Abstract
Various isoforms of myocyte enhancer factor-2 (MEF2) constitute a group of nuclear proteins found to play important roles in increasing types of cells. In neurons, MEF2s are required to regulate neuronal development, synaptic plasticity, as well as survival. MEF2s promote the survival of several types of neurons under different conditions. In cellular models, negative regulation of MEF2s by stress and toxic signals contributes to neuronal death. In contrast, enhancing MEF2 activity not only protects cultured primary neurons from death in vitro but also attenuates the loss of dopaminergic neurons in substantia nigra pars compacta in a 1-methyl 4-phenyl 1,2,3,6-tetrahydropyridine mouse model of Parkinson’s disease. In this work, the mechanisms of regulation of MEF2 function by several well-known neurotoxins and their implications in various neurodegenerative diseases are reviewed.
Keywords: myocyte enhancer factor-2, neurotoxins, neurodegenerative diseases
1. MEF2 Family of Transcription Factors
The myocyte enhancer factor-2 (MEF2) proteins are members of the MADS (MCM1-agamous-deficiens-serum response factor) family of transcription factors (Naya and Olson, 1999). A hallmark of MADS-box proteins is their combinational association with other MADS domain factors and additional heterologous classes of transcriptional regulators (Shore and Sharrocks, 1995). Vertebrate MEF2 proteins are encoded by four genes (MEF2A, B, C, and D), each of which gives rise to alternatively spliced transcripts. The MEF2 isoforms are expressed in distinct, but overlapping patterns during embryogenesis and in adult tissues. Posttranslational modification is largely responsible for the function of MEF2 proteins (McKinsey et al., 2002).
The first 56 amino acids at the N terminus, termed MADS-box, are the minimal region needed for DNA binding. The MADS box is a highly conserved structural motif involved in the regulation of homeotic fate, growth, and differentiation in many organisms. Adjacent to the MADS box is a 29 amino acid MEF2 domain that mediates high-affinity DNA binding and homo- and hetero-dimerization with other MEF2 proteins. The vertebrate MEF2 proteins share about 50% amino acid identity overall and about 95% similarity in the highly conserved MADS box and MEF2 domain (Heidenreich and Linseman, 2004). MADS box proteins generally bind A/T rich DNA sequences, whereas MEF2 binds preferentially to the consensus sequence 5′-CC(A/T)(T/A)AAATAG-3′ (Andres et al., 1995). The MADS box and MEF2 domain are necessary and sufficient for DNA binding but lack transcriptional activity on their own. The C-terminal portion of MEF2 proteins contains the transcriptional activation domain, as well as, a number of regulatory minidomains including a nuclear localization sequence and multiple phosphorylation motifs (Potthoff and Olson, 2007). The C-terminus is subject to complex patterns of alternative splicing and has relatively low amino acid homology between the various MEF2 isoforms. The structural organization of MEF2 proteins allows them to receive and respond to multiple inputs from various intracellular signaling pathways. Thus, MEF2 activity is profoundly influenced by signals from the extracellular environment.
2. Functions of MEF2 Family of Transcription Factors
The MEF2 family of transcription factors is highly expressed in cells of muscle lineage, where they have been shown to be important regulators of gene expression during development of skeletal, cardiac, and smooth muscle (Naya and Olson, 1999). In these tissues, MEF2 proteins interact with myogenic basic helix-loop-helix (bHLH) transcription factors such as Myo D to activate myogenesis (Molkentin and Olson, 1996). In addition to their critical role in muscle development, MEF2 proteins are involved in adult cardiac hypertrophy (Kolodziejczyk et al., 1999).
MEF2 proteins are also present in cells of the immune system where they mediate cell fate decisions. T-cell activation leads to MEF2-mediated transcriptional activation of Nur77, an orphan nuclear steroid receptor that activates apoptosis in these cells (Youn et al., 1999). MEF2 binds directly to regulatory elements in the Nur77 gene and cooperates with NFAT to drive Nur77 expression (Youn et al., 2000).
MEF2 proteins are highly enriched in neurons and exhibit distinct patterns of expression in different regions of the brain with highest levels in the cerebellum, cerebral cortex and hippocampus (Heidenreich and Linseman, 2004). In neurons, MEF2 transcriptional activity is tightly regulated by extracellular stimuli. MEF2 can be activated by neurotrophin stimulation as well as calcium influx. MEF2 functions as a converging factor to regulate neuronal proliferation, differentiation, survival, as well as synapse development. Deregulation MEF2 activity had been associated with many neurodegenerative diseases, including Parkinson’s disease (PD) and Alzheimer’s disease (AD) (Burton et al., 2002; Smith et al., 2006).
