Abstract
Transcriptional regulation of eukaryotic protein-coding genes requires the participation of site-specific transcription factors that bind distal regulatory elements, as well as factors which, together with RNA polymerase II, form the basal transcription machinery at the core promoter. Because gene regulation requires proper communication between promoters and enhancers, often over great distances, it is important to understand the potentially inter-related transcription factor interactions at both of these elements. How this is achieved on tissue-specific genes, such as the immunoglobulin heavy chain (IgH) in B cells remains unclear. Here we review known interactions at the Igh variable region (VH) promoters and present our perspective on promoter-enhancer interactions that are likely important for Ig gene regulation in B cells.
Regulated expression of Igh
The seminal studies of Jacob and Monod performed in E. Coli led to the idea of regulated transcription (1). Subsequent biochemical and genetic analyses established key aspects of gene regulation in prokaryotes (2). It has, however, been far more challenging to obtain a comparable understanding of the mechanisms that underlie tissue-specific gene regulation in mammals. Beyond the increased complexity of mammalian genomes and transcriptional machineries (3), this also reflects the difficulty of faithfully recapitulating, in well-defined in vitro systems, the long-range enhancer-promoter interactions that are necessary for tissue specific gene expression. While this has somewhat hampered our understanding of mechanistic details of regulatory processes, significant knowledge about tissue-specific transcriptional regulation has been derived from transgenic studies in murine models as well as cell-based transfection and/or transduction studies. Collectively, these studies have revealed that site-specific, enhancer-bound regulatory factors communicate, through various cofactors, with the basal transcriptional machinery that associates with core promoters near the transcriptional initiation site (3). However, for most genes the precise molecular mechanisms and contact points between the enhancer- and promoter-bound transcription factors remain unclear. Added to this complexity is the fact that tissue-specific genes are subject to natural constraints that often involve regulation of chromatin structure and subnuclear positioning (4–7). For instance, Ig genes have to negotiate tissue- and differentiation-specific epigenetic changes in chromatin structure (accessibility), alteration in nuclear architecture (repositioning and compaction), genetic recombination (from unrearranged to fully rearranged alleles) and transcriptional regulation before it can be expressed (4–7).
Transcription of Ig genes is ongoing throughout B cell development (8). However, the recombination process complicates the understanding of Ig gene regulation in early stages of B cell development. Therefore, studies of the recombined functional Ig genes provide a clearer mechanistic understanding of how enhancers communicate with promoters. The recombined Igh gene has a rather simple promoter (referred to as the VH promoter) that is comprised primarily of a conserved TATA box around −30 and a highly conserved DNA sequence element (called the octamer) around −70 relative to the transcription start site (4, 8). Recent studies have identified several other functional promoter elements in this region, although these may not be conserved across all gene families (9). The activity of the VH promoter is driven by downstream enhancers (reviewed in 4) and transcriptional regulation is ultimately believed to require enhancer-promoter interactions. In this review we bring together current knowledge on these interactions with emphasis on regulation of the VH promoter. We will first briefly discuss components of the general transcriptional machinery. We will then discuss functions of the Igh enhancers followed by the promoter elements and corresponding factors that might be relevant for these enhancer-promoter communications. Although various distinct mechanisms for enhancer-mediated transcriptional activation are only just becoming clear, the existing data largely points to a looping mechanism for enhancer-promoter communication necessary to regulate Igh transcription.
General transcriptional machinery and co-factors
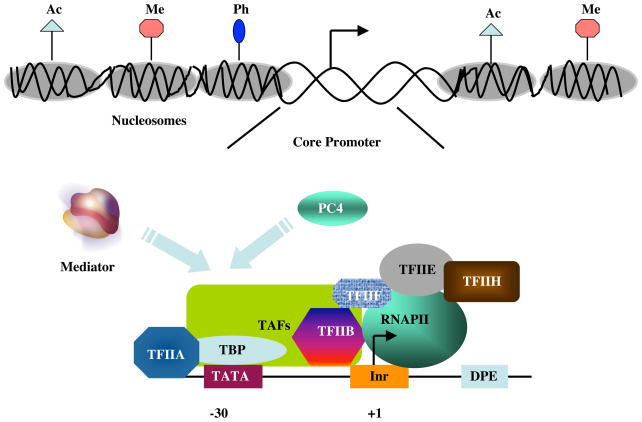
The transcriptional regulation of protein coding genes requires participation of multiple factors. These include both site-specific DNA binding proteins that interact with their cognate sequence elements either upstream or downstream of the transcription start site, and RNA polymerase II (Pol II) and cognate general transcription factors (GTFs) that comprise the general transcription machinery and represent the ultimate targets of the gene-specific factors (3). The GTFs together with Pol II are assembled into a preinitiation complex (PIC) at the “core” promoter/transcription start site (3), which predominantly includes combinations of the TATA box, Initiator (Inr) and downstream promoter (DPE) elements (ref. 10; Fig 1). The rate of formation, stability and function (initiation) of the PIC is governed by the site-specific transcription factors, usually by long-range interactions that involve numerous other transcriptional co-factors (3,11).
