for CYP2C19 model were also the worst among the five models, with the accuracy of 80.59%, when C = 2.0 and γ = 1.0. The predictive accuracy (AUC) of the CYP2C19 model against its test set was 0.89 (Fig. 2C).
CYP2C19 is the only isozyme without crystal structure available among the five most important CYP450 enzymes. However, it shares 91% amino acid sequence identity with CYP2C9.25 The high sequence similarity was reflected by the high extent of shared discriminating features between the two corresponding models -- among the top 20 atom types of the CYP2C19 model, 16 atom types overlapped with the top 20 atom types of the CYP2C9 model. The acidic atom type ranked number 7 in the CYP2C19 model. One feature that was more important for CYP2C19 model was the number of HBDs. There were 15.6% compounds with three or more HBDs in the training set showing active response in the assay, while the active rate increased to 48.3% for the compounds with two or less HBDs. This observation is in accordance with the reported trend that CYP2C19 favors more hydrophilic ligands in comparison with CYP2C9.25
CYP2D6
The CYP2D6 data was the most imbalanced, in terms of active rate, among the five isoforms of CYP450 in the study. There were less than 14% active compounds in both the training and test sets. With C setting to 2.0 and γ to 1.0, the 7-fold CV reached a high accuracy of 89.5%. The resulting model can predict the test set with an AUC of 0.85 (Fig. 2D). Screening the top 10% or 20% ranked compounds by the model will recover 47.7% and 66.7% of all the actives in the test set, respectively.
With only 4% of relative content in human liver microsome, CYP2D6 takes part in metabolizing nearly 30% of the known drugs.29 CYP2D6 catalyzes the oxidation of various classes of drugs, including antiarrhythemics, antidepressants, antipsychotics, β-blockers, and analgesics.30 The broad spectrum of CYP2D6 substrates implies an adaptive ligand binding site, capable of accommodation of structurally diverse molecules.
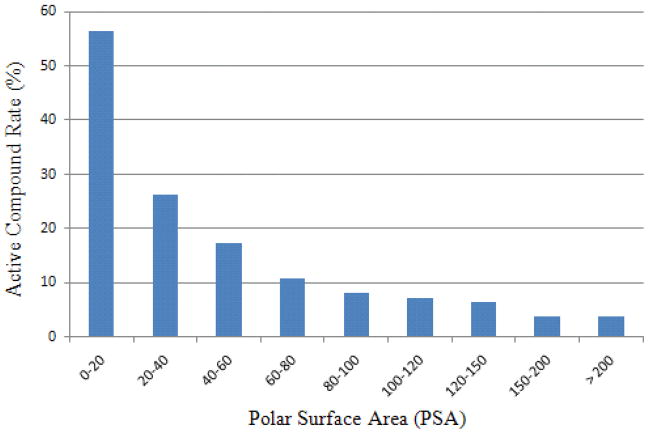
The crystal structure of CYP2D6 revealed a well defined active site formed by ASP301, GLU216, PHE483 and PHE120,6 endorsing the earlier observation that CYP2D6 substrates typically contain a basic nitrogen and a planar aromatic ring.6 The most relevant features extracted from feature selection were the PSA, the number of HBA, the count of the nitrogen atoms in a saturated ring that is not directly linked to carbonyl or aromatic atoms, the count of the nitrogen atoms in imines, and the presence of methylene carbon atom with two protons attached. Figure 7 illustrates a clear declining trend of compound active rate with increasing PSA. This observation agrees with the common strategies applied by medicinal chemists to reduce the CYP2D6 inhibitory potential of compounds. The major strategy widely adopted by medicinal chemists to avoid CYP2D6 liability is to increase the hydrophilicity of compounds by replacing aromatic rings with saturated rings or introducing hydroxyl and amide groups.30 Both hydroxyl and amide groups contribute significantly to the PSA of a molecule. Basic nitrogen atom in a ring, such as the nitrogens in a piperidine ring, also demonstrated discriminating power in the model. Compounds with and without this atom type were 25.54% and 11.34% active, respectively.
Figure 7.
The active compound rate is shown to decline with increasing PSA as observed in the CYP2D6 model.