3. Regulation of MEF2 Transcription Factors by Neurotoxins
3.1. Regulation of MEF2 by excitotoxic insults
Following membrane depolarization of cultured cerebellar granule neurons (CGNs), an established in vitro model for studying activity-dependent neuronal survival, MEF2C was shown to be phosphorylated by p38 MPAK and promote neuronal survival (Mao et al., 1999). Neuronal activity was shown to activate Ca2+-dependent protein phosphatase calcineurin, which up-regulate MEF2 transcriptional activity (Mao and Wiedmann, 1999). In contrast, lack of neuronal activity leads to hyperphosphorylation of MEF2 at unknown site(s), resulting in destabilization of MEF2 proteins. It was shown later that neurotoxicity causes cleavage of MEF2s by a caspase-dependent mechanism (Li et al., 2001). Transfection of constitutively active form of MEF2 rescued MEF2 transcriptional activity after NMDA insult and prevents neuronal apoptosis. Additionally, similar MEF2 cleavage fragments were generated in vivo during focal stroke damage. Hence, this pathway appears to have pathophysiological relevance in vivo (Okamoto et al., 2002). Using primary CGNs as a model, recent work by Gong et al revealed cyclin-dependent kinase 5 (Cdk5) as the first negative regulator of MEF2 (Gong et al., 2003). It was demonstrated that neurotoxic signals activate Cdk5, which directly phosphorylates MEF2s at their transactivation domain, leading to their destabilization and inhibition of their activity. Subsequent studies by Tang et al showed that Cdk5 phosphorylation induces MEF2 destabilization by greatly enhancing caspase-dependent cleavage of MEF2A and D (Tang et al., 2005). In contrast to MEF2A and D, MEF2C is not phosphorylated by Cdk5 after glutamate exposure and, therefore, resistant to glutamate-induced caspase-dependent degradation. Blocking Cdk5 or enhancing MEF2 activity reduced glutamate-induced apoptosis. These findings defined an important regulatory mechanism that for the first time linked pro-death activities of Cdk5 with caspase-dependent cleavage of MEF2 (Fig. 1). The convergence of Cdk5 phosphorylation-dependent caspase-mediated degradation of nuclear survival factors exemplified by MEF2 may represent a general process applicable to the regulation of other survival factors under diverse excitotoxic conditions.
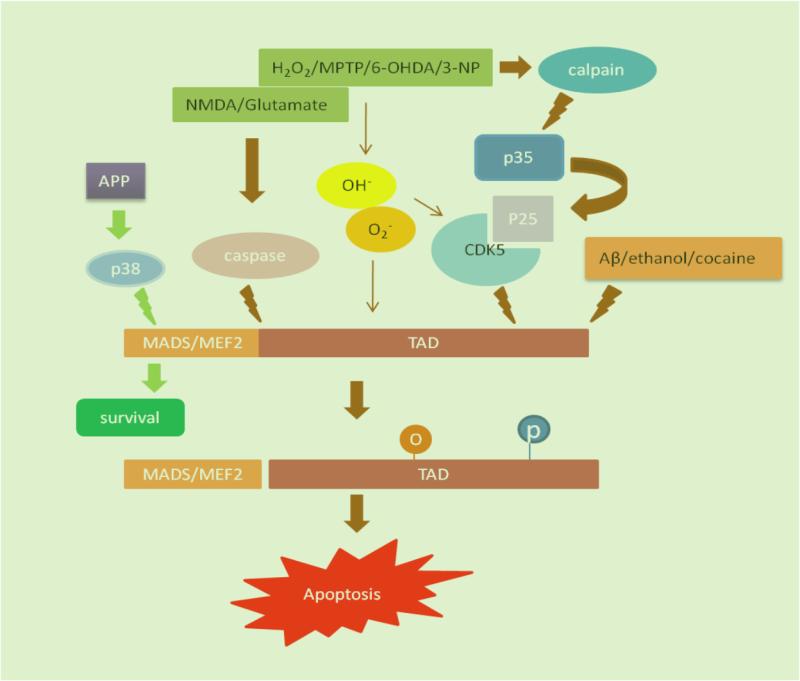
Fig. 1.