Figure 1.
Model for formation of a pre-initiation complex (PIC) on the core promoter following local chromatin modifications. a) A chromatin template that has been remodeled through the action of histone modifying and ATP-dependent nucleosome remodeling factors (not shown). Ac, Me, and Ph represent acetylation, methylation and phosphorylation marks on histone tails, respectively. It is believed that nucleosomal displacement around the core promoter facilitates interactions of the general transcription machinery that result in formation of a preinitiation complex. b) A pre-initiation complex on a core promoter containing TATA, Inr and DPE elements at the normal positions. The minimal preinitiation complex contains RNA polymerase II (RNAPII) and the six GTFs (TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH) that comprise the general transcriptional machinery, with site-specific TATA box interactions mediated exclusively by TBP and potential Inr and DPE interactions mediated by the TBP-associated factors (TAFs) that, for simplicity, are not shown individually. Following interactions with transcriptional activators at distal sites (not shown), the general transcriptional cofactors PC4 and Mediator may target multiple components of the general machinery and facilitate formation and/or function (initiation) of the pre-initiation complex.
Transcriptional co-factors can be broadly divided into two classes – those that directly link site-specific regulatory factors to the general transcription machinery (11) and those that act indirectly on the promoter/enhancer interactions of these factors through modifications of chromatin structure (12–19). Although it is highly likely that many of these factors and their cognate histone marks are involved in Ig gene transcriptional regulation, their actual functions and mechanisms of action in the regulation of Ig genes in B cells remain mostly undetermined.
The cofactors that act more directly include general co-activators such as Positive cofactor 4 (PC4) and the Mediator complex and cell-specific cofactors such as the B cell-specific OCA-B (11). The Mediator is a multiprotein complex (~ 30 subunits) that, in addition to interacting directly with Pol II, can interact with and mediate functions of numerous transcriptional cofactors, including those acting at the level of chromatin (11). While in vitro studies have shown requirements for both PC4 and Mediator/PC2 in Igh transcriptional regulation via synergism with octamer factor and cognate co-activator, OCA-B (20, 21), how these various co-factors are functionally involved in expression of the rearranged Igh gene in normal B cells is currently unknown.
Igh enhancers and transcriptional regulation
Igh transcriptional activation during B-cell differentiation occurs in discrete and independently regulated domains bearing chromatin boundaries with defined histone modification marks (22–26). The enhancers play important roles at multiple stages of Ig gene expression, including at the recombined Igh locus.
The intronic (Eμ) and 3′-RR enhancers
The best characterized Igh enhancers are the intronic (Eμ) enhancer located between the 3′-most joining (J) and (constant-mu) Cμ regions and the 3′ enhancers (located downstream of Cα, here referred to as the 3′-regulatory region or 3′-RR) that are spread over 30 kb (Fig 2A). These enhancers contain binding sites for several transcription factors that include NF-κB, E2A, BSAP/Pax5, Ets proteins (including PU.1) and octamer factors (OCTs) and both enhancers exhibit B cell-restricted as well as developmentally regulated functions (reviewed in 4). The intronic enhancer is required during early (pre-B to mature B cell) stages of differentiation where it drives Igh gene assembly and IgH Cμ gene expression (8). On the other hand the 3′-RR, appear to be required for high-level expression of class-switched Ig genes in activated B cells (27–30).
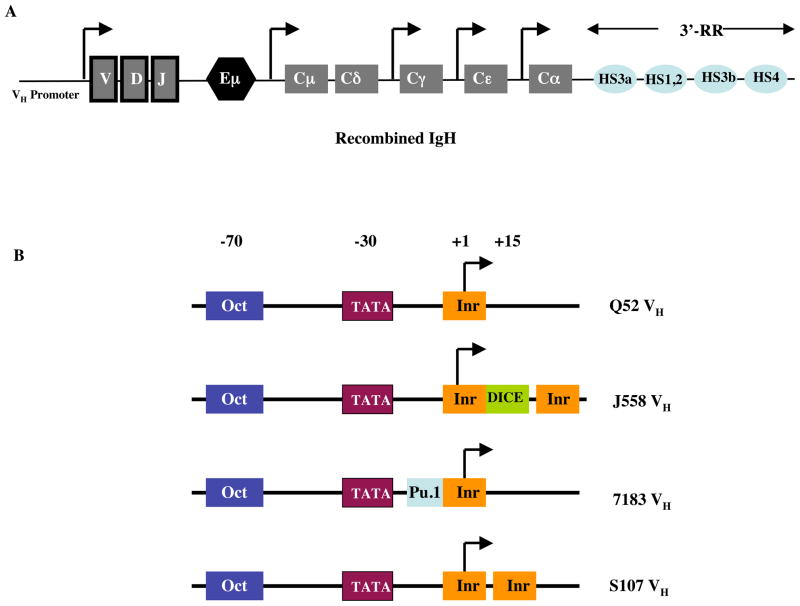
Figure 2. Location and diversity of VH promoters.
