Abstract
Learning and memory are encoded within the brain as biochemical and physical changes at synapses that alter synaptic transmission, a process known as synaptic plasticity. Although much is known about factors that positively regulate synaptic plasticity, very little is known about factors that negatively regulate this process. Recently, the signaling protein RGS14 was identified as a natural suppressor of hippocampal-based learning and memory, as well as synaptic plasticity within CA2 hippocampal neurons. RGS14 is a multifunctional scaffolding protein that integrates unconventional G protein and MAP kinase signaling pathways that are themselves key regulators of synaptic plasticity, learning, and memory. Here, we highlight known roles for RGS14 in brain physiology and unconventional G protein signaling pathways, and propose molecular models to describe how RGS14 may integrate these diverse signaling pathways to modulate synaptic plasticity in CA2 hippocampal neurons.
Regulators of G protein signaling (RGS) proteins
The RGS family of proteins is an important component of G protein coupled-receptor (GPCR) signaling pathways [1–4]. Established models propose that GPCRs activate heterotrimeric G proteins (Gαβγ) by serving as guanine nucleotide exchange factors (GEFs) to trigger GTP binding on the Gα subunit followed by Gβγ dissociation. Activated Gα-GTP and Gβγ interact with downstream effectors and signaling pathways to regulate cell and organ physiology [5]. Signaling is terminated by hydrolysis of GTP to GDP through the intrinsic GTPase activity of the Gα subunit. RGS proteins recognize both receptors [6] and activated Gα-GTP to selectively accelerate Gα GTPase activity and limit the duration of G protein signaling. This large family of nearly 40 distinct signaling proteins is classified into eight subfamilies according to sequence identities and shared functions as GTPase accelerating proteins (GAPs) [1–4]. RGS protein structures range from simple with a single domain, to highly complex with multiple protein binding domains (reviewed in [1–4]). The protein architecture of complex family members suggests that many RGS proteins serve as multifunctional integrators of G protein signaling pathways. In addition to acting as GAPs towards activated Gα subunits, certain complex RGS proteins exhibit other functions on Gα subunits and additional binding partners [2, 4]. This review will focus on the signaling roles of one particular complex RGS protein, RGS14, and its physiological functions in the hippocampus.
RGS14 is a structurally complex RGS protein
RGS14 is a 61 kDa protein classified within the D/R12 subfamily of RGS proteins. The closest relatives of RGS14 are RGS12 and RGS10, although RGS10 is much smaller and shares only a single RGS domain in common with RGS14. Besides the conserved RGS domain, RGS14 contains a second Gα binding domain (GPR/GoLoco domain) and two Ras/Rap-binding domains (RBDs) [7, 8] (Figure 1), suggesting that RGS14 serves signaling functions in addition to or distinct from the canonical GAP functions of an RGS protein. In particular, the presence of distinct binding sites on both RGS14 and RGS12 for Gα in either its active GTP-bound or inactive GDP-bound form indicates that RGS14 and RGS12 are unique among RGS proteins. Evidence to be discussed below suggests that RGS14 likely serves as a multifunctional scaffolding protein that integrates G protein and mitogen-activated protein (MAP) kinase signaling pathways. First, we will summarize what is known about the roles of RGS14 in brain physiology as a context for its known and proposed signaling functions.
