Abstract
Developmental regulatory proteins are commonly utilized in multiple cell types throughout development. The Drosophila single-minded (sim) gene acts as master regulator of embryonic CNS midline cell development and transcription. However, it is also expressed in the brain during larval development. In this paper, we demonstrate that sim is expressed in 3 clusters of anterior central brain neurons: DAMv1/2, BAmas1/2, and TRdm and in 3 clusters of posterior central brain neurons: a subset of DPM neurons, and two previously unidentified clusters, which we term PLSC and PSC. In addition, sim is expressed in the lamina and medulla of the optic lobes. MARCM studies confirm that sim is expressed at high levels in neurons but is low or absent in neuroblasts (NBs) and ganglion mother cell (GMC) precursors. In the anterior brain, sim+ neurons are detected in 1st and 2nd instar larvae but rapidly increase in number during the 3rd instar stage. To understand the regulation of sim brain transcription, 12 fragments encompassing 5’-flanking, intronic, and 3’-flanking regions were tested for the presence of enhancers that drive brain expression of a reporter gene. Three of these fragments drove expression in sim+ brain cells, including all sim+ neuronal clusters in the central brain and optic lobes. One fragment upstream of sim is autoregulatory and is expressed in all sim+ brain cells. One intronic fragment drives expression in only the PSC and laminar neurons. Another downstream intronic fragment drives expression in all sim+ brain neurons, except the PSC and lamina. Thus, together these two enhancers drive expression in all sim+ brain neurons. Sequence analysis of existing sim mutant alleles identified 3 likely null alleles to utilize in MARCM experiments to examine sim brain function. Mutant clones of DAMv1/2 neurons revealed a consistent axonal fasciculation defect. Thus, unlike the embryonic roles of sim that control CNS midline neuron and glial formation and differentiation, postembryonic sim, instead, controls aspects of axon guidance in the brain. This resembles the roles of vertebrate Sim that have an early role in neuronal migration and a later role in axonogenesis.
Keywords: Autoregulation, Axon guidance, Brain, Drosophila, Enhancer, Optic lobes, single-minded
1. Introduction
The formation of functional central nervous system (CNS) neural circuits consists of a series of events beginning with neurogenesis, followed by axonogenesis, synaptic connectivity, and differentiation. These circuits underlie the complex behaviors found throughout the animal kingdom. One key aspect of CNS development is the action of transcriptional regulatory proteins. Since their number is relatively modest, they are often used multiple times during development to regulate different sets of genes and developmental processes. Mechanistically understanding how transcriptional regulation controls neurodevelopment will ultimately provide insight into the evolutionary basis for species differences in neural circuitry and behavior.
The Drosophila and mammalian single-minded (sim) genes provide an excellent example of how transcription factors perform multiple roles during CNS development (Crews, 2003). Drosophila sim encodes a basic-helix-loop-helix-PAS (bHLH-PAS) protein that forms a DNA binding heterodimer with the Tango (Tgo) bHLH-PAS protein (Sonnenfeld et al., 1997). During embryogenesis, sim is prominently expressed in the cells that lie along the midline of the Drosophila CNS and acts as a master regulator of CNS midline cell development (Nambu et al., 1991). The midline cells consist of a diverse group of motoneurons, interneurons, neurosecretory cells, and glia (Wheeler et al., 2006). In sim mutants, the midline cells fail to form, and midline-specific transcription is generally absent (Nambu et al., 1990). Consistent with a master regulatory role, misexpression of sim throughout the neuroectoderm results in the transformation of the entire CNS into only midline cells (Nambu et al., 1990). Later in development, sim is expressed in differentiated midline glia and neurons and was shown to control midline glial transcription (Wharton et al., 1994). During larval development, sim continues to be expressed in the midline cells of the ventral nerve cord and is also expressed in the brain, in both the central brain region and the lamina and medulla of the optic lobes (Pielage et al., 2002). The role of sim in the optic lobes was functionally investigated and shown to play a role in the differentiation of laminar precursor cells into mature neurons (Umetsu et al., 2006). Mammals have two sim genes, Sim1 and Sim2 (reviewed in Fan et al., 1996). Sim1 is expressed in the developing hypothalamus, including cells of the paraventricular nucleus (PVN), anterior periventricular nucleus (aPV), and supraoptic nucleus (SON). Genetic analysis of Sim1 homozygous mutant mice revealed an absence of the PVN, aPV, and SON and implicated Sim1 in controlling the migration of PVN and SON neurons into the anterior neuroendocrine hypothalamus (Xu and Fan, 2007). Thus, Drosophila and mammalian sim genes play important roles in generating functional CNS cell types and can control precursor formation, differentiation, and migration.
Genetic and expression data have indicated additional potential roles for both Drosophila and mammalian sim. Drosophila sim is expressed in the larval central brain (Pielage et al., 2002), and in adults this region (central complex) has been implicated in coordinating movement (Strauss, 2002). Behavioral analysis of a temperature-sensitive sim allele revealed that when shifted to the non-permissive temperature after embryonic neurogenesis was complete, adult flies showed locomotor defects (Pielage et al., 2002). Morphological analysis of the adult brain indicated a disorganization of the central complex neuropil. These results suggested a defect in interhemispheric communication and a subsequent inability to coordinate movement. Recent work on murine Sim1 and Sim2 revealed that they play a role in controlling axonogenesis of mammillary body axons (Marion et al., 2005). The results from mammals indicate that sim can function in axonogenesis, and this is also a potential role for sim in central brain development given the sim disorganized neuropil phenotype.
In this paper, we further explore the expression and function of sim in the larval central brain. We demonstrate that sim is expressed in six regions in the central brain, three on the anterior side and three on the posterior side, and identify the relevant neuronal clusters with respect to existing Drosophila brain maps. Many of the sim+ neurons send their axons across the midline. Experiments involving a transgenic locus-wide survey of sim DNA fragments identified enhancers that can drive all aspects of sim larval brain gene expression. These include distinct enhancers that can drive expression in specific sim+ neuronal clusters, as well as a sim autoregulatory enhancer. We utilize sim mutant MARCM clones to show that the loss of sim in an anterior brain cluster results in a defect in the fasciculation of axons that are crossing the midline. This type of defect may result in deficiencies in interhemispheric communication, consistent with sim mutant defects in coordinating movement. These results also reveal further similarities in the function of Drosophila and mammalian sim genes, since both contribute to axonogenesis as well as neurogenesis and migration.
2. Results
2.1 Postembryonic sim+ cells predominantly utilize a single transcript
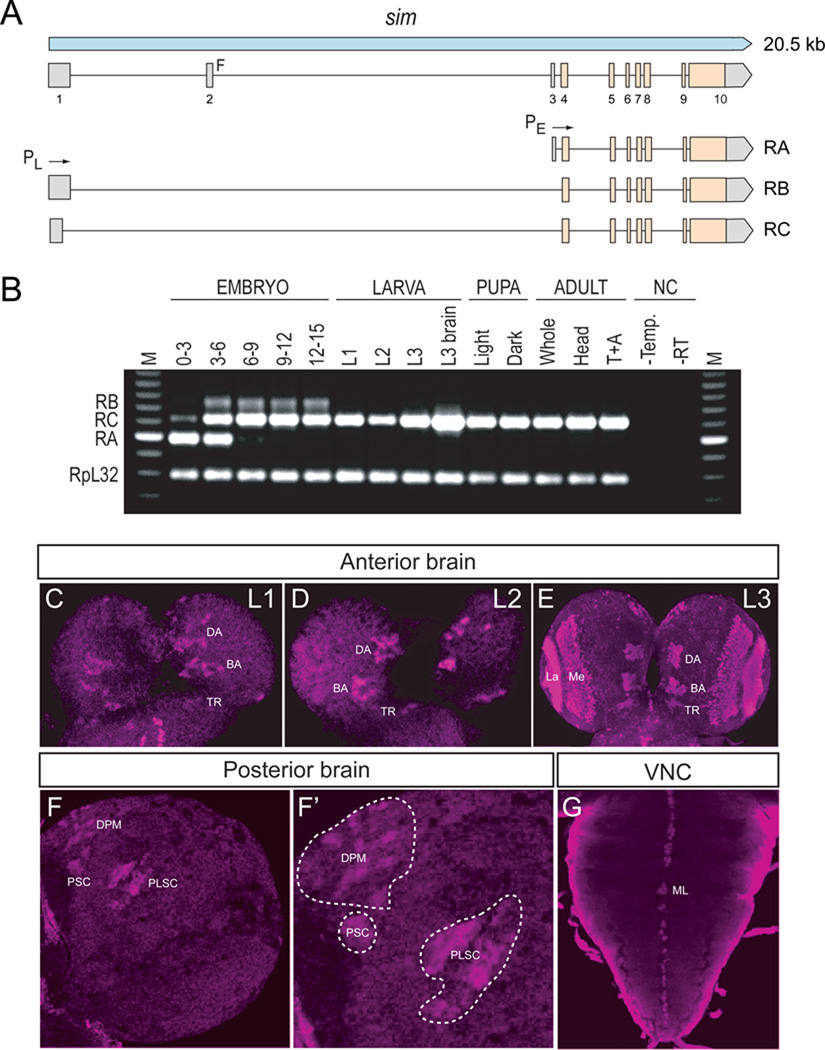
Previous sequence analysis of embryonic sim cDNA clones revealed that the sim transcription unit consists of 9 exons and 3 different embryonic transcripts (RA, RB, and RC) and spans 20.5 kb of genomic DNA (Fig. 1A) (Kasai et al., 1998). Recent RNA-Seq data further suggests that female adult sim expression may utilize an additional, previously unknown exon (exon 2 in Fig. 1A) and promoter to yield a fourth sim transcript (Graveley et al., 2011). The sim RA, RB, and RC transcripts utilize two promoters: an early promoter (PE) and a late promoter (PL). The RA transcript consists of exons 3–10 and is transcribed from PE. Transcripts RB and RC contain exons 1 and 4–10. They are transcribed from PL. The RB and RC transcripts differ in the use of alternative donor splice sites in exon 1. Since exon 1 encodes only 5’-UTR sequences, RB and RC produce the same protein. The entire protein coding sequence of RB and RC is contained within RA, but RA has a different 5’-UTR and an additional 24 aa of coding sequence at the N-terminus. The function of the additional RA protein residues is unknown.
Fig. 1. Postembryonic CNS expression of sim.