(a) Architecture of the VDJ rearranged Igh gene. The locations of Eμ and the 3′-RR enhancers are shown. The VH promoter is upstream of the recombined VDJ region. Various constant (C)-region genes are also indicated along with their promoters (distances are not to scale) (b) Diversity and architectures of various families of VH promoters. Although the majority of the family members show the depicted architecture, there still exists considerable heterogeneity (ref. 9). While the positions and functions of both TATA box and octamer (Oct) elements are experimentally determined, the positions of the Inr, DICE and Pu.1 elements are based on sequence analyses (ref. 9). Functional contributions of the DICE element, both in isolation and in conjunction with Eμ have been shown in vitro. The functional significance of the putative Inr and Pu.1 elements in Igh transcription is predicted but experimentally undetermined.
The Eμ intronic enhancer activity has been localized to a 700-bp region of the JH-Cμ intron (Fig 2A), although the bulk of this activity maps to a 220-bp “core” region that contains all the functionally characterized binding sites for transcription factors (4). The “core” enhancer is flanked by two A/T rich matrix-associated regions (MARs, DNA sequences that interact with the nuclear matrix). MARs are believed to augment linked enhancer activity and can protect associated genes from position effects by acting as boundary or insulator elements. However, although deletion of Eμ inhibits VDJ recombination with a more pronounced effect on the VDJ step compared to the DJ step (31), only the “core” enhancer is essential for these events, as deletion of MARs in the endogenous locus does not affect VDJ recombination or Igh gene expression from the altered allele (4). Despite a requirement for Eμ in VDJ rearrangement is clearly evident from gene targeting experiments, a recent study indicates that deletion of Eμ from a rearranged Igh gene has no appreciable effects on either Igh transcript levels or B cell development in mice. However, lack of Eμ impacts allelic exclusion (32). Likewise, experiments in cell lines have suggested that Igh expression is normal from rearranged alleles lacking Eμ whereas both Eμ and the 3′-enhancer appear to be important for CSR (reviewed in 4). Deletion of Eμ also leads to variegated IgH protein expression (33). Thus, IgH protein is expressed in only a subset of cells after enhancer deletion (33). However, in IgH expressing cells that lacked Eμ, the level of expression was comparable to the cells in which the enhancer was intact (33). The IgH expressing but Eμ lacking cells (“on” state) propagated only around 100 cell divisions, whereas the cells not expressing IgH (“off” state) propagated for around 40,000 cell divisions. Thus, Eμ could be regulating propagated “on” versus “off” states and thereby involved in initiation versus maintenance of Ig gene expression (33, 34).
It has also been shown that the intronic enhancer serves as a source of antisense transcripts, and that transcription extends into the VH part of the locus in pro-B cells and remodels chromatin for recombination (35). Antisense transcription is abrogated in mice lacking Eμ, suggesting Eμ controls antisense transcription and thereby recombination (35).
The 3′-RR constitutes a large and complex region that lies 200 kb downstream of Eμ and appears to be required for regulation of Ig class switch recombination (CSR) (29, 30). This 30 kb region has four functional domains designated HS1,2, HS3a, HS3b and HS4. HS3a and 3b are 97% identical in sequence but appear in opposite orientations (28, 36). HS1,2, HS3a and HS3b function primarily in activated B cells while HS4 appears to be active throughout B cell differentiation, although recent in vivo studies suggest a redundant role amongst different 3′-enhancers (28).
Relationship between Eμ and 3′-RR
The 3′-RR, as a whole, possesses locus control activity and may function cooperatively with Eμ (27). In an elegant series of experiments it was shown that in unstimulated mature B cells, the distantly located Eμ and 3′-RR associate in vivo to form a loop that encompasses the Igh constant region (CH) genes (37). It was further proposed that in unstimulated B cells, looping in the Igh locus leads to closer spatial proximity of the germline transcription promoters with the Eμ:3′-RR complex resulting in a topological structure termed synaptosome[ET1]. Synaptosome, in turn, helps form a “poised” core chromatin conformation that later facilitates germline transcription in response to isotype-specific B cell activation (37).
Information about the evolutionary hierarchy of two Ig enhancers comes from studies in bony (also known as teleost) fish, members of the earliest vertebrate lineage to have a functional IgH locus (38). Although the best-characterized teleost family, that of channel catfish, contains a functional Eμ enhancer, this species lack 3′-RR (38). Teleosts also show restricted diversity in their antibody response and lack somatic hypermutation. Therefore, the mammalian Igh locus (with both enhancers) likely arose from the primordial gene (containing only the Eμ enhancer) via multiple steps of duplication and transposition (38).