Pathways of regulation of MEF2 function by neurotoxins. Various neurotoxins modulate MEF2 activity through activating caspase and calpain/CDK5 pathways. H2O2, MPTP, 6-OHDA and 3-NP induce MEF2 oxidization through generating free radicals. Aβ, ethanol and cocaine modulate MEF2 activity through unidentified mechanism. APP enhances MEF2 function through p38MAPK pathway. O indicates oxidization; P indicates phosphorylation.
3.2. Regulation of MEF2 by MPTP and 6-OHDA
The mechanisms underlying dopamine neuron loss in Parkinson’s disease (PD) are not clearly defined. Striatal dopamine depletion profoundly reduces the density of spines and corticostriatal glutamatergic synapses formed on D(2) dopamine receptor expressing striatopallidal medium spiny neurons. This spine pruning was dependent upon Ca2+ entry through Cav1.2 L-type Ca2+ channels. Depolarization and MEF2 up-regulate the expression of two genes linked to synaptic remodeling, Nur77 and Arc. Taken together, these studies establish a framework within which striatal adaptations regulated by MEF2 activity is linked to the symptoms of PD (Tian et al., 2010).
Several neurotoxins cause PD in animals. Crocker et al reported previously that calpains play a central role in dopamine loss after 1-methyl 4-phenyl 1,2,3,6-tetrahydropyridine (MPTP) treatment (Crocker et al., 2003). Recent work by Smith et al provided further evidence that Cdk5-mediated modulation of the transcription factor MEF2 is downstream of calpain. They found that MPTP activates Cdk5 by increasing calpain-dependnet conversion of the Cdk5 activator p35 to a pathogenic p25. Hyperactivation of Cdk5 was shown to phosphorylate MEF2D on Ser444 and inactivate it. This played a key role in the loss of dopaminergic neurons since MEF2D mutant resistant to Cdk5 phosphorylation protected these neurons from MPTP toxicity in mouse model of PD (Smith et al., 2006).
Autophagy is an important process for maintaining cellular homeostasis. Deregulation of autophagy is associated with PD and other neurodegenerative diseases. Yang et al reported recently that chaperone-mediated autophagy (CMA) degrades MEF2s directly in neurons (Yang et al., 2009). This process is deregulated by 6-hydroxydopamine (6-OHDA) and MPTP, and in Parkinson’s disease (Yang et al. unpublished data), consistent with a recent report that MPTP deregulates autophagy (Dehay et al., 2010) (Fig. 1).
3.3. Regulation of MEF2 by amyloid-beta peptide
Alzheimer’s disease (AD) is characterized by the deposition of the amyloid-beta peptide (Aβ) in the brain, which is proteolytically cleaved from a large amyloid-beta precursor protein (APP) by beta- and gamma-secretases. Beta-APP cleaving enzyme (BACE1), a transmembrane aspartyl protease, has been recognized as the beta-secretase. The promoter region and 5′-untranslated region (UTR) of BACE1 contain multiple transcription factor binding sites, such as MEF2 binding site, suggesting that MEF2 activity may be associated with Aβ toxicity (Lahiri et al., 2006). In addition, Dong et al showed that Aβ inhibits MEF2 activity in SN4741 cells. Inhibition of MEF2 activity by mature Aβ peptide is dose-dependent and peptide length specific, and regulated by metal irons such as Cu2+ and Zn2+ (Dong et al., 2007) (Fig. 1). Another recent study demonstrated that a polymorphism of the MEF2A gene is associated with increased risk of developing late-onset Alzheimer’s disease (LOAD). Because MEF2 promotes neuronal survival while the P279L allele has been associated with a reduction in the transcriptional activation activity of MEF2A (Gonzalez et al., 2007), the effect of this allele could be mediated through down-regulation of anti-apoptotic activity of MEF2A.
In contrast to Aβ, Aβ precursor protein APP effectively protects against apoptosis of neuronal cells under stress. Burton et al reported that expression of wild-type human APP (hAPPwt) but not familial Alzheimer’s disease mutant APP (FAD-hAPPmut) in APP-deficient rat B103 cells increases p38 MAPK-dependent phosphorylation and activation of MEF2. Furthermore, over-expression of dominant negative MEF2 in hAPPwt-expressing cells enhanced staurosporine-induced apoptosis while MEF2wt enhanced the capacity of hAPPwt to confer resistance to apoptosis (Fig. 1). Thus, MEF2 plays a critical role in APP-mediated neuroprotection (Burton et al., 2002).