CYP3A4
The CYP3A4 data sets used in this study contained around 33% of active compounds. The 7-fold CV accuracy was optimized to 81.1% under the condition of C = 2.0 and γ = 1.0. The optimized model achieved a predictive accuracy (AUC) of 0.87 (Fig. 2E) on the test set.
CYP3A4 is the most highly expressed and the most important isoform of CYP450, responsible for metabolizing about 50% of marketed drugs.29 Therefore, CYP3A4 is also the isozyme most often involved in drug-drug interactions, and inhibition or induction of CYP3A4 is more likely to lead to unwanted accumulation of therapeutic agents.4 Human CYP3A4 is known to be capable of metabolizing both small molecules with molecular weight below 200 and large natural products.31 A better understanding of structural features of CYP3A4 and its interactions with small molecules are highly desirable. However, when the first crystal structure of CYP3A4 was solved, it brought about more questions than answers. The estimated cavity volume of CYP3A4 varies largely from 1173Å3 to 2682 Å3.10 Furthermore, it has been found to bind and metabolize multiple substrates simultaneously.32 The extreme flexibility of CYP3A4 not only challenges the efforts of structure-based modeling approaches, such as molecular docking, but also presents a tough case for QSAR studies due to the large diversity of its ligand structures.33
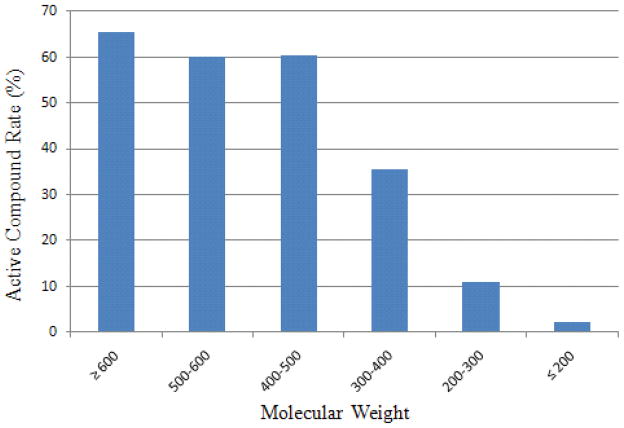
The features that displayed the highest impact on the CYP3A4 model included MW, the number of aromatic rings, the number of the branching atoms, and the number of rotatable bonds. As shown in Fig. 8, CYP3A4 seems to favor larger molecules. Compounds with molecular weight of 400 and above were 60% active in the CYP3A4 training data, while smaller molecules with molecular weight of 200 or less were only 2.1% active. Larger molecules can assume more interactions with the enzyme, while the CYP3A4 active site is extremely flexible, thus, larger molecules have an increased chance of becoming CYP3A4 ligands. Not only the size of a molecule matters, but the molecular shape is equally important. Linear molecules are less likely to bind tightly to the CYP3A4 active site.
Figure 8.
The relationship between the active compound rate and molecular weight in the CYP3A4 model.