VH Promoter Elements and Transcription Factor Interactions
Each VH gene segment contains an independent promoter that is required to express the mature recombined Ig transcript (8). In isolation, VH region promoter activity appears to be selective to B lymphoid lineage cells, although the molecular mechanisms responsible for such specificity remain largely unknown (4). It should be noted that B cell-selectivity of VH (and Vκ promoters has been only tested in transient transfection experiments in transformed cell lines and IgH and Igκ transgenes show broad expression patterns without any consistent B cell-specific expression (4). However, the transgenic experiments are potentially complicated by the fact that expression is copy number- and integration site-dependent, and generalizations should be made with caution (4).
Co-activators
In addition to the core promoter elements that include the consensus TATA box and the Inr element, the hallmark of all VH and Vκ promoters is the presence of an octamer element that is usually located within 100 bp of the transcriptional initiation site (4, 39, Fig 2). The remarkable conservation of the octamer element in both Ig light and heavy chain gene promoters[ET2], together with its appearance in both Eμ and 3′-RR, led to the expectation that transcription factor interactions with this element would be critical for B-cell specific regulation of Ig genes (4). Surprisingly, although octamer elements might contribute to B-cell specificity, the precise in vivo functions of the cognate OCT factors that bind to these sites remain unclear (40).
The B cell-specific co-activator OCA-B (also known as OBF-1 or Bob-1) was discovered in biochemical complementation assays (20). OCA-B can interact directly with either OCT-1 or OCT-2 and can be co-recruited with either transcription factor to the octamer element (20, 21). In addition, OCA-B, in conjunction with OCT-1, also appears to interact with negative co-regulator SMRT, thereby indicating its potential for the negative regulation of non-Ig genes (41). Gene targeting experiments in mice revealed that OCA-B is not essential for Igh transcription or early B cell differentiation, although it is essential both for antigen-dependent B cell differentiation (including germinal center formation) and for normal expression of secondary Ig isotypes (42, 43). These results further underscore the complications surrounding the octamer element and suggest that there is perhaps a high degree of redundancy in octamer-mediated interactions involving OCT-1, OCT-2 and OCA-B and, further, that the B cell specificity likely involves other regulatory elements and corresponding factors (40).
DICE, Inr and PAIR elements
Recently, the Igh promoter element Downstream Immunoglobulin Control Element (DICE) that forms B cell-specific DNA-protein complexes was identified. DICE is located immediately downstream of the transcription initiation site of a majority of the murine heavy chain variable promoters (44). DICE appears to be required both for optimal activity of a VH promoter and for stimulation of VH promoter activity by the intronic enhancer in transfection experiments (44). Unlike the ubiquitous presence of the Octamer motif in all VH promoters, a complete annotated map of the murine IgH V region sequence reveals that the best DICE matches are found in J558-class promoters, the most distal VH family with the most intrinsically active promoters (9, 44).
Interestingly, the J558 VH family members appear to have two Inr elements flanking a DICE around the transcription start site (Fig 2B, and ref. 9). Even though it remains unknown how specific differences in promoter architecture contribute to the transcriptional regulation of rearranged IgH genes, a J558 promoter has been shown to be stronger than a S107 promoter in in vitro transcriptional assays (45). Although the mechanism responsible for this difference is unknown at present, in vitro experiments have indicated that the multifunctional transcription factor TFII-I can bind to both DICE and Inr elements (44, 46). Thus, TFII-I may potentially enhance the strength of J558 VH promoters, compared to other VH promoters, by virtue of binding to both Inr and DICE and its associated communication with the basal transcriptional machinery.
More recently, a novel promoter element was identified within the distal VH cluster (47). This element, termed PAIR (Pax-5 activated intergenic repeat), is interspersed in the distal VH gene region and binds several factors that include Pax-5, E2A, CTCF and Rad21 (a component of the cohesin complex). Interestingly, whereas Pax-5 binding to these elements is restricted to pro-B cells, E2A and CTCF binding persists even at later stages of B cell development (47). Given that both CTCF and cohesins are required for cell type-specific chromatin organization (48, 49), and that cohesin has been implicated in enhancer-promoter interactions (50) these observations suggest that PAIR elements are likely to be important for Igh transcriptional regulation and may play a role in enhancer-promoter communication.
Igh locus enhancer-promoter communication
A number of studies of enhancer function, including ones involving B and T cell antigen receptor loci, have indicated a looping model in which enhancer-bound factors “contact” promoter-bound factors such as Pol II and components of the basal machinery (34, 49–54). However, it is becoming clear that enhancers activate transcription through additional mechanisms, such as formation of so-called “transcription factories” (54, 55). According to this model, active gene loci can co-localize with transcription factors and Pol II within the nucleus in discrete “spots” termed as transcription factories and enhancer-promoter communication occurs within these factories (54, 55). It has also come to light that the number of enhancers in the mammalian genome appears to be rather large and exhibit characteristic chromatin features (54). These features consist of not only histone 3 lysine 4 mono-methylation (H3K4me1) mark but also histone 3 lysine 27 acetylation (H3K27ac) and histone 3 lysine 27 tri-methylation (H3K27me3) marks (56, 57). Moreover, enhancer effects on histone modifications can be gene-specific. For instance, while deletion of Eμ results in a significant reduction in histone acetylation over a several hundred kilobase region, deletion of the β-globin locus control region (LCR) does not affect histone acetylation (reviewed in 34). In addition, it was shown that whereas deletion of the T cell specific E4p enhancer does not affect CD4 expression in resting T cells, this E4p-independent CD4 expression is lost upon T-cell proliferation (58). Thus, the E4p enhancer is not required for the initiation of expression but is necessary for the propagation of an epigenetic state during activation (34, 58).