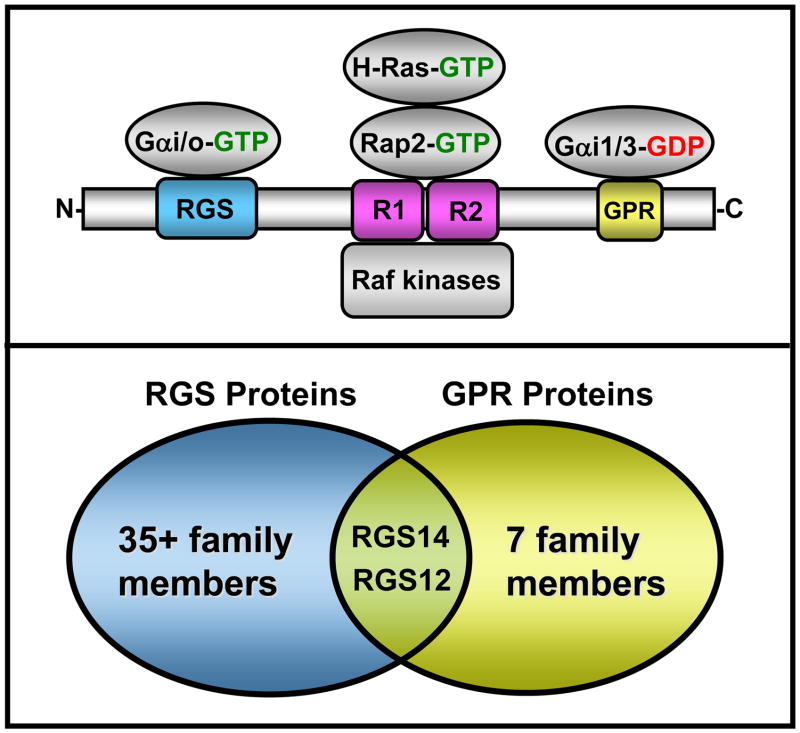
Figure 1. RGS14 domain structure and its identified binding partners.
Top: RGS14 directly binds activated Gαi family members and Gαo through its RGS domain, and it also specifically binds inactive Gαi1 and Gαi3 via its GPR domain. Activated H-Ras, Rap2, and Raf kinases directly interact with the Ras/Rap-binding domains (R1 and R2). Bottom: RGS14 is structurally and functionally unique in that it shares both an RGS domain and a GPR domain that places it and its closest relative RGS12 into both the RGS protein and the Group II AGS protein (GPR domain-containing) subfamilies.
RGS14 is a brain protein enriched in hippocampal CA2 neurons
In efforts to discern functional roles for RGS14 in cell and organ physiology, earlier studies found that RGS14 protein is expressed primarily in the brain, spleen, thymus, and lymphocytes of rodents [7–10]. Of these tissues, RGS14 is most abundant in adult brain, specifically within terminally differentiated neurons [9]. Although RGS14 certainly must have key functions in immunology based on this tissue distribution pattern, this review will focus only on its roles in the central nervous system (CNS). Within the rodent brain, RGS14 is expressed almost exclusively in neurons of the CA2 subregion of the hippocampus (Figure 2) [11], and appears to be enriched in dendrites and spines, with much less protein present in cell bodies and synaptic terminals [11]. These findings suggest that RGS14 is trafficked within its host neurons to regulate primarily postsynaptic signaling events.
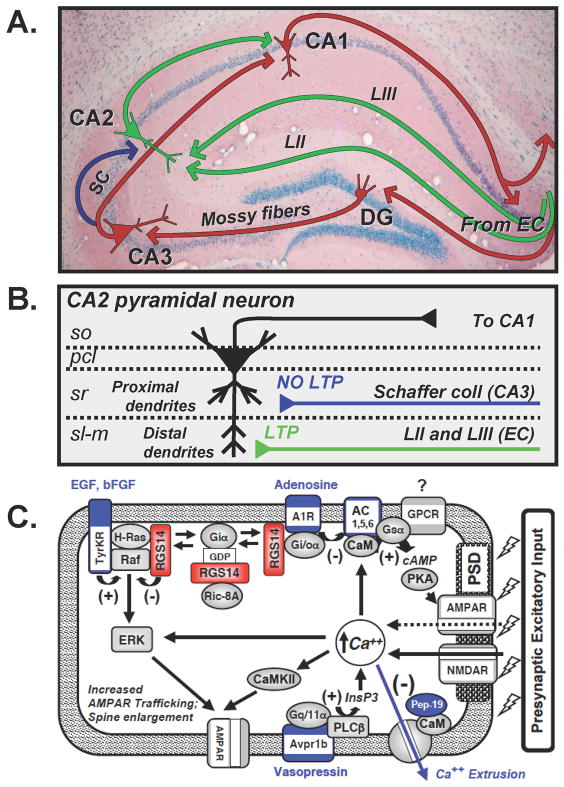
Figure 2. Hippocampal circuitry and possible roles for RGS14 in the suppression of LTP in CA2 neurons.