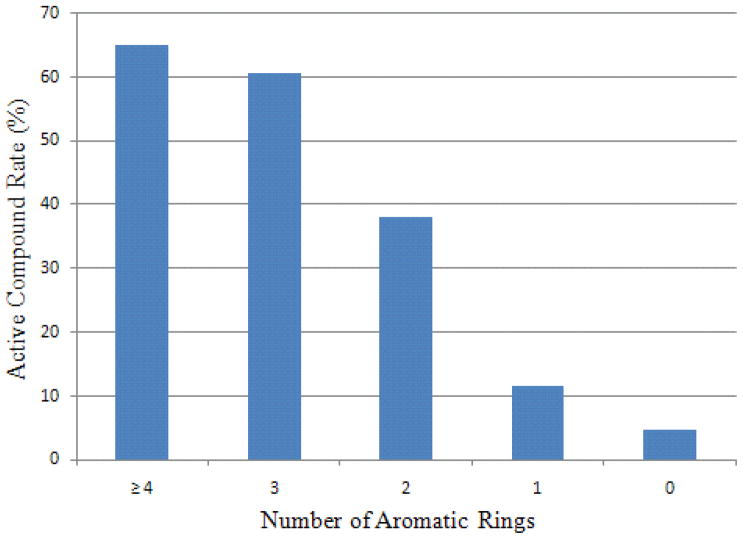
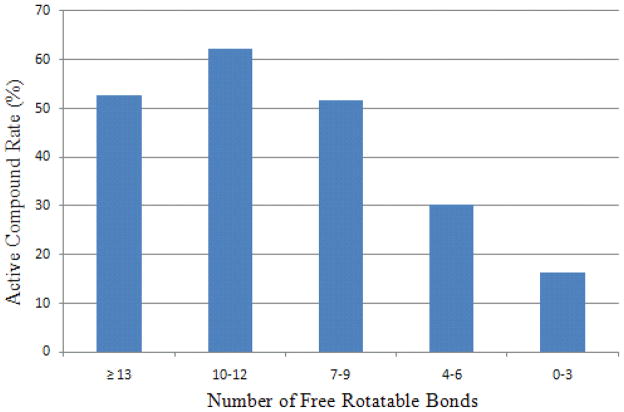
Figure 9 illustrates a trend of decreasing compound active rate associated with a decreasing number of aromatic rings in the CYP3A4 training data. Compounds with no aromatic ring are the least likely to be CYP3A4 active, while three or more aromatic rings in a molecule greatly increase the chance for the molecule to be CYP3A4 active. The results are consistent with the general observation that CYP3A4 is in charge of metabolizing large, neutral, and greasy compounds.25 Although CYP3A4 itself is extremely flexible, small molecules with higher flexibility tend to bind to CYP3A4 tighter. Interestingly, those extremely flexible small molecules with more than 13 rotatable bonds (average molecular weight of 321.8) showed a 10% decrease in active rate compared to the peak. (Fig. 10) This phenomenon could partly be attributed to the fact that the unfavorable entropic contributions overwhelmed the enthalpy gains.
Figure 9.
The compound active rate is shown to decline with decreasing number of aromatic rings in the CYP3A4 model.
Figure 10.
Compound active rate as a function of the number of free rotatable bonds in the CYP3A4 model.
Comparison with other molecular descriptors
A parallel model construction was carried out for the same data sets using the extended connectivity fingerprints (ECFP_6),17 MOE 2D descriptors, and Daylight fingerprints (FPs) as molecular descriptors. Both ECFP_6 and MOE 2D descriptors offered predictive performances comparable to that of atom types, as shown in Table 2, yet ECFP_6 is less interpretable than atom types while the MOE 2D descriptors contain largely whole molecule properties. The 186 MOE 2D descriptors performed as well as atom types on 4 of 5 CP450 isozymes, with CYP2C19 as the exception. A sharp decrease in AUC-ROC was observed for CYP2C19 (Table 2), implying that the MOE 2D descriptors are less tolerate to noisy datasets. Unexpectedly, the performance of Daylight FPs dropped dramatically in predicting the test sets of all five isoforms of CYP450 (Table 2). The Daylight FPs determine connectivity pathways in molecules and map them to overlapping bit segments using a hash function.34 We used a Daylight FP version consisting of 2048 bit positions and monitoring pathways of length 0–7. The 2048-bit FP was computed for each compound in the data sets using Daylight toolkits.34 Although the models offered nearly perfect predictions for the training sets with only one to two misclassified compounds, the predictive power of the all five FP-based models was much weaker than the atom type-based models. One possible explanation for the poor predictive performance of the Daylight FP models is that the Daylight FPs carry so much specific structural information that the resulting classifier has a limited applicability domain (AD),35 in other words, the descriptor space of the test sets is not well covered by that of the training data. On the contrary, atom typing breaks a molecule to fragments, which enhances the extrapolation power by expanding the coverage of the chemical space, while at the same time, the 218 atom types are far less specific than the 2048-bit Daylight FPs, such that the model is less likely to confer an activity to the features that it has never learned.
Table 2.