In light of these observations, it is pertinent to ask how downstream enhancers communicate with the VH promoters to regulate Igh transcription. In transfection assays, a distant Igh 3′κenhancer cannot function properly in the absence of at least one proximal promoter element positioned between the core promoter and the enhancer, which might explain the existence of the positionally conserved octamer element (59). A mechanistic explanation is provided by the fact that while Mediator (and TFIIB a GTF) interactions at the core promoter were dependent upon enhancers, the binding of an activator to a proximal element(s) (such as octamer) is necessary for efficient nucleation of the transcription pre-initiation complex by enhancing the recruitment of TATA binding protein (59).
Other studies have addressed physical interactions between the Igh 3′-RR and promoters (60, 61). The t(14:18) lymphomas are characterized by translocation of the anti-apoptotic gene Bcl2 from chromosome 18q21 to the Igh locus on chromosome 14q32, which results in aberrant activation of the translocated Bcl2 allele (60). Using chromosome conformation capture (3C) assays, a functional and long-range (over a 350 kb genomic region) interaction between the Igh 3′-RR and Bcl2 promoter was shown (60). Interestingly, both OCT-2 and OCA-B were implicated in mediating such interactions (60). Likewise, the 3′-RR physically interacts with the VH promoter (61). In addition, in plasmacytomas harboring a reciprocal translocation between the Myc and the Igh locus, a direct interaction of the 3′-RR with the Myc promoter has also been observed (61). Together with the finding that looping in the Igh locus leads to closer spatial proximity of the germline transcription promoters with the Eμ:3′-RR complex (37), these observations strongly point to a looping mechanism for promoting promoter-enhancer communication in regulating Ig gene transcription.
Recent work has provided important biochemical insights that might explain 3′-RR (HS4) and VH promoter interactions (62). The Inr- and DICE-interacting transcription factor TFII-I was found to physically and functionally interact with OCA-B, to regulate Igh expression in two ways (62). First, OCA-B and HDAC3 bound directly and competitively to TFII-I, such that OCA-B relieves HDAC3-mediated Igh promoter repression by competing with HDAC3 for binding to promoter-bound TFII-I. Second, using 3C assays, it was shown that Igh 3′-RR-bound OCT1/2-OCA-B and core promoter (DICE)-bound TFII-I mediate promoter-enhancer interactions in both cis and trans and that these interactions are important for Igh transcription in plasmacytoma cells (62). Importantly, deficiencies in either factor led to significant reductions not only in endogenous Igh enhancer-promoter interactions but also in transcription both in plasmacytoma cells and in primary murine spleen cells (62). Consistent with the studies of Igh 3′-RR and Bcl2 promoter interactions (60), these observations clearly indicate an enhancer-promoter communication mechanism involving looping facilitated by OCT- OCA-B and TFII-I (Fig 3). It will be interesting to determine whether 3′-RR communication with the Bcl2 (60) promoter and/or Myc (61) promoter involves TFII-I, which might indicate a general role for TFII-I in mediating such long-range interactions.
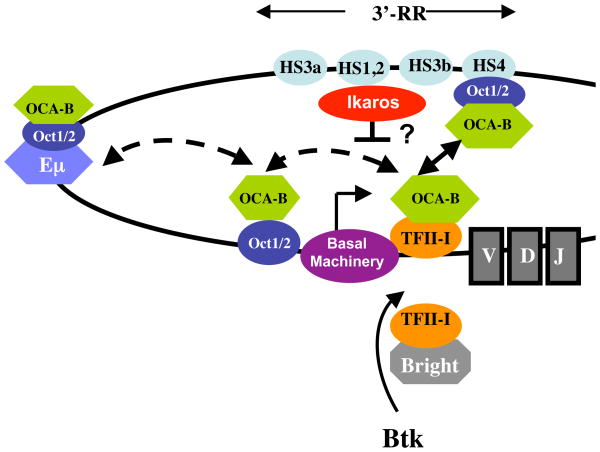
Figure 3. Model for Igh promoter-enhancer communication.