(A) Diagram of hippocampal circuitry. Red arrows indicate the classical dentate gyrus (DG)-CA3-CA1 trisynaptic circuit. Input from the entorhinal cortex (EC) synapse on granule neurons in the DG. Mossy fiber projections from DG synapse on CA3 pyramidal neurons. Schaffer Collateral (SC) projections from CA3 (red pathway) synapse on CA1 neurons to complete the trisynaptic circuit. The blue arrow indicates CA3 Schaffer collaterals distinct from the trisynaptic circuit that synapse on CA2 pyramidal neurons. Green arrows indicate separate circuits in which distinct EC inputs (LII and LIII) synapse on CA2 dendrites. These CA2 neurons subsequently project to CA1. (B) Differential synaptic plasticity on CA2 pyramidal neurons is elicited by distinct synaptic inputs. Layers of the hippocampus are shown at left; Stratum Oriens (so), Pyramidal Cell Layer (pcl), Stratum Radiatum (sr), and Stratum-lacunosum moleculare (sl-m). CA3 Schaffer collateral (blue) synapse on proximal apical dendrites of CA2 neurons within the Stratum Radiatum. High frequency stimulation of Schaffer collaterals generates no LTP in CA2 proximal dendrites. Projections from layer II and III of the EC (green) synapse on distal apical dendrites within the sl-m layer. Stimulation of EC inputs generates LTP in CA2 distal dendrites. (C) Cartoon model of a dendritic spine from CA2 neurons that express RGS14, and potential roles for RGS14 in the negative regulation of CA2 synaptic plasticity. Shown are distinct properties and signaling proteins that are uniquely or highly expressed in CA2 neurons (blue), additional signaling proteins that are involved in synaptic plasticity (gray), and proposed roles for RGS14 (red).
RGS14 is important for the acquisition of hippocampal-based spatial learning and object memory
The highly restricted brain distribution pattern for RGS14 suggests it serves a key role in hippocampal function. The hippocampus is a well-defined center important for spatial learning and memory. In tests of wild-type mice and their littermates lacking the RGS14 gene/protein, we found that the presence of RGS14 within CA2 neurons appears to suppress hippocampal-based spatial learning and object memory [11]. RGS14 knockout (RGS14-KO) mice learn more quickly to navigate a water-maze and locate a submerged escape platform, indicating that loss of RGS14 significantly enhances the acquisition rate of spatial learning. Additional tests of hippocampal memory with these mice showed that loss of RGS14 improved novel-object memory without altering other behaviors not directly associated with the hippocampus, such as open field locomotor activity, startle response, and anxiety [11]. Taken together, these findings suggest that RGS14 naturally inhibits certain forms of hippocampal-based learning and memory.
RGS14 is a natural suppressor of synaptic plasticity in CA2 neurons
Memory is thought to be encoded within the brain as biochemical and physical changes at synapses leading to alterations in neurotransmission, a process known as synaptic plasticity. One such form of synaptic plasticity is the long lasting increase in the strength of excitatory glutamatergic synaptic transmission (long-term potentiation; LTP) that can be induced with high frequency afferent stimulation. LTP has been best characterized within the well-defined dentate gyrus (DG)-CA3-CA1 trisynaptic circuit of the hippocampus [12–14], with the overwhelming majority of studies performed in the CA1 region.
The DG-CA3-CA1 pathway (Figure 2A, red) is a three-synapse (trisynaptic) circuit consisting of: 1) input from the entorhinal cortex (EC), forming synapses on granule neurons in the DG; 2) axons originating from the DG that project to area CA3, forming large “mossy fiber” synapses on dendrites of CA3 pyramidal neurons; and 3) CA3 axons, also known as the Schaffer collateral (SC) fibers, connecting to the dendrites of CA1 pyramidal neurons in an area known as the Stratum Radiatum. Area CA2 is almost always underrepresented in the literature, likely due to its earlier controversial status as a transition zone between CA1 and CA3 rather than as a separate area. In fact, the CA2 had been clearly defined decades earlier [15] as an area consisting of unique neurons that are similar in size to the large CA3 neurons, and that resemble CA1 neurons in that they receive no mossy fiber synaptic input from the DG. Instead, CA2 neurons receive their main input from the CA3 Schaffer collaterals, like CA1 (Figure 2A, blue).