Comparison of CYP450 models constructed from atom types and other descriptors.
| AUC-ROC | P450 Isoforms | ||||
|---|---|---|---|---|---|
| 1A2 | 2C19 | 2C9 | 2D6 | 3A4 | |
| Atom Types | 0.93 | 0.89 | 0.89 | 0.85 | 0.87 |
| ECFP_6 | 0.92 | 0.88 | 0.88 | 0.83 | 0.87 |
| MOE 2D | 0.92 | 0.79 | 0.88 | 0.85 | 0.87 |
| Daylight FP | 0.63 | 0.61 | 0.62 | 0.59 | 0.68 |
The highly predictive models presented in this study add new evidence to the conclusion that the optimized atom types are more interpretable at the structural level and are capable of generating reliable and robust QSAR models, when combined with a high-quality dataset and a powerful machine learning algorithm.
Conclusion
SVM classification models have been built for the five most important isoforms of CYP450 (1A2, 2C9, 2C19, 2D6, and 3A4) based on a large qHTS data set with over 6000 compounds available for both model training and testing. The five CV optimized SVC models built by using the atom typing molecular descriptors exhibited consistently high predictive power when applied to the equally-populated test sets with accuracies between 0.85 and 0.93, as measured by the AUC of ROC plots. The results indicated that the atom typing descriptors generated from a large, high quality data set were capable of feeding information rich learning materials to the SVM learner. Useful information of structural features was derived from feature importance analysis for each isozyme of CYP450. The privileged structural features that could result in inhibitory and stimulatory activity against different CYP450 isozymes can serve as valuable guidelines in the drug discovery process.
Acknowledgments
This work was supported by the Intramural Research Programs of the National Human Genome Research Institute, National Institutes of Health. We thank in particular Rena Zheng for helpful comments and suggestions during the preparation of this manuscript.
References
- 1.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–91. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 2.Roy K, Roy PP. QSAR of cytochrome inhibitors. Expert Opin Drug Metab Toxicol. 2009;5:1245–66. doi: 10.1517/17425250903158940. [DOI] [PubMed] [Google Scholar]
- 3.Bailey DG, Spence JD, Edgar B, Bayliff CD, Arnold JM. Ethanol enhances the hemodynamic effects of felodipine. Clin Invest Med. 1989;12:357–62. [PubMed] [Google Scholar]
- 4.Arimoto R. Computational models for predicting interactions with cytochrome p450 enzyme. Curr Top Med Chem. 2006;6:1609–18. doi: 10.2174/156802606778108951. [DOI] [PubMed] [Google Scholar]
- 5.Wolf CR, Smith G, Smith RL. Science, medicine, and the future: Pharmacogenetics. BMJ. 2000;320:987–90. doi: 10.1136/bmj.320.7240.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, Lewis CJ, Tennant MG, Modi S, Eggleston DS, Chenery RJ, Bridges AM. Crystal structure of human cytochrome P450 2D6. J Biol Chem. 2006;281:7614–22. doi: 10.1074/jbc.M511232200. [DOI] [PubMed] [Google Scholar]
- 7.Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem. 2007;282:14348–55. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 8.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–6. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 9.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–8. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 10.Gay SC, Roberts AG, Halpert JR. Structural Features of Cytochromes P450 and Ligands that Affect Drug Metabolism as Revealed by X-ray Crystallography and NMR. Future Med Chem. 2:1451–68. doi: 10.4155/fmc.10.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pochapsky TC, Kazanis S, Dang M. Conformational plasticity and structure/function relationships in cytochromes P450. Antioxid Redox Signal. 13:1273–96. doi: 10.1089/ars.2010.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox T, Kriegl JM. Machine learning techniques for in silico modeling of drug metabolism. Curr Top Med Chem. 2006;6:1579–91. doi: 10.2174/156802606778108915. [DOI] [PubMed] [Google Scholar]