(A) Schematic of the sim locus shows the embryonic alternative splice variants of sim: RA, RB, and RC. The sim transcription unit (blue; 20.5 kb) consists of 10 exons, and embryonic expression derives from two promoters: PL and PE. The coding sequence of each transcript is filled tan, and the untranslated regions are filled gray. modENCODE RNA-Seq data (Graveley et al., 2011) revealed a novel female-specific adult exon (F; exon 2), which is likely transcribed from a third promoter. (B) Developmental profile of sim splice variants determined by RT-PCR of total RNA. Multiplex RT-PCR was employed using total RNA from each developmental stage and primers specific for each splice variant and the RpL32 gene, which is uniformly expressed throughout development (Graveley et al., 2011; O'Connell and Rosbash, 1984). Times of embryonic RNA indicates the collection times in hr after egg laying. Larval RNA was prepared from 1st instar (L1), 2nd instar (L2), 3rd instar (L3) larvae, and L3 brain. Pupal RNA was isolated from light pupae representing the first two days of pupation and dark pupae from the third and fourth days of pupation. Adult collections were from 2-day-old adults (equal numbers of males and females), 2-day-old adult heads, and 2-day-old adult thoraces and abdomens (T+A). No PCR products were observed in the negative controls (NC): (1) PCR reaction without template (-Temp) and (2) PCR reaction without reverse transcriptase (RT). M lanes are molecular weight markers, which confirmed that the RT-PCR products were the expected size. (C–E) Dissected brains stained with anti-Sim from (C) 1st instar (L1), (D) 2nd instar (L2), and (E) 3rd instar (L3) larvae show Sim localization (magenta) in three paired clusters of cells in the anterior central brain throughout larval development. The number of Sim+ central brain cells is relatively similar in 1st and 2nd instar larvae but increases significantly between the 2nd instar and late 3rd instar larval stages. The Sim+ anterior central brain cells are: (DA) DAMv1/2, (BA) BAmas1/2, and (TR) TRdm neurons. Sim is also present in the lamina (La) and medulla (Me) of the optic lobes. (F) On the posterior side of the brain, Sim is present in neurons in the DPMm1–3 and DPMpm1/2 neurons (DPM), as well as two previously undescribed clusters of neurons that we tentatively refer to as (PSC) Posterior Sim+ Cluster and (PLSC) Posterior Sim+ Lateral Cluster. (F’) Enlarged view of (F) with each neuronal group outlined with a white dotted line. (G) Sim is present in the midline cells (ML) of the 3rd instar larval ventral nerve cord (VNC).
To examine the utilization of the three different embryonic transcripts (RA, RB, and RC) during development, RT-PCR was carried out using transcript-specific primers with RNA from the embryonic, larval, pupal, and adult stages (Fig. 1B). RA is present in only the 0–3 and 3–6 hr embryonic collections. The RB transcript is utilized from 3 to 15 hr of embryonic development but not later. The RC transcript is observed weakly at 0–3 hr but then strongly at all stages of embryonic and postembryonic development. These results closely match Northern blot and RNA-Seq experiments (Crews et al., 1988; Graveley et al., 2011). sim is expressed in a variety of embryonic and postembryonic cell types – while different embryonic cell types may express RA, RB, RC, or multiple isoforms, the RT-PCR results suggest that the majority of postembryonic cell types express only RC.
2.2 Identification of sim+ neurons in the larval brain
Clusters of sim+ neurons (Pielage et al., 2002) in the anterior brain are present in 1st, 2nd, and 3rd instar larvae, and their number significantly increases by the 3rd instar larval stage (Fig. 1C–E). These cells include 3 paired clusters of neurons on the anterior side of the brain (Fig. 1C–E), neurons on the posterior of the brain (Fig. 1F), and the medulla and lamina of the optic lobes (Fig. 1E). Recent work has provided a comprehensive description of the organization of the 3rd instar larval brain (Pereanu and Hartenstein, 2006). Combining anti-Sim staining with either MAb BP106 (anti-Neurotactin) staining or MARCM allowed us to identify the sim+ brain cells as described below. The 3 paired sim+ neuronal clusters on the anterior side of the brain correspond to DAMv1/2, BAmas1/2, and TRdm (Fig. 1E,2A). Along the dorsal and posterior sides of the brain, there are another 3 groups of sim+ neurons (Fig. 1F). One group of sim+ neurons is derived from the DPMm1–3 and DPMpm2 cell lineages (Fig. 1F; referred to collectively as DPM neurons). The remaining two groups are previously undescribed and are tentatively designated as the Posterior Sim+ Cluster (PSC) and Posterior Lateral Sim+ Cluster (PLSC). Expression of sim is also present in the cells along the midline of the 3rd instar larval ventral nerve cord (Fig. 1G).
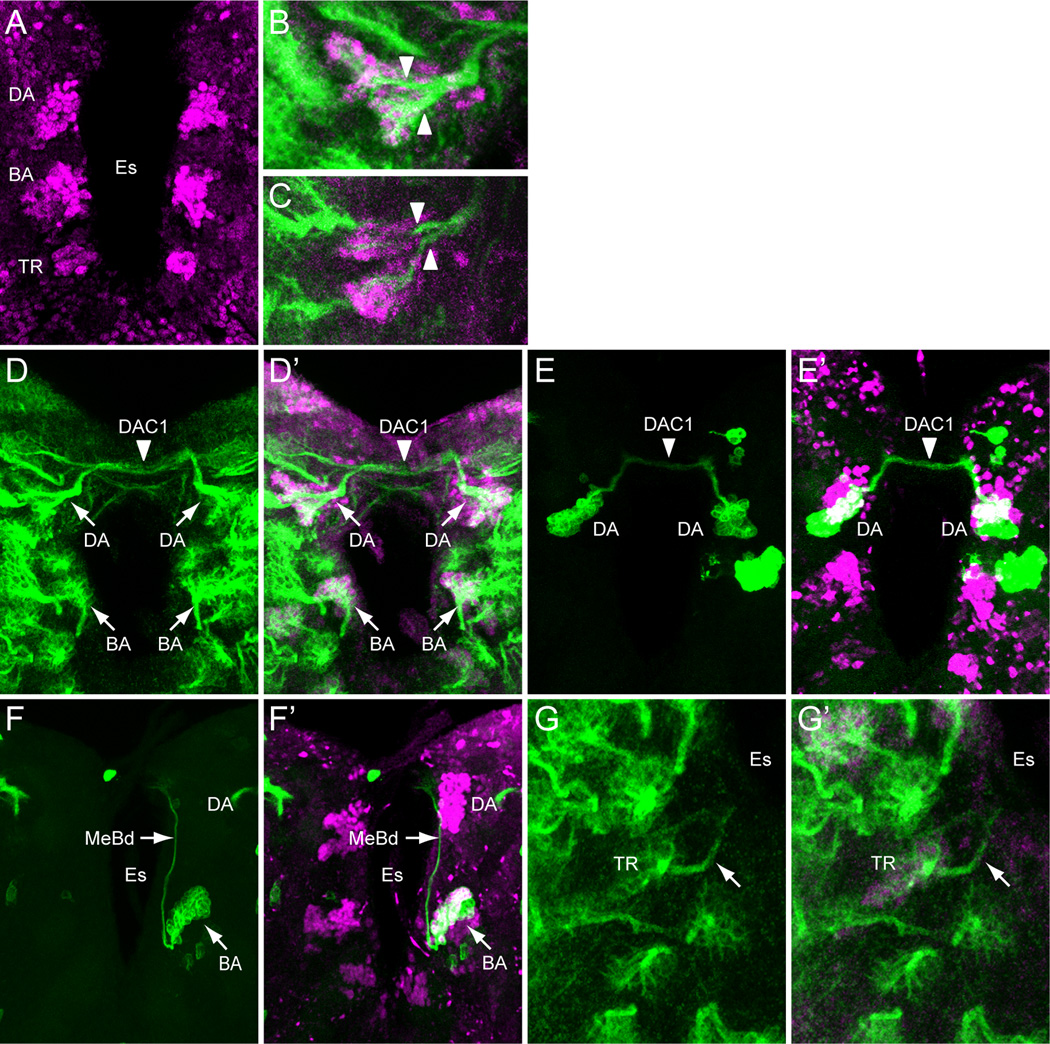
Fig. 2. Identification of sim+ larval brain neurons.
(A) 3rd instar larval brain stained with anti-Sim to show the 3 paired clusters of sim+ neurons: (DA) DAMv1/2, (BA) BAmas1/2, and (TR) TRdm neurons. Esophageal opening (Es) lies between the brain hemispheres. (B,C) Brains stained with anti-Sim (magenta) and MAb BP106 (green) showing (B) DAMv1/2 neurons and (C) BAmas1/2 neurons. Each cluster consists of two groups of neurons that extend an axon (arrowheads) that converge into a single axon tract. (D,D’) Brain stained with anti-Sim (magenta) and MAb BP106 (green) showing neuronal cell bodies and axon tracts. The characteristic tracts of the DAMv1/2 (DA) and BAmas1/2 (BA) neurons are shown (arrows). The DAMv1/2 axons can be seen crossing the midline in the DAC1 axon tract (arrowhead). (E,E’) Brain visualized for GFP (green) and Sim (magenta) showing two DAMv1/2 (DA) MARCM clones that fasciculate together in DAC1 (arrowhead). (F,F’) The characteristic ascending MeBd axon tract (arrow) is shown emanating from a BAmas1/2 (BA) MARCM clone. (G,G’) Brain stained with MAb BP106 (green) and anti-Sim (magenta) illustrating that the sim+ TRdm neurons extend a characteristic short projection (arrow) toward the neuropil near the ventral esophagus (Es).
DAMv1/2 sim+ cells
Co-staining between anti-Sim and MAb BP106 (anti-Neurotactin) and comparison of their location and axonal morphology (Fig. 2A,B,D) to the data of Pereanu and Hartenstein, 2006 indicates that the dorsal-most anterior sim+ cells are DAMv1,2 neurons. In addition, using MARCM, we obtained a number of clones for these sim+ cells that revealed axonal processes characteristic of the DAMv1/2 cells (Fig. 2E). The DAMv1 and DAMv2 cells are derived from two Type I NBs whose progeny are adjacent (Pereanu and Hartenstein, 2006) (Fig. 2B). The axonal processes of each NB fasciculate together and then combine into a single tract that migrates in a posteriormedial direction and crosses the midline in the dorsal anterior commissure (DAC1) (Fig. 2D,E). All of the neuronal progeny of each NB are sim+, although co-staining MARCM clones (Lee and Luo, 1999) for Elav, a neuronal marker (Berger et al., 2007), indicates that the NB and GMC are sim− (Supp. Fig. 1). In one fortuitous example, DAMv1/2 cells were MARCM-labeled on both sides of the brain, clearly showing their axons fasciculating together across the midline (Fig. 2E).
BAmas1/2 sim+ neurons
The medial anterior group of sim+ cells (Fig. 1E,2A) is the BAmas1/2 neurons. These cells were distinguishable based on their position relative to MAb BP106 staining (Fig. 2C,D). The BAmas1/2 neurons are derived from two Type I NBs, and their axons fasciculate together into a tract that rises vertically within the MeBd tract and then crosses the midline in the DAC1 tract. MARCM clones also confirmed the identity of these cells based on their characteristic axonal trajectories (Fig. 2F). All of the BAmas1/2 neurons are sim+.
TRdm neurons
The smallest and ventral-most of the anterior sim+ clusters are the TRdm neurons that form part of the tritocerebrum. These cells lie at the ventral base of the esophageal opening (Fig. 1E,2A) and can be recognized based on their position and characteristic neurite bifurcation (Pereanu and Hartenstein, 2006) as visualized with MAb BP106 staining (Fig. 2G). All of the TRdm neurons are sim+.
DPMm1–3 and DPMpm2 neurons
The dorsal-most sim+ cells are members of the DPM group of neurons (Pereanu and Hartenstein, 2006). These cells are derived from Type II NBs and are dispersed throughout the dorso-posterior brain and cross the midline via a number of commissural tracts (Izergina et al., 2009). sim is expressed in only a subset of DPM neurons. The identification of the sim+ subset of DPMm1–3 and DPMpm2 neurons is based on their location in the brain (Fig. 1F).