Due to potential interactions of TFII-I with both Inr and DICE (particularly in J558 VH promoters), TFII-I is likely to have stable interactions with the core promoter. Because TFII-I also interacts with OCA-B, the latter is recruited to the VH promoter as well. In this model, TFII-I also interacts with MAR-bound Bright transcription factor. By virtue of its interaction with both TFII-I and OCT1, OCA-B can facilitate communication between the core promoter, upstream octamer and the downstream 3′-RR, resulting in looping and juxtaposition of upstream and downstream elements. Although an interaction between the VH promoter and the intronic/Eμ enhancer has not yet been shown, the fact that that Eμ harbors octamer sites that can bind OCT1/2-OCA-B raises the possibility that core promoter-bound TFII-I–OCA-B can also interact with Eμ-bound OCT1/2-OCA-B during early stages of B cell development. Likewise, proximal promoter-bound OCT1/2-OCA-B can interact with core promoter-bound TFII-I–OCA-B. Dotted arrows depict the presumptive interactions, while the solid arrow indicates the demonstrated interaction. Ikaros interacts with the 3′-RR and inhibits transcription, although its role in promoter-enhancer communication is untested. In addition, both TFII-I and OCA-B appear to be downstream of the BCR/Btk signaling cascade, thereby potentially connecting BCR signaling to IgH transcription.
The presence of octamer elements in the Eμ enhancer (4) raises the possibility that similar enhancer-promoter interactions between Eμ and VH could be mediated by TFII-I and OCA-B. Because the Eμ and 3′-RR function in distinct stages of B cell development, such interactions would likely be regulated in a stage-specific fashion as well. Moreover, since OCT1/2-OCA-B also binds to the proximal promoter octamer site, we conjecture that multiple OCT1/2-OCA-B-TFII-I interactions govern Igh transcription at distinct stages of B cell development (Fig 3).
An evolutionary connection between TFII-I and OCA-B can also be envisioned. Thus, whereas catfish OCA-B has a very well conserved POU domain, it appears to lack a conserved C-terminal trans-activation domain akin to that in mammalian OCA-B (63). TFII-I interacts with OCA-B through a central region that seems to be relatively well conserved between mammals and catfish (62). Thus, it is likely that TFII-I interacts with OCA-B in catfish and provides the activation potential that is missing from the teleost OCA-B. Given that the catfish Igh contains only an Eμ-related enhancer that harbors OCT1 and OCT2, it is possible that OCT1/2-OCA-B-TFII-I interactions arose first in conjunction with Eμrelated functions and, during evolution, eventually became relevant for the 3′-RR. Conversely, it is also possible that these interactions were not relevant for Eμ and later co-evolved only with the 3′-RR.
In addition to the factors that positively influence 3′-RR interactions, an interesting recent observation is that the transcription factor Ikaros negatively controls isotype gene transcription during Ig CSR by binding to the 3′-RR (64). Ikaros binding controls transcription by suppressing active chromatin marks and accessibility of activation-induced cytidine deaminase (AID) to the γ2b and γ2a genes. Importantly, such repression promotes switching to other isotype genes by allowing them to compete for AID-mediated recombination (64). However, it is not yet known whether binding of Ikaros to the 3′-RR might regulate looping and alter communication with the VH promoter.
Igh transcription and connection to B cell signaling
Genetic ablation of TFII-I in mice results in early embryonic lethality, thus precluding the assessment of its precise role in B cell function (65). However, TFII-I is a target of Bruton’s tyrosine kinase (Btk), and genetic ablation of Btk or natural mutations in Btk lead to a B cell phenotype known as x-linked immune-deficiency (xid) (46, 66). TFII-I is downstream of the BCR-Btk signaling pathway in B cells (46), and the finding that TFII-I together with OCA-B control Igh transcription via looping (62) also raise the exciting possibility that BCR-signaling is connected, through TFII-I, to OCT1/2-OCA-B functions and regulates enhancer-promoter communication. This notion has been further substantiated by recent observations connecting OCA-B to BCR signaling via Syk (67).
TFII-I is also linked to Igh transcriptional regulation because it physically and functionally interacts with Bright, a MAR-interacting factor implicated in Ig gene expression (68). Moreover, TFII-I connects Bright to the BCR-Btk pathway as well (69). Hence, TFII-I may prove to be a missing piece of the puzzle that, along with Oct-1/2-OCA-B, is important for establishing proper communication between VH promoters and Igh enhancers in a BCR-Btk signal-dependent fashion (Fig 3).
Concluding remarks
Given the diversity in VH promoter architecture with newly discovered DNA sequence elements, as well as multiple enhancers that act during distinct stages of B cell development, it is a significant challenge to understand how these enhancers communicate with diverse VH promoters in a stage-specific fashion. The notion that enhancer effects on histone modifications and chromatin structure can be gene-specific and/or stage-specific also adds to the complexity. Recent studies have begun to address these issues and identify transcription factors that are likely to be involved in complex regulation of Ig genes during development. Although distinct mechanisms of enhancer-mediated transcriptional regulation of eukaryotic genes are being discovered, recombined Igh transcriptional regulation primarily involves a looping mechanism for enhancer-promoter communication. The technical advances in studying promoter-enhancer communication, including Hi-C (a genome-wide high resolution variation of 3C) and 4C (circular chromosome conformation capture) assays (70, 71) in conjunction with new imaging techniques (reviewed in 26), will certainly aid in our understanding of this process in far greater detail in the near future.