- 13.Arimoto R, Prasad MA, Gifford EM. Development of CYP3A4 inhibition models: comparisons of machine-learning techniques and molecular descriptors. J Biomol Screen. 2005;10:197–205. doi: 10.1177/1087057104274091. [DOI] [PubMed] [Google Scholar]
- 14.Veith H, Southall N, Huang R, James T, Fayne D, Artemenko N, Shen M, Inglese J, Austin CP, Lloyd DG, Auld DS. Comprehensive characterization of cytochrome P450 isozyme selectivity across chemical libraries. Nat Biotechnol. 2009;27:1050–5. doi: 10.1038/nbt.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–8. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cali JJ, Ma D, Sobol M, Simpson DJ, Frackman S, Good TD, Daily WJ, Liu D. Luminogenic cytochrome P450 assays. Expert Opin Drug Metab Toxicol. 2006;2:629–45. doi: 10.1517/17425255.2.4.629. [DOI] [PubMed] [Google Scholar]
- 17. [accessed Aug 24, 2011];Pipeline Pilot. http://accelrys.com/products/pipeline-pilot/
- 18.Sun H. A universal molecular descriptor system for prediction of logP, logS, logBB, and absorption. J Chem Inf Comput Sci. 2004;44:748–57. doi: 10.1021/ci030304f. [DOI] [PubMed] [Google Scholar]
- 19.Vapnik V. Statistical Learning Theory. John Wiley and Sons; New York: 1998. [Google Scholar]
- 20.Noble WS. What is a support vector machine? Nat Biotechnol. 2006;24:1565–7. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 21.Vapnik V. The Nature of Statistical Learning Theory. Pringer-Verlag; New York: 1995. [Google Scholar]
- 22.Cristianini N, Shawe-Taylor J. An Introduction to Support Vector Machines. Cambridge University Press; Cambridge: 2005. [Google Scholar]
- 23.Cortes C, Vapnik V. Support-vector networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- 24.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. 2001. [Google Scholar]
- 25.Lewis DF, Ito Y. Human P450s involved in drug metabolism and the use of structural modelling for understanding substrate selectivity and binding affinity. Xenobiotica. 2009;39:625–35. doi: 10.1080/00498250903000255. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y-W, Lin C-J. Combining SVMs with various feature selection strategies. In: Guyon I, Gunn S, Nikravesh M, Zadeh L, editors. Feature extraction, foundations and applications. Springer; 2006. [Google Scholar]
- 27.Byvatov E, Schneider G. SVM-based feature selection for characterization of focused compound collections. J Chem Inf Comput Sci. 2004;44:993–9. doi: 10.1021/ci0342876. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa M, Hashimoto Y. Improvement in aqueous solubility in small molecule drug discovery programs by disruption of molecular planarity and symmetry. J Med Chem. 54:1539–54. doi: 10.1021/jm101356p. [DOI] [PubMed] [Google Scholar]
- 29.Wang JF, Zhang CC, Chou KC, Wei DQ. Structure of cytochrome p450s and personalized drug. Curr Med Chem. 2009;16:232–44. doi: 10.2174/092986709787002727. [DOI] [PubMed] [Google Scholar]
- 30.Le Bourdonnec B, Leister LK. Medicinal chemistry strategies to reduce CYP2D6 inhibitory activity of lead candidates. Curr Med Chem. 2009;16:3093–121. doi: 10.2174/092986709788803033. [DOI] [PubMed] [Google Scholar]
- 31.Kenworthy KE, Bloomer JC, Clarke SE, Houston JB. CYP3A4 drug interactions: correlation of 10 in vitro probe substrates. Br J Clin Pharmacol. 1999;48:716–27. doi: 10.1046/j.1365-2125.1999.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shou M, Grogan J, Mancewicz JA, Krausz KW, Gonzalez FJ, Gelboin HV, Korzekwa KR. Activation of CYP3A4: evidence for the simultaneous binding of two substrates in a cytochrome P450 active site. Biochemistry. 1994;33:6450–5. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- 33.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci U S A. 2006;103:13682–7. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. [accessed Aug 24, 2011];Daylight Toolkits. http://www.daylight.com/products/toolkit.html.
- 35.Weaver S, Gleeson MP. The importance of the domain of applicability in QSAR modeling. J Mol Graph Model. 2008;26:1315–26. doi: 10.1016/j.jmgm.2008.01.002. [DOI] [PubMed] [Google Scholar]