PSC and PLSC neurons
This small cluster of sim+ cells, tentatively named PSC, was previously unidentified, most likely because they stain poorly with MAb BP106 in the 3rd instar larvae. The lack of BP106 staining indicates that these are primary neurons, and not secondary neurons (Pereanu and Hartenstein, 2006). When Gal4 lines expressed in the PSC (see below) are crossed to UAS-tau-GFP, an occasional neurite is observed that crosses the midline (Fig. 4E). The PLSC group of sim+ neurons is slightly ventro-lateral to the PSC cells and consists of more neurons. These cells are located more posterior than lineages previously described.
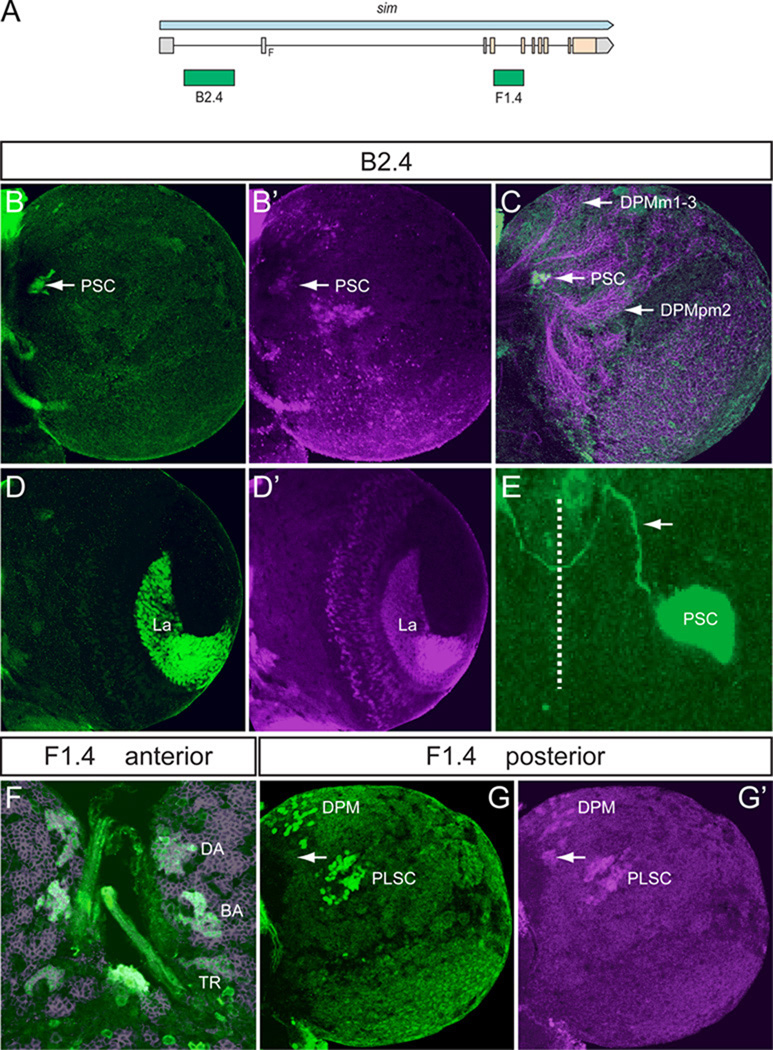
Fig. 4. Two distant fragments recapitulate central brain expression.
(A) Schematic depicting the location of the B2.4 and F1.4 fragments with respect to the sim gene. B2.4 is located within the first intron 10.4 kb away from the F1.4 fragment, which spans the entire third intron. (B,B’) B2.4-Gal4 UAS-nuc-GFP drives GFP expression in the PSC neurons (arrow) on the posterior side of the larval brain. Brains stained with anti-GFP (green) and anti-Sim (magenta). (C) Staining B2.4-Gal4 UAS-nuc-GFP with MAb BP106 and anti-GFP illustrates axon tracts characteristic of the DPMm1–3 and DPMpm2 nerve cell clusters (BP106) and indicates that the PSC neurons reside near the site where some posterior axonal tracts enter the commissure. (D,D’) B2.4-Gal4 UAS-nuc-GFP drives GFP expression in Sim+ optic lobe laminar neurons (La). (E) B2.4-Gal4 UAS-tau-GFP stained with anti-Tau2 (green) reveals an axonal projection (arrow) emanating from the PSC that crosses the midline (dotted line) and fasciculates with contralateral PSC axons. (F) F1.4-Gal4 UAS-mCD8-GFP reveals GFP expression (green) in the Sim+ DAMv1/2 (DA), BAmas1/2 (BA), and TRdm (TR) neurons. Brains were stained with anti-GFP and MAb BP106 (magenta). (G,G’) On the posterior side of the brain, F1.4-Gal4 UAS-nuc-GFP expresses GFP (green) in the Sim+ DPM neurons and PLSC neurons but not in the PSC neurons (arrows).
2.3 Transgenic screen for postembryonic sim-specific brain enhancers
Identifying regulatory DNA fragments that drive expression in specific cells corresponding to the gene’s normal expression pattern can enhance insight into the expression and function of a gene. This is commonly carried out in Drosophila using the Gal4/UAS transgenic approach (Brand and Perrimon, 1993) in which fragments of DNA are coupled to promoter-Gal4, and that transgenic strain is crossed to a reporter (e.g. UAS-GFP) for expression studies or crossed to a UAS transgenic line that allows cell ablation or reduction/enhancement of neural transmission. Our goals were to identify larval brain enhancers to better understand brain gene expression and to potentially isolate Gal4 strains that could target Gal4 to specific subsets of brain cells for future functional studies.
We initially assayed 12 fragments spanning 29.6 kb of sim genomic DNA, including 1.2 kb 5’ to sim, introns 1, 3, and 7 (with respect to RC) and 9.6 kb 3’ to sim (Fig. 3A). The piccolo (pic) gene is 1.0 kb 5’ to sim, and the timeout gene lies 10.4 kb 3’ to sim. Thus, the extent of the sim gene is potentially 31.9 kb, assuming that no sim regulatory elements reside within or distal to pic and timeout. Fragments were PCR-amplified, cloned into the pBCGw-UCP PhiC31 Gal4 vector, and introduced into the attP2 site at 68A4 using germline transformation (Groth et al., 2004), with the exception of the existing sim3.7 Gal4 line (Nambu et al., 1991). These fragments were assayed in vivo for their ability to drive postembryonic larval CNS expression.
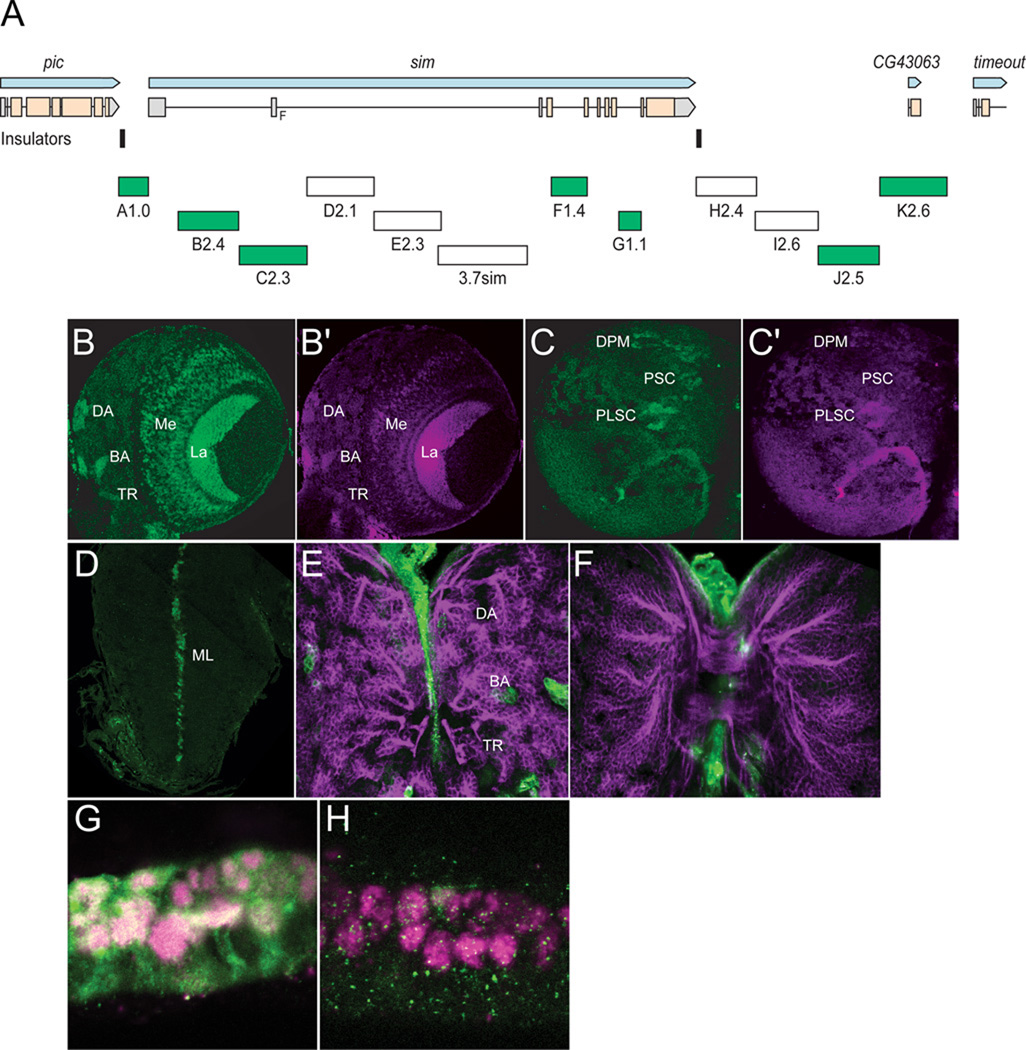
Fig. 3. Transgenic analysis of the sim regulatory region.
(A) Schematic of a 37.8 kb genomic region that includes the sim gene and neighboring pic CG43063, and timeout genes. Two insulator protein binding sites surrounding sim are indicated by vertical lines. Fragments that were analyzed by Gal4 transgenesis are labeled A–K and include the fragment size in kb. The 3.7sim fragment has been previously described (Nambu et al., 1991). Green boxes indicate fragments that drive postembryonic expression, while fragments with no postembryonic expression are unfilled. (B,B’) A1.0-GFP.nls 3rd instar larval brains were stained with anti-GFP (green) and anti-Sim (magenta) to assess whether GFP co-localizes with Sim+ brain cells. On the anterior side of the brain, A1.0-GFP.nls drives expression in DAMv1/2 (DA), BAmas1/2 (BA), and TRdm (TR). In addition, GFP expression is detected in two optic lobe ganglia, the lamina (La) and medulla (Me). (C,C’) On the posterior side of the brain, A1.0-GFP.nls drives GFP expression in DPMm1–3 neurons and DPMpm2 neurons (DPM), PLSC neurons, and PSC neurons. (D) A1.0-GFP.nls drives GFP expression in the midline cells (ML) of the larval ventral nerve cord cells. (E,F) Mutant version of A1.0-GFP.nls, in which both Sim:Tgo binding sites were mutated, was analyzed. The larval brain was stained for GFP (green) and MAb BP106 (magenta). (E) On the anterior side of the brain, the BP106 staining indicates the location of the DAMv1/2 (DA), BAmas1/2 (BA), and TRdm (TR) neurons, and these cells were GFP−. (F) On the posterior side of the same brain, there was an absence of GFP in brain neurons. (G,H) Sagittal views of stage 11 A1.0-GFP.nls embryos with (G) unmutated A1.0-GFP.nls and (H) a version of A1.0-GFP.nls with the Sim:Tgo sites mutated. Single segment is shown with internal up and anterior to the left. Embryos are stained for GFP (green) and anti-Sim (magenta), which stains all midline cells. The unmutated A1.0-GFP.nls drives robust expression in all midline cells, whereas expression is absent in the mutated version.