3.4. Regulation of MEF2 by 3-nitropropionic acid
The toxin 3-nitropropionic acid (3-NP) produced by certain plants and fungi is a specific inhibitor of succinate dehydrogenase (SDH), a mitochondrial complex II respiratory enzyme required for energy production. Its administration in vivo induces selective striatal pathology similar to that observed in Huntington’s disease (HD), thus providing a useful experimental model for this disease (Brouillet et al., 1999). In primary hippocampal neurons and rats, 3-NP activates calpain and Cdk5, and induces neuronal death. This correlated with a decrease in MEF2 level and activity (Fig. 1). Thus, inhibition MEF2 function may underlie 3-NP induced neurotoxicity (Crespo-Biel et al., 2009; Crespo-Biel et al., 2007).
3.5. Regulation of MEF2 by oxidative stress
Oxidative stress plays a central role in mediating the toxic effects of many neurotoxins (Roberts et al., 2009). For example, MPTP, 6-OHDA, and dopamine have all been shown to induce cell death through generating excessive oxidative stress in cells (Blum et al., 2001). To investigate whether oxidative stress may induce cell death through inhibition of MEF2, Gong et al treated cortical neurons with hydrogen peroxide and showed that hydrogen peroxide markedly increases nuclear and cytoplasmic Cdk5 kinase activity (Gong et al., 2003). They identified the prosurvival transcription factor MEF2 as a direct nuclear target of Cdk5 and showed that Cdk5 phosphorylated MEF2D at Ser444 in its transactivation domain to inhibit MEF2 activity (Fig. 1). MEF2 mutants resistant to Cdk5 phosphorylation restored MEF2 activity and protected primary neurons from Cdk5 and neurotoxin-induced apoptosis.
3.6. Regulation of MEF2 by addictive drugs
Abuse of addictive drugs has been a big burden for modern society. It is known that repeated exposure to cocaine increases dendritic spines on medium spiny neurons of the nucleus accumbens (NAc) and causes sensitized behavioral responses. Pulipparacharuvil et al found that cocaine regulates MEF2 transcription factors to control these two processes in vivo (Pulipparacharuvil et al., 2008). It was shown that cocaine suppresses striatal MEF2 activity in part through a mechanism involving cAMP, the regulator of calmodulin signaling (RCS), and calcineurin. Reducing MEF2 activity in the NAc in vivo was required for the cocaine-induced increases in dendritic spine density, while surprisingly increasing MEF2 activity in the NAc enhanced sensitized behavioral responses to cocaine. Together, these findings implicate MEF2 as a key regulator of structural synapse plasticity and sensitized responses to cocaine and suggest that reducing MEF2 activity in NAc may be a compensatory mechanism to limit long-lasting maladaptive behavioral responses to cocaine.
A recent work showed that chronic ethanol feeding impairs MEF2 expression and is associated with glucose transporter 4 (GLUT4) decrease in rat myocardium. Ethanol reduced the mRNA and protein levels of MEF2A and 2D, reduced GLUT4 transcription, and elevated serum TNFα level. Taken together, chronic ethanol exposure decreased the expression of AMPKα and MEF2, and was associated with GLUT4 decline in rat myocardium (Chen et al., 2010). It would be of great interest to test whether ethanol regulates MEF2 function in the brain.
One of the major functions of MEF2 in neuron is controlling the development of synapse. Several MEF2 targets are mutated in human neurological disorders resulted from synaptic defect including epilepsy and autism spectrum disorders, suggesting that these disorders may be caused by disruption of an activity-dependent gene program that controls synapse development (Flavell et al., 2008). Fragile X syndrome (FXS), the most common genetic form of mental retardation and autism, is caused by loss-of-function mutations in an RNA-binding protein, fragile X mental retardation protein (FMRP). FMRP is required for synapse elimination by the activity-dependent transcription factor MEF2 (Pfeiffer et al., 2010). In addition, MEF2C deficiency has also been associated with Rett syndrome, an autism-related disorder with severe defect in synapse development (Li et al., 2008). It would important to determine whether neurotoxins affect synapse development through modulating MEF2 functions in the brain.