Most commonly, LTP is robust and studied in CA1 neurons that receive the CA3-derived Schaffer collaterals. Unlike synapses on their CA1 neighbors, though, the Schaffer collateral synapses on CA2 pyramidal neurons do not typically exhibit LTP in response to high frequency stimulation [16, 17]. This lack of LTP in CA2 pyramidal neurons is attributed to increased calcium buffering capacity and increased calcium extrusion [16]. We found that even with the very active calcium handling in CA2 neurons, loss of RGS14 permits Schaffer collateral synapses in CA2 to now exhibit robust LTP, strongly suggesting that RGS14 is a natural suppressor of LTP in most CA2 synapses [11]. Given the distribution of RGS14 almost entirely within CA2, it is not surprising that LTP in CA1 was unaffected in slices from the RGS14-KO mice. These findings provide a direct link between RGS14 expression and hippocampal learning, memory, and synaptic plasticity that may depend on region-specific protein expression.
In addition to the Schaffer collateral input from CA3, CA2 neurons also receive distinct synaptic input from layer II (LII) and III (LIII) neurons of the EC (Figure 2A, green). Neurons of the CA1, CA2, and CA3 region are large and pyramidal in shape, and have dendrites that extend from the apex of the cell body (apical dendrites) that are either proximal (close to the cell body) or distal (extend far away) (Figure 2B). Unlike the CA3-Schaffer collateral input to CA2 neurons that synapse on the proximal and middle sections of the dendrites, the CA2 input from the EC form synapses on the distal portion of the dendrites in a region known as the Stratum Lacunosum Moleculare. Recently, Chevaleyre and Siegelbaum found that both LII and LIII EC pathways are capable of exhibiting robust NMDA receptor-dependent LTP in CA2 neurons, in contrast to the synapses from CA3 that fail to express LTP [18]. These findings suggest that regional differences or compartmentalization of molecular signaling machinery within CA2 neurons may provide distinct synaptic outputs in response to input and activity from CA3 or from the different layers of the EC. Consistent with this idea, we have found that RGS14 protein is differentially localized to a subset of CA2 dendritic spines and spine necks [11]. However, we see no obvious exclusion of RGS14 expression from the Stratum Lacunosum Moleculare, suggesting the involvement of other modulators at these more distal synapses.
Thus, our data showing enhancement of both memory and Schaffer collateral LTP in mice lacking RGS14 strongly suggest that the role of CA2 in learning and memory is likely dependent on RGS14-containing dendritic spines of CA2 synapses, which does not necessarily include the trisynapic DG-CA3-CA1 circuit. Dendritic spines act to limit the synaptic microenvironments with distinct protein expression profiles, calcium handling properties, or other synaptic properties. Of note, we find that a subset of RGS14 protein appears to localize to the PSD of dendritic spines [9, 11], indicating that RGS14 is well positioned to modulate signaling events important for synaptic plasticity. We will next discuss how RGS14 may act as a multifunctional integrator of signaling pathways important for synaptic plasticity.
RGS14 is a scaffold that binds G proteins and components of the MAP kinase signaling pathway that are important for synaptic plasticity
RGS14 interacts directly with and regulates the function of signaling proteins that are critically important for synaptic plasticity, learning, and memory. RGS14 was first discovered as a Rap1/2 binding partner [8], and each of the identified RGS14 binding partners Gαi/o, H-Ras, Rap2, and Raf kinases has been shown to control various aspects of synaptic plasticity within hippocampal neurons [19–24]. Following an initial report that one of the isolated purified RBDs of RGS14 can interact with H-Ras in vitro [25], we and others demonstrated that RGS14 binds both activated H-Ras and Raf-1 in cells [26, 27] to inhibit ERK-mediated MAP kinase signaling by platelet-derived growth factor (PDGF) [26]. Activated H-Ras recruits RGS14 to the plasma membrane in the absence of exogenous Gαi1, allowing RGS14 to bind Raf-1 and regulate PDGF-induced signaling [26]. Co-expressed wild-type Gαi1 reverses the ability of RGS14 to inhibit PDGF-induced ERK phosphorylation because, while bound to Gαi1, RGS14 can no longer bind Raf-1 [26]. This indicates that RGS14 may act as a molecular switch between binding/regulating Gαi1 and binding/regulating Raf-1 and subsequent Raf-1-induced ERK phosphorylation.