Immunostaining with anti-Sim was used to identify sim+ cells. As described below, we have identified enhancers that drive GFP in all known sim+ postembryonic cell types, and thus all sim postembryonic enhancers may reside within the 31.9 kb sim locus.
2.4 The A1.0 fragment is an autoregulatory enhancer that drives expression in all postembryonic sim+ cells
The A1.0 fragment contains PL and 1.0 kb of 5’-flanking genomic DNA. Previous work had shown that this region contained an enhancer that drove embryonic midline expression (Muralidhar et al., 1993), and this expression was genetically dependent on sim (Nambu et al., 1991). We confirmed that A1.0-GFP.nls drives embryonic midline expression (Fig. 3G). When postembryonic expression was assayed, we observed that GFP reporter expression was present in all sim+ brain cells and the midline cells of the ventral nerve cord. This included the sim+ anterior brain neurons (Fig. 3B), the medulla and lamina of the optic lobes (Fig. 3B), the posterior brain neurons (Fig. 3C), and larval VNC midline cells (Fig. 3D) (Table 1).
Table 1.
Summary of sim transgenic line larval CNS expression. A summary of transgenic lines that drive reporter gene expression in sim+ neurons in the larval brain. An “X” indicates expression in a particular cell type, while a blank square signifies absence of expression.
| Transgene | DAMv1/2 BAmas1/2 |
TRdm | DPM | PLSC | PSC | Optic lobe Lamina |
Optic lobe Medulla |
VNC midline |
Pan- neural |
Non-Sim+ Brain |
None |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1.0-GFP | X | X | X | X | X | X | X | X | |||
| B2.4-Gal4 | X | X | |||||||||
| C2.3-Gal4 | X | ||||||||||
| D2.1-Gal4 | X | ||||||||||
| E2.3-Gal4 | X | ||||||||||
| 3.7sim-Gal4 | X | ||||||||||
| F1.4-Gal4 | X | X | X | X | X | ||||||
| L1.0-Gal4 | X | X | X | ||||||||
| M582-Gal4 | X | ||||||||||
| N494-Gal4 | X | X | X | X | |||||||
| O501-Gal4 | X | ||||||||||
| P261-Gal4 | X | X | X | X | X | ||||||
| Q255-Gal4 | X | X | X | ||||||||
| G1.1-Gal4 | X | ||||||||||
| H2.4-Gal4 | X | ||||||||||
| I2.6-Gal4 | X | ||||||||||
| J2.5-Gal4 | X | ||||||||||
| K2.6-Gal4 | X |
The Sim:Tgo heterodimeric transcription factor binds to ACGTG sequences (Sonnenfeld et al., 1997). The A1.0 fragment has two ACGTG putative Sim:Tgo binding sites. When these sites were mutated together in A1.0-Gal4 and tested after germline line transformation, all embryonic midline and postembryonic expression was absent (Fig. 3E,F,H). This is consistent with a requirement for sim autoregulation in A1.0-driven CNS and midline expression. However, it also suggests that the subdivision of A1.0 is unlikely to lead to transgenic lines that drive expression in subsets of sim+ brain cells, and no further subdivision of A1.0 was undertaken.
2.5 Two complementary sim midline enhancers recapitulate sim expression in the central brain and optic lobes
Two fragments, B2.4 and F1.4, each drive gene expression in non-overlapping subsets of sim+ central brain and optic lobe neurons. B2.4 expression is present in the PSC and lamina of the optic lobes (Fig. 4A–E). Crossing B2.4-Gal4 to UAS-tau-GFP reveals an outgrowth of axons from the PSC neurons that project dorsally and then cross the midline (Fig. 4E). The F1.4 fragment drives expression in the central brain in all sim+ anterior and posterior clusters, except PSC neurons (Fig. 4F,G). In addition, F1.4 drives expression in the medulla of the optic lobes (see below). Thus, the B2.4 and F1.4 fragments contain enhancers that together control the expression of all sim brain expression; they are complementary and control non-overlapping aspects of sim expression (Table 1).
2.6 Molecular dissection of distinct sim+ brain enhancers
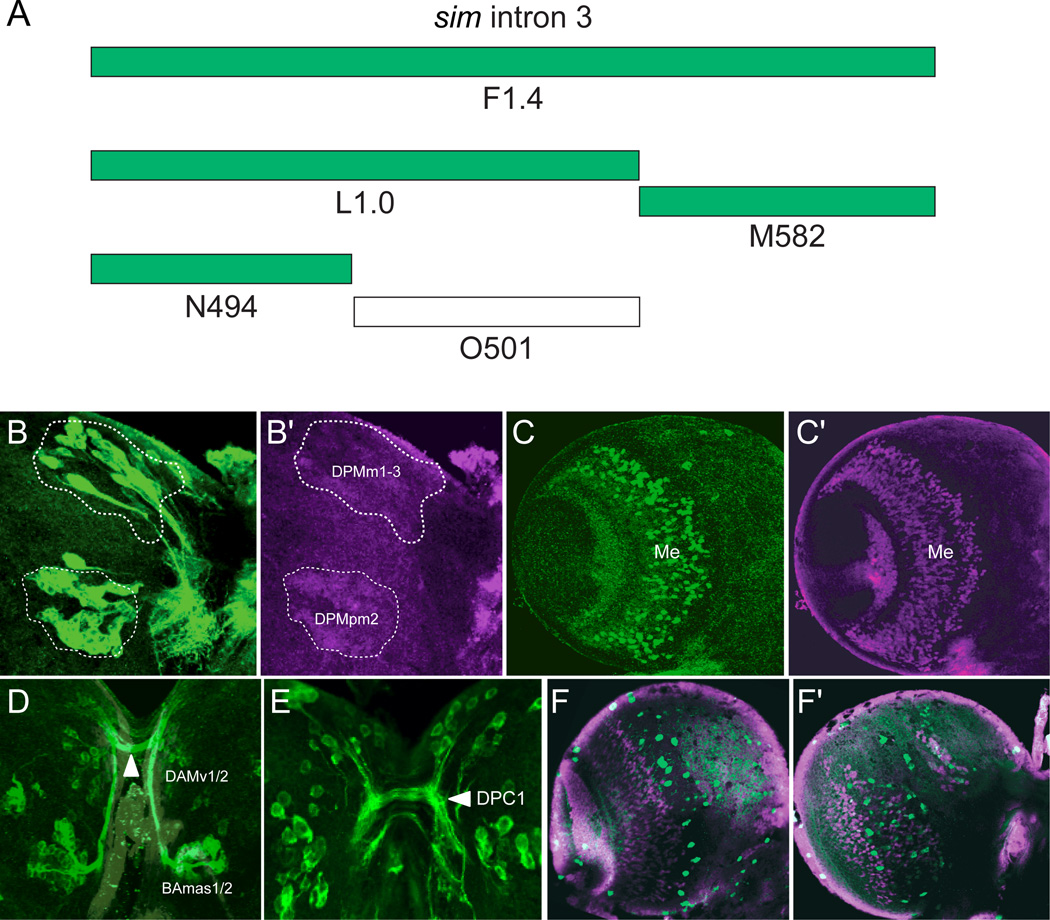
One issue concerning how sim postembryonic brain expression is controlled is the nature of the brain enhancers: is expression of each sim+ neuronal cluster driven by a separate enhancer, or do individual enhancers drive expression in multiple clusters? To begin to address this question, the F1.4 fragment was divided into 4 subfragments, and each was tested for brain expression (Fig. 5A; Table 1). The L1.0 fragment drives expression only in the sim+ DPM, PLSC, and medulla neurons (Fig. 5B) but not in the anterior brain neurons. The adjacent M582 fragment is expressed in only the medulla (Fig. 5C). The number of medullary neurons is reduced compared to F1.4. Although L1.0 and M582 together include the complete F1.4 fragment, which is expressed in the anterior brain neurons, neither L1.0 nor M582 drives expression in these cells. However, one L1.0 subfragment, N494, drives expression in the DAMv1/2 and BAmas1/2 neurons but not the TRdm neurons (Fig. 5D), so that anterior brain enhancers reside in L1.0. On the posterior side of the brain, N494 drives expression in the DPM and PLSC neurons (Fig. 5E), and in a subset of medulla neurons. The O501 fragment has only non-specific expression in scattered brain cells (Fig. 5F,G). Thus, the N494 fragment has enhancers for all of the sim+ brain clusters found in F1.4, expect TRdm, which was not expressed by any of the F1.4 subfragments. There are two distinct enhancers that contribute to medulla expression: L494 and M582. Oddly, the L1.0 fragment, which encompasses N494, only drives posterior, but not anterior, brain expression.
Fig. 5. Intron 3 subfragments drive expression in subsets of Sim+ cells.
(A) The schematic illustrates subfragments of the F1.4 fragment tested for postembryonic brain expression. Green blocks indicate Gal4 constructs that drive GFP in sim+ brain cells, and the unfilled block indicates an absence of Sim+ brain expression. (B,B’) L1.0-Gal4 drives UAS-mCD8-GFP expression in the DPMm1–3, DPMpm2, and PLSC neurons (not shown) on the posterior side of the brain. The brain was stained for GFP (green) and Sim (magenta), and the DPM clusters are circled. (C,C’) M582-Gal4 UAS-nuc-GFP expression drives GFP expression (green) in a subset of medullary neurons (Me). (D) N494-Gal4 UAS-tau-GFP reveals GFP expression (green) in the DAMv1/2 and BAmas1/2 Sim+ neurons on the anterior side of the brain. The BAmas1/2 axons project ipsilaterally before crossing the midline in the commissural tract (arrowhead) along with axons from DAMv1/2. (E) GFP+ cells with N494-Gal4 UAS-tau-GFP expression in the posterior brain overlap with DPM and PLSC neurons. Characteristic of the DPM neurons, the GFP-labeled neurons send their axons across the DPC1 commissure (arrowhead). (F,G) O501-Gal4 UAS-mCD8-GFP shows sporadic GFP+ cells on both the (F) anterior and (G) posterior sides of the brain that only coincidentally overlap with Sim+ neurons.