Acknowledgments
R.S. is supported by the Intramural Research Program of the National Institute on Aging (Baltimore), R.G.R is supported by National Institutes of Health Grant CA113872, and A.L.R is supported by National Institutes of Health grant 1R56AI079206.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Ptashne M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem Sci. 2005;30:275–279. doi: 10.1016/j.tibs.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Roeder RG. Lasker Basic Medical Research Award. The eukaryotic transcriptional machinery: complexities and mechanisms unforeseen. Nat Med. 2003;9:1239–1244. doi: 10.1038/nm938. [DOI] [PubMed] [Google Scholar]
- 4.Clame K, Sen R. Transcription of Immunoglobulin Genes. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular biology of B cells. Elsevier; Amsterdam; Boston: 2004. pp. 83–99. [Google Scholar]
- 5.de Pooter RF, Kee BL. E proteins and the regulation of early lymphocyte development. Immunol Rev. 2010;238:93–109. doi: 10.1111/j.1600-065X.2010.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruault M, et al. Re-positioning genes to the nuclear envelope in mammalian cells: impact on transcription. Trends Genet. 2008;24:574–581. doi: 10.1016/j.tig.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Takizawa T, et al. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yancopoulos GD, Alt FW. Regulation of the assembly and expression of variable-region genes. Ann Rev Immunol. 1986;4:339–368. doi: 10.1146/annurev.iy.04.040186.002011. [DOI] [PubMed] [Google Scholar]
- 9.Johnston CM, et al. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- 10.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2011;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, et al. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 15.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 16.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan F, et al. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 20.Luo Y, Roeder RG. B-cell-specific coactivator OCA-B: biochemical aspects, role in B-cell development and beyond. Cold Spring Harb Symp Quant Biol. 1999;64:119–131. doi: 10.1101/sqb.1999.64.119. [DOI] [PubMed] [Google Scholar]
- 21.Blazek E, et al. The mediator of RNA polymerase II. Chromosoma. 2005;113:399–408. doi: 10.1007/s00412-005-0329-5. [DOI] [PubMed] [Google Scholar]
- 22.Chakraborty T, et al. A 220-nucleotide deletion of the intronic enhancer reveals an epigenetic hierarchy in immunoglobulin heavy chain locus activation. Exp Med. 2009;206:1019–1027. doi: 10.1084/jem.20081621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty T, et al. Repeat organization and epigenetic regulation of the DH-Cmu domain of the immunoglobulin heavy-chain gene locus. Mol Cell. 2007;27:842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Sen R, Oltz E. Genetic and epigenetic regulation of IgH gene assembly. Curr Opin Immunol. 2006;18:237–242. doi: 10.1016/j.coi.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Bergman Y, et al. Epigenetic mechanisms that regulate antigen receptor gene expression. Curr Opin Immunol. 2003;15:176–181. doi: 10.1016/s0952-7915(03)00016-5. [DOI] [PubMed] [Google Scholar]
- 26.Jhunjhunwala S, et al. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138:435–448. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khamlichi AA, et al. The 3′ IgH regulatory region: a complex structure in a search for a function. Adv Immunol. 2000;75:317–345. doi: 10.1016/s0065-2776(00)75008-5. [DOI] [PubMed] [Google Scholar]
- 28.Bébin AG, et al. In vivo redundant function of the 3′ IgH regulatory element HS3b in the mouse. J Immunol. 2010;184:3710–3717. doi: 10.4049/jimmunol.0901978. [DOI] [PubMed] [Google Scholar]
- 29.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 30.Cogne M, Birshtein BK. Regulation of class Switch Recombination. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular Biology of B Cells. Elsevier; Amsterdam; Boston: 2004. pp. 289–305. [Google Scholar]
- 31.Serwe M, Sablitzky F. V(D)J recombination in B cells is impaired but not blocked by targeted deletion of the immunoglobulin heavy chain intron enhancer. EMBO J. 1993;12:2321–2327. doi: 10.1002/j.1460-2075.1993.tb05886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Eckhardt LA. A role for the IgH intronic enhancer E mu in enforcing allelic exclusion. J Exp Med. 2009;206:153–167. doi: 10.1084/jem.20081202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronai D, et al. The epigenetic stability of the locus control region-deficient IgH locus in mouse hybridoma cells is a clonally varying, heritable feature. Genetics. 2004;167:411–421. doi: 10.1534/genetics.167.1.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen R, Grosschedl R. Memories of lost enhancers. Genes Dev. 2010;24:973–979. doi: 10.1101/gad.1930610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolland DJ, et al. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Mol Cell Biol. 2007;27:5523–5533. doi: 10.1128/MCB.02407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madisen L, Groudine M. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt’s lymphoma cells. Genes Dev. 1994;8:2212–2226. doi: 10.1101/gad.8.18.2212. [DOI] [PubMed] [Google Scholar]