4. Perspective
The MEF2 family of transcription factors acts as effectors of diverse signaling pathways to regulate fundamental cellular processes in neurons. The function of MEF2 in neurons and its modulation by neurotoxins are just start to be revealed. Many areas remain unexplored. For instance, it is not clear if the regulatory mechanisms identified to control MEF2 in one cell type may also play an important role in another type of cell; if neurotoxins known to modulate MEF2 in neurons may also dysregulate MEF2s in other cellular systems; and if isoform-specific regulation of MEF2 by neurotoxins underlies some aspects of the pathologic processes in diseases. Answers to these questions will surely increase our understanding of MEF2 function in the nervous system.
Acknowledgements
This work was partially supported by NIH grants ES015317, AG023695, NS048254, (Z.M.), ES016731 (G.M., Z.M.) and Michael J. Fox Foundation (Z.M.).
Footnotes
Conflict of interest: The authors declare that no conflict of interest exists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andres V, Cervera M, Mahdavi V. Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissue-specific sequence constraints. J Biol Chem. 1995;270:23246–23249. doi: 10.1074/jbc.270.40.23246. [DOI] [PubMed] [Google Scholar]
- Blum D, Torch S, Lambeng N, Nissou M, Benabid AL, Sadoul R, Verna JM. Molecular pathways involved in the neurotoxicity of 6-OHDA, dopamine and MPTP: contribution to the apoptotic theory in Parkinson’s disease. Prog Neurobiol. 2001;65:135–172. doi: 10.1016/s0301-0082(01)00003-x. [DOI] [PubMed] [Google Scholar]
- Brouillet E, Conde F, Beal MF, Hantraye P. Replicating Huntington’s disease phenotype in experimental animals. Prog Neurobiol. 1999;59:427–468. doi: 10.1016/s0301-0082(99)00005-2. [DOI] [PubMed] [Google Scholar]
- Burton TR, Dibrov A, Kashour T, Amara FM. Anti-apoptotic wild-type Alzheimer amyloid precursor protein signaling involves the p38 mitogen-activated protein kinase/MEF2 pathway. Brain Res Mol Brain Res. 2002;108:102–120. doi: 10.1016/s0169-328x(02)00519-3. [DOI] [PubMed] [Google Scholar]
- Chen L, Wang F, Sun X, Zhou J, Gao L, Jiao Y, Hou X, Qin CY, Zhao J. Chronic ethanol feeding impairs AMPK and MEF2 expression and is associated with GLUT4 decrease in rat myocardium. Exp Mol Med. 2010;42:205–215. doi: 10.3858/emm.2010.42.3.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-Biel N, Camins A, Pallas M, Canudas AM. Evidence of calpain/cdk5 pathway inhibition by lithium in 3-nitropropionic acid toxicity in vivo and in vitro. Neuropharmacology. 2009;56:422–428. doi: 10.1016/j.neuropharm.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Crespo-Biel N, Camins A, Pelegri C, Vilaplana J, Pallas M, Canudas AM. 3-Nitropropionic acid activates calpain/cdk5 pathway in rat striatum. Neurosci Lett. 2007;421:77–81. doi: 10.1016/j.neulet.2007.05.038. [DOI] [PubMed] [Google Scholar]
- Crocker SJ, Smith PD, Jackson-Lewis V, Lamba WR, Hayley SP, Grimm E, Callaghan SM, Slack RS, Melloni E, Przedborski S, Robertson GS, Anisman H, Merali Z, Park DS. Inhibition of calpains prevents neuronal and behavioral deficits in an MPTP mouse model of Parkinson’s disease. J Neurosci. 2003;23:4081–4091. doi: 10.1523/JNEUROSCI.23-10-04081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Bove J, Rodriguez-Muela N, Perier C, Recasens A, Boya P, Vila M. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci. 2010;30:12535–12544. doi: 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Canfield JM, Mehta AK, Shokes JE, Tian B, Childers WS, Simmons JA, Mao Z, Scott RA, Warncke K, Lynn DG. Engineering metal ion coordination to regulate amyloid fibril assembly and toxicity. Proc Natl Acad Sci U S A. 2007;104:13313–13318. doi: 10.1073/pnas.0702669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Tang X, Wiedmann M, Wang X, Peng J, Zheng D, Blair LA, Marshall J, Mao Z. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron. 2003;38:33–46. doi: 10.1016/s0896-6273(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez P, Alvarez V, Menendez M, Lahoz CH, Martinez C, Corao AI, Calatayud MT, Pena J, Garcia-Castro M, Coto E. Myocyte enhancing factor-2A in Alzheimer’s disease: genetic analysis and association with MEF2A-polymorphisms. Neurosci Lett. 2007;411:47–51. doi: 10.1016/j.neulet.2006.09.055. [DOI] [PubMed] [Google Scholar]