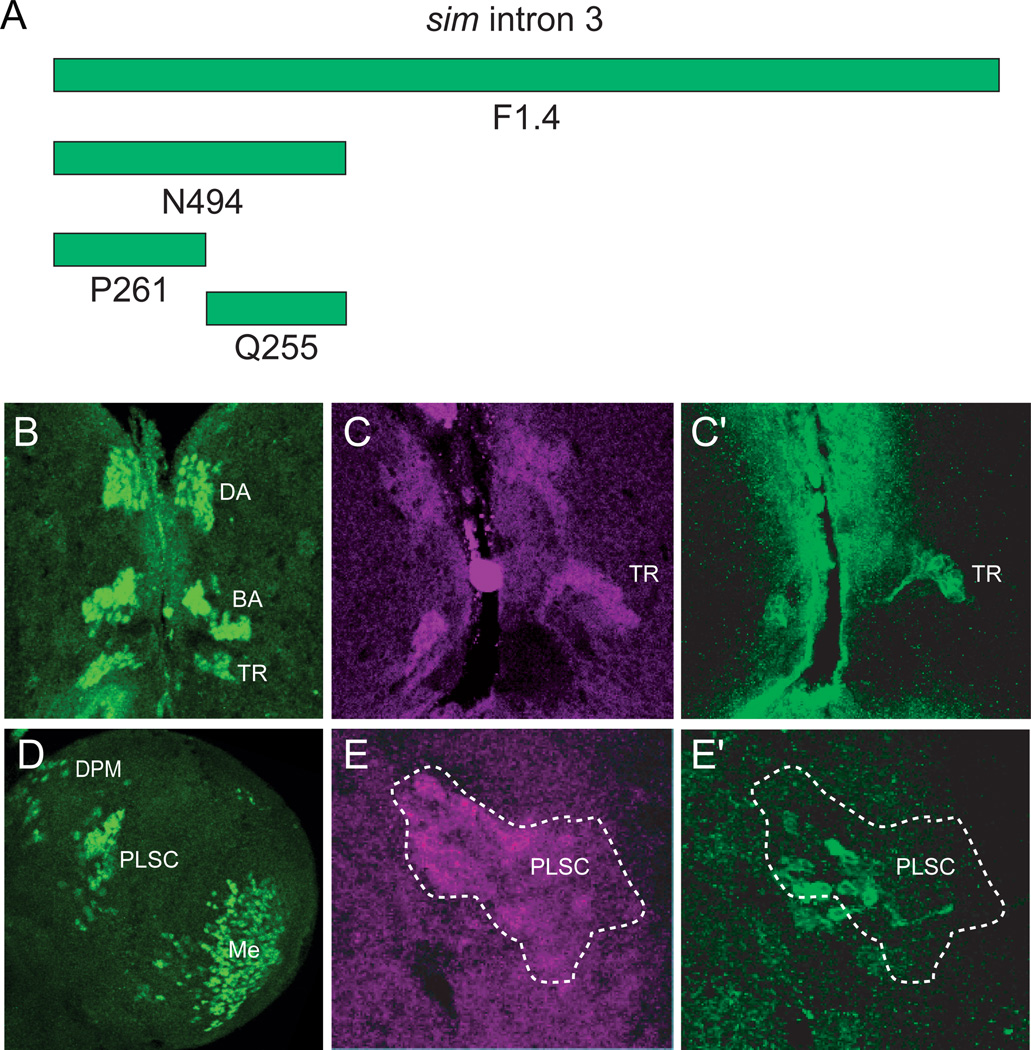
The N494 fragment, which is expressed in most anterior and posterior Sim+ neurons, was further subdivided into two fragments, P261 and Q255 (Fig. 6A; Table 1), to investigate sim brain enhancers. Expression driven by the P261 fragment closely resembled N494-driven expression. P261-Gal4 UAS-nuc-GFP showed GFP expression in all anterior Sim+ brain regions (Fig. 6B), the DPM and PLSC posterior brain clusters and the medulla (Fig. 6D). Interestingly, P261 showed GFP expression in the TRdm neurons, even though expression was absent in these cells in N494-Gal4. The Q255 fragment drove expression in TRdm (Fig. 6C) and weakly in a scattered subset of PLSC (Fig. 6E) and medulla neurons.
Fig. 6. P261 drives expression in most central brain cells.
(A) The N494 fragment from intron 3 was further subdivided into P261 and Q255. (B,D) P261-Gal4 UAS-nuc-GFP revealed GFP expression in: (B) all anterior Sim+ central brain neurons, including TRdm, and (D) the medulla and all posterior Sim+ neurons, except PSC. (C,E) Q255-Gal4 UAS-nuc-GFP was expressed in (C,C’) TRdm neurons and (E,E’) weakly in the PLSC neurons. Brains were stained with anti-GFP (green) and anti-Sim (magenta).
In summary, a single 261 bp fragment, P261 that is derived from F1.4, contains enhancers for all sim+ brain regions, except PSC and the optic lobe lamina. However, the adjacent Q255 fragment also includes elements driving expression in TRdm and weakly in the PLSC and medulla. In addition, another non-overlapping fragment, M582, has elements driving medulla expression. The B2.4 fragment has enhancers that drive PSC and lamina expression, which complements F1.4 expression. The A1.0 fragment has expression in all sim+ brain cell types and is driven by sim autoregulation. Thus, sim central brain expression consists of a combination of distinct enhancers that together initiate and maintain brain expression. In some cases (e.g. medulla), there are multiple enhancers that contribute to expression. It also is possible that enhancers driving expression in some sim+ cells (e.g. DAMv1/2 and BAmas1,2) are not separable.
2.7. Identification of a sim pan-neuronal enhancer
Additional fragments had varying patterns of expression that did not match known sim expression. Three fragments (G1.1, J2.5, and K2.6; Supp. Fig. 2A) drove expression in the brain but not in the cells that overlap with sim expression (Supp. Fig. 2B–H); Table 1. Five fragments (D2.1, E2.3, sim3.7, H2.4, and I2.6) did not drive relevant brain expression (Table 1). Two of the fragments with expression in brain cells that do not overlap with sim, J2.5 and K2.6, reside in the sim 3’-flanking regions and are the most distal fragments from the sim transcription unit that were tested. These fragments may contribute to the brain expression of the adjacent timeout or CG43063 genes. Another fragment, C2.3 (Supp. Fig. 3A), produces unusual expression in all NBs, GMCs, and neurons in the 3rd instar larval brain (Supp. Fig. 3B). The highest expression is present in NBs and GMCs, while expression in neurons is weaker (Supp. Fig. 3C). Expression is also observed in the optic lobe, although at low levels, and in the ventral nerve cord, with the exception of the posterior-most abdominal segments. Multiple overlapping transformants employing different promoters and vectors all showed the same pattern, indicating that the expression was not due to a cloning artifact. While C2.3 pan-neuronal expression overlaps with Sim+ neurons in the brain, the C2.3 pattern is highly distinct from sim, and the C2.3 enhancer presumably reflects the existence of a pan-neuronal enhancer normally repressed in the CNS.
2.8 sim functions in controlling central brain axon guidance
To assess the genetic role of sim in central brain development, MARCM (Lee and Luo, 1999) was carried out with multiple sim alleles to generate mutant clones. Since sim mutant strains with severe embryonic phenotypes had not previously been sequenced nor their corresponding molecular defects identified, we sequenced all 7 coding sequence exons of 12 sim alleles (Supp. Fig. 4A, Supp. Table 1; another sim allele, simJ1–47, was sequenced previously (Pielage et al., 2002)). Three mutants (sim2, sim8, and simBB68) predicted to produce truncated proteins were selected for MARCM, since they are likely to be amorphic. Mutant sim embryos from the 3 selected strains were stained with anti-Sim raised against a bacterially synthesized protein fragment that is predicted to lack all 3 mutant proteins, and no Sim immunoreactivity was observed in homozygous mutant embryos (Supp. Fig. 4B). In addition, all 3 showed severe sim collapsed axon scaffold phenotypes (Thomas et al., 1988).
Mutant clones were analyzed for the DAMv1/2 neurons, since they could be readily identified based on their location adjacent to Sim+ neurons from the adjoining DAMv1/2 NB progeny and they have observable axonal projections in the 3rd instar larval brain (Fig. 7). As a control, we analyzed MARCM larvae that did not harbor a mutant sim allele (Fig. 7A,B). Each wild-type and sim clone possessed a NB, associated GMCs, and Elav+ neurons. Clones mutant for sim had similar numbers of neurons (N=41.4±5.2, n=10) (Fig. 7C–G) as wild-type clones (N=36.0±4.0, n=8) (Fig. 7A,B). Each wild-type and mutant clone contained ~50% of the DAMv1/2 neurons, indicating that most of the progeny of one of the two DAMv1/2 NBs were GFP+. Thus, in the case of the sim mutant clones, most, if not all, of the neurons from a single NB were mutant. These results indicate that the role of sim in DAMv1/2 neurons does not greatly affect neurogenesis, unlike the role of sim in controlling embryonic CNS midline cell formation (Crews, 2003). Instead, axonal fasciculation defects were apparent in sim larval brain mutant MARCM clones.
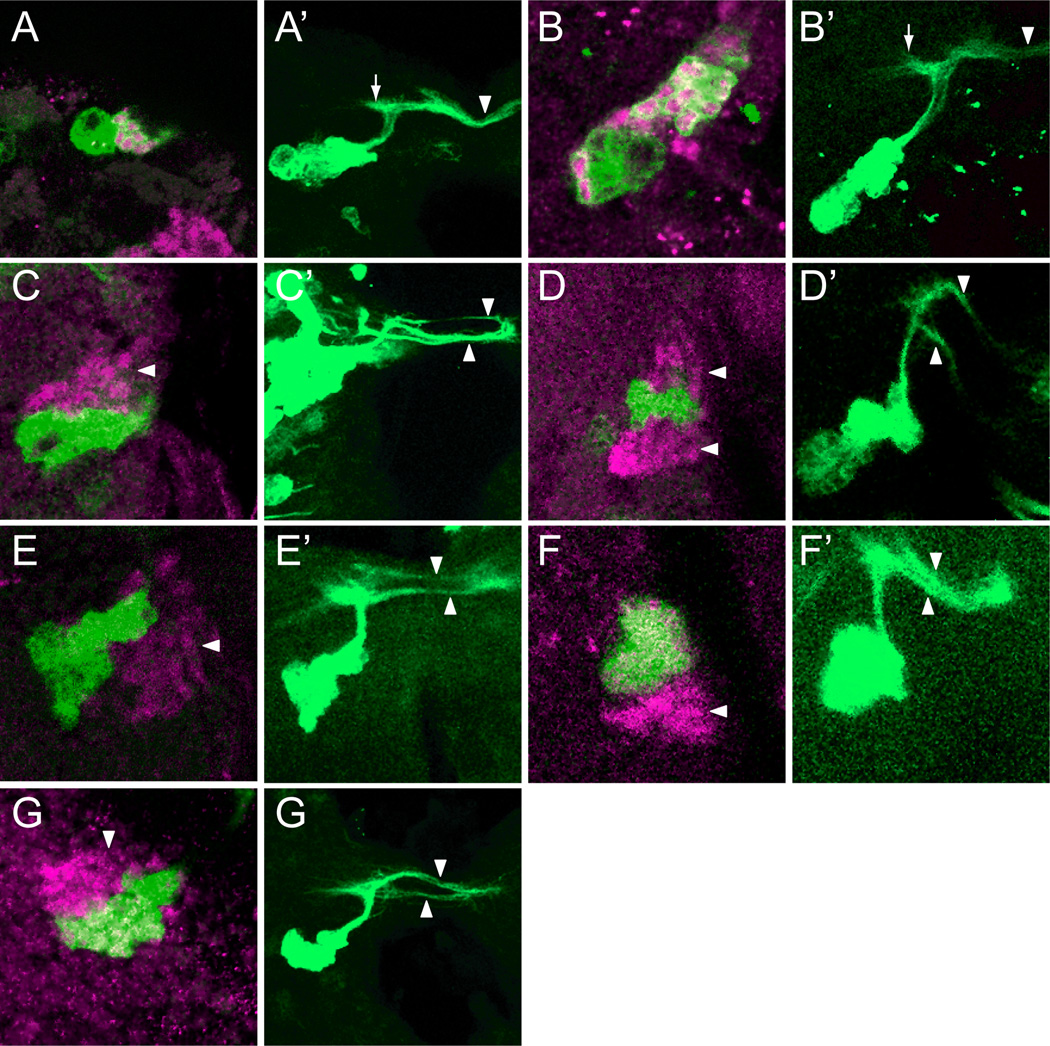
Figure 7. sim mutant clones show axon fasciculation defects.