- 37.Wuerffel R, et al. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magor BG, et al. Transcriptional enhancers and the evolution of the IgH locus. Immunol Today. 1999;20:13–17. doi: 10.1016/s0167-5699(98)01380-2. [DOI] [PubMed] [Google Scholar]
- 39.Brekke KM, Garrard WT. Assembly and analysis of the mouse immunoglobulin kappa gene sequence. Immunogenetics. 2004;56:490–505. doi: 10.1007/s00251-004-0659-0. [DOI] [PubMed] [Google Scholar]
- 40.Matthias P. Lymphoid specific transcription mediated by the conserved octamer site: Who is doing what? Sem. Immunol. 1998;10:111–163. doi: 10.1006/smim.1998.0117. [DOI] [PubMed] [Google Scholar]
- 41.Kakizawa T, et al. Silencing mediator for retinoid and thyroid hormone receptors interacts with octamer transcription factor-1 and acts as a transcriptional repressor. J Biol Chem. 2001;276:9720–9725. doi: 10.1074/jbc.M008531200. [DOI] [PubMed] [Google Scholar]
- 42.Kim U, et al. The B cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 1996;383:542–547. doi: 10.1038/383542a0. [DOI] [PubMed] [Google Scholar]
- 43.Schubart DB, et al. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 44.Tantin D, et al. Regulation of immunoglobulin promoter activity by TFII-I-Class transcription factors. J Biol Chem. 2004;279:5460–5469. doi: 10.1074/jbc.M311177200. [DOI] [PubMed] [Google Scholar]
- 45.Buchanan KL, et al. Differential transcription efficiency of two Ig VH promoters in vitro. J Immunol. 1995;155:4270–4277. [PubMed] [Google Scholar]
- 46.Roy AL. Signal-induced functions of the transcription factor TFII-I. Biochim Biophys Acta. 2007;1769:613–621. doi: 10.1016/j.bbaexp.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ebert A, et al. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Hou C, et al. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci, U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Degner-Leisso SC, Feeney AJ. Epigenetic and 3-dimensional regulation of V(D)J rearrangement of immunoglobulin genes. Semin Immunol. 2010;22:346–352. doi: 10.1016/j.smim.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oestreich KJ, et al. Regulation of TCRbeta gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24:381–391. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 52.Sekimata M, et al. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity. 2009;31:551–564. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee GR, et al. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cook PR. A model for all genomes: the role of transcription factories. J Mol Biol. 2010;395:1–10. doi: 10.1016/j.jmb.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 56.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chong MM, et al. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes Dev. 2010;24:659–669. doi: 10.1101/gad.1901610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bertolino E, Singh H. POU/TBP cooperativity: A mechanism for enhancer action from a distance. Mol Cell. 2002;10:397–407. doi: 10.1016/s1097-2765(02)00597-x. [DOI] [PubMed] [Google Scholar]
- 60.Duan H, et al. Functional long-range interactions of the IgH 3′ enhancers with the bcl-2 promoter region in t(14;18) lymphoma cells. ncogene. 2008;27:6720–6728. doi: 10.1038/onc.2008.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ju Z, et al. Evidence for physical interaction between the immunoglobulin heavy chain variable region and the 3′ regulatory region. Biol Chem. 2007;282:35169–25178. doi: 10.1074/jbc.M705719200. [DOI] [PubMed] [Google Scholar]
- 62.Ren X, et al. Direct interactions of OCA-B and TFII-I regulate immunoglobulin heavy-chain gene transcription by facilitating enhancer-promoter communication. Mol Cell. 2011;42:342–355. doi: 10.1016/j.molcel.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richard ML, et al. BOB.1 of the channel catfish, Ictalurus punctatus: not a transcriptional coactivator? Mol Immunol. 2009;46:481–491. doi: 10.1016/j.molimm.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sellars M, et al. Ikaros controls isotype selection during immunoglobulin class switch recombination. J Exp Med. 2009;206:1073–1087. doi: 10.1084/jem.20082311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Enkhmandakh B, et al. Essential functions of the Williams-Beuren syndrome-associated TFII-I genes in embryonic development. Proc Natl Acad Sci U S A. 2009;106:181–186. doi: 10.1073/pnas.0811531106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conley ME, et al. Primary B cell immunodeficiencies: comparisons and contrasts. Annu Rev Immunol. 2009;27:199–227. doi: 10.1146/annurev.immunol.021908.132649. [DOI] [PubMed] [Google Scholar]
- 67.Siegel R, et al. Nontranscriptional regulation of SYK by the coactivator OCA-B is required at multiple stages of B cell development. Cell. 2006;125:761–774. doi: 10.1016/j.cell.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 68.Rajaiya J, et al. Induction of immunoglobulin heavy-chain transcription through the transcription factor Bright requires TFII-I. Mol Cell Biol. 2006;26:4758–4768. doi: 10.1128/MCB.02009-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt C, et al. Signalling of the BCR is regulated by a lipid rafts-localised transcription factor, Bright. EMBO J. 2009;28:711–724. doi: 10.1038/emboj.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]