- Heidenreich KA, Linseman DA. Myocyte enhancer factor-2 transcription factors in neuronal differentiation and survival. Mol Neurobiol. 2004;29:155–166. doi: 10.1385/MN:29:2:155. [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk SM, Wang L, Balazsi K, DeRepentigny Y, Kothary R, Megeney LA. MEF2 is upregulated during cardiac hypertrophy and is required for normal post-natal growth of the myocardium. Curr Biol. 1999;9:1203–1206. doi: 10.1016/S0960-9822(00)80027-5. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Ge YW, Rogers JT, Sambamurti K, Greig NH, Maloney B. Taking down the unindicted co-conspirators of amyloid beta-peptide-mediated neuronal death: shared gene regulation of BACE1 and APP genes interacting with CREB, Fe65 and YY1 transcription factors. Curr Alzheimer Res. 2006;3:475–483. doi: 10.2174/156720506779025224. [DOI] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, Okamoto S, Roberts AJ, Schwarz JJ, Lipton SA. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Linseman DA, Allen MP, Meintzer MK, Wang X, Laessig T, Wierman ME, Heidenreich KA. Myocyte enhancer factor 2A and 2D undergo phosphorylation and caspase-mediated degradation during apoptosis of rat cerebellar granule neurons. J Neurosci. 2001;21:6544–6552. doi: 10.1523/JNEUROSCI.21-17-06544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- Mao Z, Wiedmann M. Calcineurin enhances MEF2 DNA binding activity in calcium-dependent survival of cerebellar granule neurons. J Biol Chem. 1999;274:31102–31107. doi: 10.1074/jbc.274.43.31102. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci U S A. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Olson E. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr Opin Cell Biol. 1999;11:683–688. doi: 10.1016/s0955-0674(99)00036-8. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Li Z, Ju C, Scholzke MN, Mathews E, Cui J, Salvesen GS, Bossy-Wetzel E, Lipton SA. Dominant-interfering forms of MEF2 generated by caspase cleavage contribute to NMDA-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 2002;99:3974–3979. doi: 10.1073/pnas.022036399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Zang T, Wilkerson JR, Taniguchi M, Maksimova MA, Smith LN, Cowan CW, Huber KM. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron. 2010;66:191–197. doi: 10.1016/j.neuron.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, Greengard P, Nairn AC, Nestler EJ, Cowan CW. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RA, Laskin DL, Smith CV, Robertson FM, Allen EM, Doorn JA, Slikker W. Nitrative and oxidative stress in toxicology and disease. Toxicol Sci. 2009;112:4–16. doi: 10.1093/toxsci/kfp179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- Smith PD, Mount MP, Shree R, Callaghan S, Slack RS, Anisman H, Vincent I, Wang X, Mao Z, Park DS. Calpain-regulated p35/cdk5 plays a central role in dopaminergic neuron death through modulation of the transcription factor myocyte enhancer factor 2. J Neurosci. 2006;26:440–447. doi: 10.1523/JNEUROSCI.2875-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Wang X, Gong X, Tong M, Park D, Xia Z, Mao Z. Cyclin-dependent kinase 5 mediates neurotoxin-induced degradation of the transcription factor myocyte enhancer factor 2. J Neurosci. 2005;25:4823–4834. doi: 10.1523/JNEUROSCI.1331-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Kai L, Hockberger PE, Wokosin DL, Surmeier DJ. MEF-2 regulates activity-dependent spine loss in striatopallidal medium spiny neurons. Mol Cell Neurosci. 2010;44:94–108. doi: 10.1016/j.mcn.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, Mao Z. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn HD, Chatila TA, Liu JO. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J. 2000;19:4323–4331. doi: 10.1093/emboj/19.16.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn HD, Sun L, Prywes R, Liu JO. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science. 1999;286:790–793. doi: 10.1126/science.286.5440.790. [DOI] [PubMed] [Google Scholar]