Wild-type and mutant MARCM GFP+ (green) DMAv1/2 clones were stained with anti-Sim (magenta). For each pair of images, the left panel shows a single optical slice showing the GFP+ cell bodies, and the right panel is a projection that illustrates the axonal morphology. (A,B) Two wild-type GFP+ clones that overlap Sim+ neurons. The NB and GMC are Sim−. (A’,B’) The characteristic DAMv1/2 axon tract is apparent and extends centro-dorsally toward the neuropil, then elaborates filopodia (arrow) before projecting contralaterally (arrowhead) across the supraesophageal commissure. (C,D) Two simBB68 mutant clones that are adjacent to DMAv1/2 Sim+ neurons (arrowheads). (C’,D’) The axons split into multiple fascicles (arrowheads) rather than traverse the supraesophageal commissure as a single, tight fascicle. (E–F’) Two sim2 clones and a (G,G’) sim8 clone that also show multiple branches.
All wild-type MARCM clones within the DAMv1/2 cells (n=8) extended axons with a similar morphology (Fig. 7A’,B’). The GFP+ axons leave the soma dorsally as a common fascicle, elaborate filopodia ipsilaterally (Pereanu and Hartenstein, 2006), and make a sharp turn toward the contralateral side via the supraesophageal commissure. In the commissure, contralateral Sim+ axons form a common tract. MARCM sim mutant clones often showed axon defects (Fig. 7C’–G’). Of the 12 sim mutant DAMv1/2 MARCM clones, 7 showed a mutant phenotype, and 5 appeared wild type. All 3 alleles had at least one clone with a mutant phenotype, and all alleles showed a similar defect. In general, the axons of sim mutant clones extended a tract centro-dorsally, formed an ipsilateral filopodial protuberance, and then turned toward the supraesophageal commissure, resembling wild-type axons. However, a subset of mutant axons aberrantly defasciculated from the main bundle and continued across the midline. While the phenotypes differed in individual clones, they consistently revealed fasciculation defects. In summary, the mutant clones of all 3 sim alleles showed similar axon fasciculation defects, while neurogenesis was unaffected.
3. Discussion
3.1 The Drosophila sim gene is expressed in multiple clusters of identifiable larval brain neurons
Previously, we demonstrated that sim is expressed in 3 anterior clusters of neurons in the 3rd instar larval brain. In this paper, we show that these three clusters are the DAMv1/2, BAmas1/2, and TRdm neurons. Neurons that are Sim+ are present in 1st, 2nd, and 3rd instar larvae at 3 discrete positions in the anterior brain (Fig. 1A–C). It is likely that the cells in 1st and 2nd instar larvae correspond to the identified 3rd instar clusters, but this has not been directly shown. All 5 NBs that give rise to these neurons are Class I NBs, in which the NB generates GMCs, each of which divides once to give rise to two neurons. Axons of the DMAv1/2 and BAmas1/2 neurons ultimately cross the midline, whereas the TRdm axons project ipsilaterally and do not cross. The specific larval and adult functions of these neurons are unknown.
In this paper, we describe three additional clusters of Sim+ neurons that reside on the dorsal/posterior side of the brain. The dorsal-most cluster can be identified as a subset of DPM neurons, which are derived from a Type II NB that generates transit-amplifying progenitors. Another cluster of neurons could not be unambiguously identified, and we tentatively name it PLSC. These cells are also likely to be derived from Type II NBs, since they are relatively dispersed. The third cluster is a small group of neurons, which also were not previously identified, and are tentatively named PSC. The Sim+ DPM, PLSC, and PSC neurons all send axonal projections across the midline.
3.2 sim mutant DAMv1/2 neurons have axon fasciculation defects
Previous analysis of the simJ1–47 allele showed defects in adult walking behavior (Pielage et al., 2002). These defects were interpreted as an inability to coordinate movement and are consistent with a lack of interhemispheric communication. Consistent with this notion is the occurrence of 5 clusters of sim+ neurons in the central brain that cross the midline and could be communicating information that coordinates movement. Analysis of the simJ1–47 adult central complex neuropil revealed a disorganization of the axons that cross the midline (Pielage et al., 2002), providing a cellular rationale for the behavioral defects. However, it is unclear whether this phenotype reflects an absence of sim function or a hypomorphic condition. In addition, it is unknown whether the underlying phenotype affects neurogenesis or axonogenesis, since either could result in the observed adult phenotype. The simJ1–47 phenotype may also be due to an indirect non-autonomous effect of sim on other neurons or axons. In this paper, we used MARCM to examine the sim null mutant phenotype in DAMv1/2 neurons. After sequencing exonic DNA from 13 sim mutants, we selected three that were derived from different genetic backgrounds and are likely to be null mutants. All three mutants possess in–frame stop codons that should produce truncated Sim proteins. sim8 is predicted to produce a protein only 12 amino acids long, whereas sim2 and simBB68 should produce proteins that terminate in the PAS-2 domain and lack Sim activation domains. Previous work has shown that the absence of the sim activation domains results in a protein that cannot activate transcription in vivo (Franks and Crews, 1994). Consistent with these molecular defects, all 3 mutants showed similar defects. In all mutant clones, the neurons and initial axonal projections appeared normal, indicating no obvious effects of sim on neurogenesis and neurite outgrowth. However, in 7/12 mutant clones, there were clear axon fasciculation defects. In wild-type clones, the axons from all DAMv1/2 neurons extended across the midline as a single, tightly fasciculated bundle, whereas in sim mutant larvae there were more than one bundle and the axons appeared frayed.
While provocative, the sim axonal and behavioral phenotypes raise a number of issues. The first is that while axon guidance defects are observed for the DAMv1/2 neurons, it is unknown whether the behavioral defects are the result of this defect, since the simJ1–47 mutation may also affect sim function in the other anterior and posterior Sim+ brain neurons, the optic lobes, and the midline cells of the ventral nerve cord. It is also is possible that reductions in sim could control additional aspects of terminal differentiation and neurotransmission, which could also contribute to the behavioral phenotype. Another potential developmental role is that the sim+ cells themselves do not physiologically contribute to locomotion, but their axons may pioneer the axons of other neurons that control movement. Finally, since only about half of the cells in each cluster were mutant, the presence of genetically wild-type axons mixed with sim mutant axons could mask the severity of the phenotype. These issues can ultimately be resolved using targeted expression of various transgenes affecting sim function, differentiation, and neurotransmission. Axon guidance defects can be assayed in the other sim+ neurons using MARCM, but behavioral phenotypes will need to be addressed by targeting disruptions of sim function specifically to each neuronal cluster. The ability to do this was one of the goals of this study, although the Gal4 lines we generated still generally lack sufficient specificity to fully address this issue. Nevertheless, what is clear is that sim controls proper axonal patterning, but not neurogenesis, of the sim+ DAMv1/2 neurons.
Mammalian Sim1 and Sim2 and Drosophila sim play multiple roles in development, both in the CNS and in other cell types (Crews, 2003; Fan, 2003). Within the CNS, each plays a role in neurogenesis or neural migration and later in axonogenesis. Drosophila sim controls neurogenesis of embryonic CNS midline cells (Nambu et al., 1991) and differentiation of the optic lobe laminar neurons (Umetsu et al., 2006). In mammals, Sim1 plays a prominent role in neuronal migration in the hypothalamus (Xu and Fan, 2007). Additionally, the murine Sim1 and Sim2 genes are expressed in the mammillary body and control axonogenesis (Marion et al., 2005). In wild-type mice, the Sim1+ Sim2+ mammillary body cells extend axons along the principal mammillary tract (PMT) that project to the thalamus and tegmentum via the mammillotegmental (MTEG) and mammillothalamic (MTT) tracts. Genetic experiments indicated that the MTEG and MTT are greatly reduced in Sim1 Sim2 double mutant embryos and, to a lesser degree, in Sim1 single mutant embryos. Normally, the PMT extends along the ipsilateral side of the developing brain, but in Sim1 Sim2 mutant embryos, the axons abnormally cross the midline. This suggests that the mammillary body axons no longer respond to a midline-directed repellent in Sim mutant embryos. Consistent with this interpretation, Sim was shown to normally repress expression of Rig-1/Robo3, a gene that antagonizes Slit-mediated repulsion (Marillat et al., 2004; Sabatier et al., 2004). Consequently, upregulation of Rig-1/Robo3 in Sim mutant embryos results in the loss of PMT repulsion by the midline.
The Drosophila sim DAMv1/2 axonal defect differs from the mammalian Sim mutant defect in that the DAMv1/2 axons show fasciculation defects. Significantly, targeting appears roughly correct, since mutant axons branch and migrate toward the midline. Presumably, sim regulates the expression of one or more genes involved in controlling axonal fasciculation, although the identities of those genes are unknown. There are a number of Drosophila cell adhesion proteins that have been implicated in axon fasciculation (Van Vactor, 1998), including Fasciclin II, Roughest, and Cadherin-N. One possible explanation for the sim phenotype is that Sim positively regulates the levels of cell adhesion/fasciculation proteins, and when their levels drop below a certain point, defasciculation can occur. Alternatively, there exists a class of genes that are anti-adhesive, such as beaten path (Fambrough and Goodman, 1996) and protein tyrosine phosphatases (Desai et al., 1996), which may normally be repressed or silenced by Sim, and, thus, in sim mutants become active and promote defasciculation. Further insights into the mechanisms that govern axon guidance of Sim+ cells will require identifying the relevant transcriptional targets of Sim.
3.3 Multiple enhancers control sim postembryonic expression
Comprehensive transgenic analysis of the sim regulatory region identified enhancers for all postembryonic sim+ brain neurons (Table 1). The P261 fragment drives expression in the anterior central brain, DPM, PLSC, and medulla neurons. The B2.4 fragment drives expression in the PSC and lamina. Thus, these two fragments, which don’t overlap in expression, account for all of the central brain and optic lobe expression. In addition to the P261 enhancer, an adjacent fragment, Q255, drives expression in TRdm, DPM, and the medulla, and another proximate fragment, M582, drives medulla expression. Thus, in some cases, there are multiple enhancers that contribute to expression in specific brain neurons. However, we have not yet sufficiently subdivided the sim enhancer fragments into subfragments capable of driving the expression of each individual brain cell type. In some cases, this may not be possible if expression in different cell types share transcription factor binding sites. In addition, another enhancer in the A1.0 fragment, which resides in the 5’-flanking sequences, is autoregulatory (Crews and Pearson, 2009) and drives expression in all larval brains cells.
The genomic arrangement of the sim brain enhancers provides insights into the mechanisms that control sim expression. RT-PCR data strongly suggest that the brain enhancers function through PL. While the upstream A1.0 autoregulatory enhancer interacts with PL in a straightforward manner, the B2.4 and F1.4 enhancers are downstream of PL and PE and must skip over PE to interact with PL. No relevant brain enhancers were found 3’ to the sim transcription unit, although enhancers in fragments J2.5 and K2.6 with brain expression patterns that do not overlap with sim+ neurons are present in the 3’-flanking region. They may control expression of genes 3’ to sim. ChIP-chip embryonic protein analysis of Drosophila insulators by modENCODE has revealed strong binding of the CTCF, Mdg4, and Su(Hw) proteins to a site just 3’ of the sim transcription unit (Negre et al., 2010) (Fig. 3A). These proteins may act as insulators to block enhancers 3’ to sim from interacting with upstream sim promoters. Similarly, an insulator site just 3’ to the pic gene (Fig. 3A) may act to insulate pic enhancers from acting on sim promoters, and vice-versa.
4. Experimental procedures
4.1 Drosophila strains
Thirteen sim alleles were analyzed from 5 sources: sim1, sim2, sim5, sim6, sim7 (Hilliker et al., 1980); sim8 (Mayer and Nusslein-Volhard, 1988), simBB68, simJJ22, simM55, simTT63, simW3 (J. Skeath and C. Doe; unpubl.), simJ1–47 (Pielage et al., 2002), and simU (unknown). All strains were tested for noncomplementation with the sim2 null mutant at 25°C. Mutant stocks were balanced over TM3 Sb P[w+; Krüppel-Gal4] P[w+; UAS-GFP] (Casso et al., 2000) or TM3 Sb P[ry+; ftz-lacZ]. The marked balancer chromosomes allowed identification of homozygous mutant embryos. The amorphic alleles sim2, sim8, and simBB68 were recombined onto a chromosome bearing FRT82B for use in MARCM analyses. These chromosomes were combined with w; tub-Gal4 FRT82B tub-Gal80/TM3 (J. Treisman) and elav-Gal4 UAS-mCD8-GFP hs-FLP w (Bloomington Drosophila Stock Center). UAS lines employed for assaying transgenic Gal4 expression included UAS-mCD8-GFP (Lee and Luo, 1999), UAS-tau-GFP (Brand, 1995), and UAS-nuc-GFP (UAS-LacZ::GFP.nls) (Shiga et al., 1996).
4.2 RT-PCR
Total RNA was extracted from 0–3, 3–6, 6–9, 9–12, and 12–15 hr (after egg laying, AEL) embryos as well as from first, second, and third instar larvae, light (1–2 day) and dark (3–4 day) pupae, and 2-day-old adults using QIAshredder and RNeasy kits (Qiagen). Samples were treated with RQ1 RNase-free DNase (Promega) and converted to single stranded cDNA using SuperScript II First-Strand Synthesis System for RT-PCR (Invitrogen) and oligo-d(T). Transcripts were then detected by PCR in the linear range using the following primers:
sim ForE 5’-TGG ATG CTG GTT GAT GTG CGG −3’
sim ForL 5’-CAG GGA TAT GAG CAA GTG CTG AGA A −3’
sim Rev 5’-GCC CAA GTG CCA TAA ACG CAA T −3’
RpL32 For 5’-ATC CGC CCA GCA TAC AGG −3’
RpL32 Rev 5’-CTC GTT CTC TTG AGA ACG CAG −3’
Use of the primer pair, sim ForE and Rev, detected transcripts derived from PE (sim RA), whereas use of the primer pair, sim ForL and Rev, detected PL transcripts (sim RB, RC). RT-PCR products were sequenced to confirm their identity.
4.3 Transgene construction
DNA fragments containing intragenic and intronic sequences from the sim locus were PCR-amplified and subcloned into a spectinomycin-resistant pCR8/GW/TOPO Gateway-compatible vector (Invitrogen) and transformed into E. coli. The primer pairs used for PCR amplification are listed in Supp. Table 2. The fragments in pCR8, except A1.0, were then cloned via Gateway LR recombination into an ampicillin-resistant PhiC31 transformation vector, pBPGw-UCP (J. Pearson). The A1.0 fragment was cloned into Mintgate (Jiang et al., 2010) to generate A1.0-GFP.nls. Each sim transgenic construct was microinjected into Drosophila embryos that express germline-localized PhiC31 integrase and contain the PhiC31 genomic destination site attP2 (68A4) (Groth et al., 2004). PCR-based site-directed mutagenesis was used to mutate both A1.0 Sim:Tgo consensus binding sites (NACGTG) to GGATCC. Primers used are listed in Supp. Table 2.
4.4 Immunostaining
Embryos were collected, fixed, and stained using standard procedures (Patel et al., 1987). Larval brains were dissected in 1X PBS, fixed in 4% formaldehyde in 1X PBS for 20 min, and stained using the following antibodies and reagents: rat anti-Sim (1:100) (Ward et al., 1998), guinea pig anti-Sim (1:200) (Wheeler et al., 2008), murine MAb anti-BP106 (1:100, Developmental Studies Hybridoma Bank, Iowa) (Hortsch et al., 1990), murine anti-Tau2 (1:500; Sigma), rabbit anti-β-galactosidase (1:2000; Cappel), murine MAb 9F8A9 anti-Elav (1:100; Developmental Studies Hybridoma Bank, Iowa) (O'Neill et al., 1994), rabbit anti-GFP (1:1000; Abcam), Cy3-, Cy5-, Alexa350-, Alexa488-, Alexa568-conjugated secondary antibodies (1:1000; Invitrogen), HRP-conjugated secondary antibodies (Jackson ImmunoResearch), and 1.0 mg/ml DAPI (1:1000; Sigma). Fluorescently stained specimens were mounted in Aqua-Poly/Mount (Polysciences) or glycerol and imaged on Zeiss LSM510 and Pascal confocal microscopes using 40× objectives.
4.5 MARCM
hs-Flp elav-Gal4 UAS-mCD8-GFP; tub-Gal4 FRT82B tub-Gal80/+ males were crossed to virgin females bearing either FRT82B sim2; FRT82B sim8; FRT82B simBB68; or FRT82B P[w+] chromosomes (Lee and Luo, 1999). Embryos were collected for 3 hr and aged for 24 hr at 25°C, then heat shocked for 1.5 hr at 37°C, followed by aging at 25°C until they became wandering 3rd instar larvae. Brains were isolated, fixed, stained, and analyzed as described above.
4.6 Sequencing of sim mutant DNA
Genomic DNA was isolated from stage 14–15 homozygous sim mutant embryos that were identified based on the absence of balancer chromosome GFP expression. Sequencing of the sim gene was performed using DNA fragments isolated by touchdown PCR (tdPCR) (Don et al., 1991; Hecker and Roux, 1996). Seven sets of primer pairs were used to amplify exons 4–10, which comprise most of the sim coding sequence and corresponding splice sites (Supp. Table 3). PCR products were purified and sequenced at the UNC-CH Genome Analysis Facility.
Highlights.
The Drosophila single-minded regulatory gene is expressed in the central brain and optic lobes
We assigned the single-minded+ central brain cells to 6 clusters of identified neurons
Multiple enhancers functionally combine to drive expression in single-minded+ brain cells
single-minded mutants reveal a defect in central brain cell axon guidance
single-minded may direct the formation of brain circuits that control locomotion
Supplementary Material
Sim is present in brain neurons. (A–E) Wild-type 3rd instar larval brain showing that Sim protein is not present in neural precursor cells but only in neurons. Shown is a MARCM clone stained with anti-Sim (A; red), anti-Elav (B; magenta), DAPI (C; blue), and anti-GFP (D; green); merge is shown in E. The GFP+ cells include a NB (arrowhead), 2–3 putative GMCs (white arrows), and ~10 neurons (yellow arrow). Elav immunoreactivity is absent from the NB, GMCs have a low level of Elav, and neurons have high Elav levels. Sim protein is only detectable in neurons and is absent from the NB and GMCs.
Gal4 lines with postembryonic brain expression not in Sim+ cells. (A) Schematic showing the location of 3 fragments within or near the sim gene that drive expression in the postembryonic larval brain but not in Sim+ cells. G1.1 encompasses sim intron 7, and J2.5 and K2.6 lie 3’ to the sim transcription unit. (B,B’) G1.1-Gal4 UAS-mCD8-GFP shows GFP expression in anterior brain cells that are predominantly Sim−, although there is some overlap with the Sim+ DAMv1/2 (DA) neurons. Anti-Sim labeling is in magenta, and GFP is green. (C) G1.1-Gal4 UAS-mCD8-GFP is expressed sporadically on the posterior side of the brain with relatively little overlap in Sim+ cells. (D–D’’) Enlarged view of the PLSC and PSC neurons showing only a few GFP+ cells that overlap with Sim+ cells. (E,F) Brains from J2.5-Gal4 UAS-nuc-GFP larvae revealing GFP expression (green) in cells of the (E) anterior and (F) posterior brain. GFP expression does not overlap with Sim (magenta). (G,H) K2.6-Gal4 UAS-mCD8-GFP expression in neurons (green) in the (G) anterior and (H) posterior brain with minimal overlap with Sim (magenta).
C2.3 drives expression in neuroblasts and GMCs. (A) Schematic indicating the location of C2.3 in sim intron 1. (B) Expression of UAS-nuc-GFP driven by C2.3-Gal4 labels all NBs and newly born GMCs on the anterior side of the brain. (C–C’’) Enlarged image of the Sim+ (magenta) BAmas1/2 cells. GFP co-localizes with the Sim+ neurons, although expression is highest in the Sim− NB (yellow arrow) and GMCs (white arrow) and weaker in the neurons (arrowhead). In contrast, within the brain, Sim is generally highest in the neurons and lowest in the NB (Supp. Fig. 1).
Structure of Sim wild-type and mutant proteins. (A) The sequence structure of the Sim protein is shown. The length of wild-type Sim is 673 aa (Nambu et al., 1991); the Sim-PB isoform is shown. At the top are the boundaries between defined regions: basic region (b), HLH domain, PAS-1, PAS-2, AQ-rich region, and three transcriptional activation domains (A1–3) (Franks and Crews, 1994). The sites of exon junctions are shown below the sequence, with the exons (E) separated by a vertical line and the residue where the splice occurs below the line. The antigen used to generate the Sim antibody used in this work spans aa residues 413 to 650 and is indicated by a brown bar. Predicted proteins of each mutant strain are shown to the right of each allele name. The green bar indicates the predicted extent of the protein, if translated, and the length of the predicted protein in amino acids is indicated to the right. The orange vertical lines for sim7, simJ1-47, and simU indicate the sites of their missense mutations. The orange region for simW3 indicates the distance from the splice site mutation to the first in-frame termination codon. The orange region for sim1 indicates the distance from the site of the frameshift mutation to the first in-frame termination codon. (B) Stage 10 embryos from wild type, sim2, sim8, and simBB68 were stained with anti-Sim to assay expression. None of the mutants showed detectable embryonic Sim protein using the antisera generated against Sim 413–650.
Acknowledgements
The authors would like to thank Volker Hartenstein, Jim Skeath, Chris Doe, Christian Klämbt, Jessica Treisman, FlyBase, the Developmental Studies Hybridoma Bank, and the Bloomington Stock Center for Drosophila stocks and advice. This project was funded by grants from the National Institutes of Health (NICHD and NCRR) to STC. The UNC Developmental Biology NIH training grant, T32 HD046369, provided support to JCP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berger C, Renner S, Luer K, Technau GM. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev. Dyn. 2007;236:3562–3568. doi: 10.1002/dvdy.21372. [DOI] [PubMed] [Google Scholar]
- Brand A. GFP in Drosophila. Trends Genet. 1995;11:324–325. doi: 10.1016/s0168-9525(00)89091-5. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Casso D, Ramirez-Weber F, Kornberg TB. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech Dev. 2000;91:451–4. doi: 10.1016/s0925-4773(00)00248-3. [DOI] [PubMed] [Google Scholar]
- Crews ST. Drosophila bHLH-PAS Developmental Regulatory Proteins. In: Crews ST, editor. PAS Proteins: Regulators and Sensors of Development and Physiology. Kluwer; 2003. pp. 69–108. [Google Scholar]
- Crews ST, Pearson JC. Transcriptional autoregulation in development. Curr. Biol. 2009;19:241–6. doi: 10.1016/j.cub.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews ST, Thomas JB, Goodman CS. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell. 1988;52:143–151. doi: 10.1016/0092-8674(88)90538-7. [DOI] [PubMed] [Google Scholar]
- Desai CJ, Gindhart JG, Jr, Goldstein LS, Zinn K. Receptor tyrosine phosphatases are required for motor axon guidance in the Drosophila embryo. Cell. 1996;84:599–609. doi: 10.1016/s0092-8674(00)81035-1. [DOI] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes P, Mosher J, Crews ST. Drosophila single-minded represses gene transcription by activating the expression of repressive factors. Dev Biol. 2001;232:157–175. doi: 10.1006/dbio.2001.0174. [DOI] [PubMed] [Google Scholar]
- Fambrough D, Goodman CS. The Drosophila beaten path gene encodes a novel secreted protein that regulates defasciculation at motor axon choice points. Cell. 1996;87:1049–1058. doi: 10.1016/s0092-8674(00)81799-7. [DOI] [PubMed] [Google Scholar]
- Fan CM. Hormones, Obesity, Learning, and Breathing - the Many Functions of the Mammalian Single-Minded Genes. In: Crews ST, editor. PAS Proteins: Regulators and Sensors of Development and Physiology. Kluwer; 2003. pp. 205–230. [Google Scholar]
- Fan CM, Kuwana E, Bulfone A, Fletcher CF, Copeland NG, Jenkins NA, Crews S, Martinez S, Puelles L, Rubenstein JL, Tessier-Lavigne M. Expression patterns of two murine homologs of Drosophila single-minded suggest possible roles in embryonic patterning and in the pathogenesis of Down syndrome. Mol. CellNeurosci. 1996;7:1–16. doi: 10.1006/mcne.1996.0001. [DOI] [PubMed] [Google Scholar]
- Franks RG, Crews ST. Transcriptional activation domains of the Single-minded bHLH protein are required for CNS midline cell development. Mech Dev. 1994;45:269–277. doi: 10.1016/0925-4773(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker KH, Roux KH. High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. BioTechniques. 1996;20:478–485. doi: 10.2144/19962003478. [DOI] [PubMed] [Google Scholar]
- Hilliker AJ, Clark SH, Chovnick A, Gelbart WM. Cytogenetic analysis of the chromosomal region immediately adjacent to the rosy locus in Drosophila melanogaster. Genetics. 1980;95:95–110. doi: 10.1093/genetics/95.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortsch M, Patel NH, Bieber AJ, Traquina ZR, Goodman CS. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases, is dynamically expressed during embryogenesis. Development. 1990;110:1327–1340. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Izergina N, Balmer J, Bello B, Reichert H. Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 2009;4:44. doi: 10.1186/1749-8104-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Pearson JC, Crews ST. Diverse modes of Drosophila tracheal fusion cell transcriptional regulation. Mech Dev. 2010;127:265–280. doi: 10.1016/j.mod.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai Y, Stahl S, Crews S. Specification of the Drosophila CNS midline cell lineage: direct control of single-minded transcription by dorsal/ventral patterning genes. Gene Expr. 1998;7:171–189. [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Marillat V, Sabatier C, Failli V, Matsunaga E, Sotelo C, Tessier-Lavigne M, Chedotal A. The slit receptor Rig-1/Robo3 controls midline crossing by hindbrain precerebellar neurons and axons. Neuron. 2004;43:69–79. doi: 10.1016/j.neuron.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Marion JF, Yang C, Caqueret A, Boucher F, Michaud JL. Sim1 and Sim2 are required for the correct targeting of mammillary body axons. Development. 2005;132:5527–5537. doi: 10.1242/dev.02142. [DOI] [PubMed] [Google Scholar]
- Mayer U, Nusslein-Volhard C. A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev. 1988;2:1496–1511. doi: 10.1101/gad.2.11.1496. [DOI] [PubMed] [Google Scholar]
- Muralidhar MG, Callahan CA, Thomas JB. Single-minded regulation of genes in the embryonic midline of the Drosophila central nervous system. Mech Dev. 1993;41:129–138. doi: 10.1016/0925-4773(93)90043-w. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Franks RG, Hu S, Crews ST. The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell. 1990;63:63–75. doi: 10.1016/0092-8674(90)90288-p. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Lewis JL, Wharton KA, Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein which acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, Stein L, Henikoff S, Kellis M, White KP. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell PO, Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Patel NH, Snow PM, Goodman CS. Characterization and cloning of fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielage J, Steffes G, Lau DC, Parente BA, Crews ST, Strauss R, Klambt C. Novel behavioral and developmental defects associated with Drosophila single-minded. Dev Biol. 2002;249:283–299. doi: 10.1006/dbio.2002.0770. [DOI] [PubMed] [Google Scholar]
- Sabatier C, Plump AS, Le M, Brose K, Tamada A, Murakami F, Lee EY, Tessier-Lavigne M. The divergent Robo family protein rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell. 2004;117:157–169. doi: 10.1016/s0092-8674(04)00303-4. [DOI] [PubMed] [Google Scholar]
- Shiga Y, Tanaka-Matakatsu M, Hayashi S. A nuclear GFP/b-galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev Growth Diff. 1996;38:99–106. [Google Scholar]
- Sonnenfeld M, Ward M, Nystrom G, Mosher J, Stahl S, Crews S. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development. 1997;124:4571–4582. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- Strauss R. The central complex and the genetic dissection of locomotor behaviour. Curr. Opin Neurobiol. 2002;12:633–638. doi: 10.1016/s0959-4388(02)00385-9. [DOI] [PubMed] [Google Scholar]
- Thomas JB, Crews ST, Goodman CS. Molecular genetics of the single-minded locus: a gene involved in the development of the Drosophila nervous system. Cell. 1988;52:133–141. doi: 10.1016/0092-8674(88)90537-5. [DOI] [PubMed] [Google Scholar]
- Umetsu D, Murakami S, Sato M, Tabata T. The highly ordered assembly of retinal axons and their synaptic partners is regulated by Hedgehog/Single-minded in the Drosophila visual system. Development. 2006;133:791–800. doi: 10.1242/dev.02253. [DOI] [PubMed] [Google Scholar]
- Van Vactor D. Adhesion and signaling in axonal fasciculation. Curr.Opin Neurobiol. 1998;8:80–86. doi: 10.1016/s0959-4388(98)80011-1. [DOI] [PubMed] [Google Scholar]
- Ward MP, Mosher JT, Crews ST. Regulation of bHLH-PAS protein subcellular localization during Drosophila embryogenesis. Development. 1998;125:1599–1608. doi: 10.1242/dev.125.9.1599. [DOI] [PubMed] [Google Scholar]
- Wharton JKA, Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymmetric E-box elements: similarity to xenobiotic responsive regulation. Development. 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Kearney JB, Guardiola AR, Crews ST. Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev Biol. 2006;294:509–524. doi: 10.1016/j.ydbio.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SR, Stagg SB, Crews ST. Multiple Notch signaling events control Drosophila CNS midline neurogenesis, gliogenesis and neuronal identity. Development. 2008;135:3071–3079. doi: 10.1242/dev.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan CM. Allocation of paraventricular and supraoptic neurons requires Sim1 function: a role for a Sim1 downstream gene PlexinC1. Mol Endocrinol. 2007;21:1234–1245. doi: 10.1210/me.2007-0034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sim is present in brain neurons. (A–E) Wild-type 3rd instar larval brain showing that Sim protein is not present in neural precursor cells but only in neurons. Shown is a MARCM clone stained with anti-Sim (A; red), anti-Elav (B; magenta), DAPI (C; blue), and anti-GFP (D; green); merge is shown in E. The GFP+ cells include a NB (arrowhead), 2–3 putative GMCs (white arrows), and ~10 neurons (yellow arrow). Elav immunoreactivity is absent from the NB, GMCs have a low level of Elav, and neurons have high Elav levels. Sim protein is only detectable in neurons and is absent from the NB and GMCs.
Gal4 lines with postembryonic brain expression not in Sim+ cells. (A) Schematic showing the location of 3 fragments within or near the sim gene that drive expression in the postembryonic larval brain but not in Sim+ cells. G1.1 encompasses sim intron 7, and J2.5 and K2.6 lie 3’ to the sim transcription unit. (B,B’) G1.1-Gal4 UAS-mCD8-GFP shows GFP expression in anterior brain cells that are predominantly Sim−, although there is some overlap with the Sim+ DAMv1/2 (DA) neurons. Anti-Sim labeling is in magenta, and GFP is green. (C) G1.1-Gal4 UAS-mCD8-GFP is expressed sporadically on the posterior side of the brain with relatively little overlap in Sim+ cells. (D–D’’) Enlarged view of the PLSC and PSC neurons showing only a few GFP+ cells that overlap with Sim+ cells. (E,F) Brains from J2.5-Gal4 UAS-nuc-GFP larvae revealing GFP expression (green) in cells of the (E) anterior and (F) posterior brain. GFP expression does not overlap with Sim (magenta). (G,H) K2.6-Gal4 UAS-mCD8-GFP expression in neurons (green) in the (G) anterior and (H) posterior brain with minimal overlap with Sim (magenta).
C2.3 drives expression in neuroblasts and GMCs. (A) Schematic indicating the location of C2.3 in sim intron 1. (B) Expression of UAS-nuc-GFP driven by C2.3-Gal4 labels all NBs and newly born GMCs on the anterior side of the brain. (C–C’’) Enlarged image of the Sim+ (magenta) BAmas1/2 cells. GFP co-localizes with the Sim+ neurons, although expression is highest in the Sim− NB (yellow arrow) and GMCs (white arrow) and weaker in the neurons (arrowhead). In contrast, within the brain, Sim is generally highest in the neurons and lowest in the NB (Supp. Fig. 1).
Structure of Sim wild-type and mutant proteins. (A) The sequence structure of the Sim protein is shown. The length of wild-type Sim is 673 aa (Nambu et al., 1991); the Sim-PB isoform is shown. At the top are the boundaries between defined regions: basic region (b), HLH domain, PAS-1, PAS-2, AQ-rich region, and three transcriptional activation domains (A1–3) (Franks and Crews, 1994). The sites of exon junctions are shown below the sequence, with the exons (E) separated by a vertical line and the residue where the splice occurs below the line. The antigen used to generate the Sim antibody used in this work spans aa residues 413 to 650 and is indicated by a brown bar. Predicted proteins of each mutant strain are shown to the right of each allele name. The green bar indicates the predicted extent of the protein, if translated, and the length of the predicted protein in amino acids is indicated to the right. The orange vertical lines for sim7, simJ1-47, and simU indicate the sites of their missense mutations. The orange region for simW3 indicates the distance from the splice site mutation to the first in-frame termination codon. The orange region for sim1 indicates the distance from the site of the frameshift mutation to the first in-frame termination codon. (B) Stage 10 embryos from wild type, sim2, sim8, and simBB68 were stained with anti-Sim to assay expression. None of the mutants showed detectable embryonic Sim protein using the antisera generated against Sim 413–650.