Although RGS14 regulates PDGF-induced ERK phosphorylation in an H-Ras- and Gαi1-dependent manner [26], how this occurs remains unknown. Various groups have reported unconventional roles for G proteins and interactions of G proteins with receptors that are not GPCRs (for a recent review, see [28]). Relevant to RGS14, recent studies have examined the role of Gαi in directly regulating PDGF receptor/ERK-mediated MAP kinase signaling. Pertussis toxin (PTX) treatment of cells prevents Gαi/o-coupling to receptors, which subsequently blocks c-Src activation and ERK phosphorylation by PDGF, indicating a possible role for Gαi in PDGF receptor regulation of c-Src signaling [29]. Though speculative, it is also possible that pertussis toxin may inhibit the function of non-receptor GEFs (e.g. Resistance to Inhibitors of Cholinesterase-8A, also known as Ric-8A) on Gαi [30] to reverse the effects of Gαi on c-Src activation, although there is no evidence yet to support this idea. The PDGFβ receptor is also shown to induce phosphorylation of Gαi upon stimulation, which enhances ERK phosphorylation [31].
A key element to the involvement of Gαi in this process is the potential role of a GPCR. Germane to this point was the discovery that the PDGFβ receptor interacts with the EDG1 receptor (also known as S1P1), a Gαi-linked GPCR [31]. Co-expression of both the PDGFβ receptor and EDG1 stimulates an increase in both Gαi phosphorylation and subsequent ERK activation following PDGF treatment [31]. How or even if RGS14 participates in PDGFβ/EDG1 receptor signaling is not known, but these studies highlight potential mechanisms for how RGS14 may switch from binding Gαi to binding activated H-Ras and regulating MAP kinase signaling. Stimulation of a GPCR may induce Gαi activation, subsequently influencing RGS14 localization and its preference for binding partners (e.g. Gαi vs. Raf-1). Formation of a GPCR:Gαi:RGS14 complex may also sequester RGS14 from binding its MAP kinase signaling partners. Additional studies will be necessary to distinguish between these possibilities, though our recent preliminary studies [32] indicate that RGS14 can functionally interact with GPCR:Gαi pairs. Taken together, these findings suggest that RGS14 may engage in both conventional and unconventional G protein signaling mechanisms to integrate G protein and MAP kinase signaling pathways.
RGS14 participates in unconventional G protein signaling
In addition to the RGS domain, RGS14 also contains a second Gα binding domain, the G protein regulatory (GPR) motif (also known as GoLoco, but henceforth referred to as GPR motif), which binds only inactive Gαi1-GDP and Gαi3-GDP [9, 33–35]. Growing evidence suggests that proteins containing GPR motifs participate in newly appreciated “unconventional” G protein signaling cascades [36, 37]. Unlike the well-established “conventional” G protein systems that involve a GPCR, a heterotrimeric Gαβγ complex, an effector, and an RGS protein, these unconventional pathways involve a Gα protein and other proteins that substitute for the receptor, effector, and Gβγ in a functional signaling complex (Figure 3). Though little is known about the physiological roles of unconventional G protein signaling complexes, evidence suggests that these proteins and their linked signaling pathways regulate key aspects of cell division in lower and higher eukaryotes and synaptic signaling in mammalian brain [36–42]. At the center of these unconventional complexes is a family of proteins that share one or more GPR motifs [36, 37, 43]. The Gαi-interacting GPR motif that is present in RGS14 is a shared and defining feature of the Group II Activator of G protein Signaling (AGS) proteins [36]. Of note, RGS14 and its closest relative RGS12 are the only RGS proteins (excluding splice variants) among the nearly 40 family members that contain a GPR motif. This attests to the unique multifunctional nature of these two proteins, and also highlights the fact that RGS14 and RGS12 alone sit at the interface of the very distinct mammalian RGS family and the Group II AGS family of signaling proteins (Figure 1).
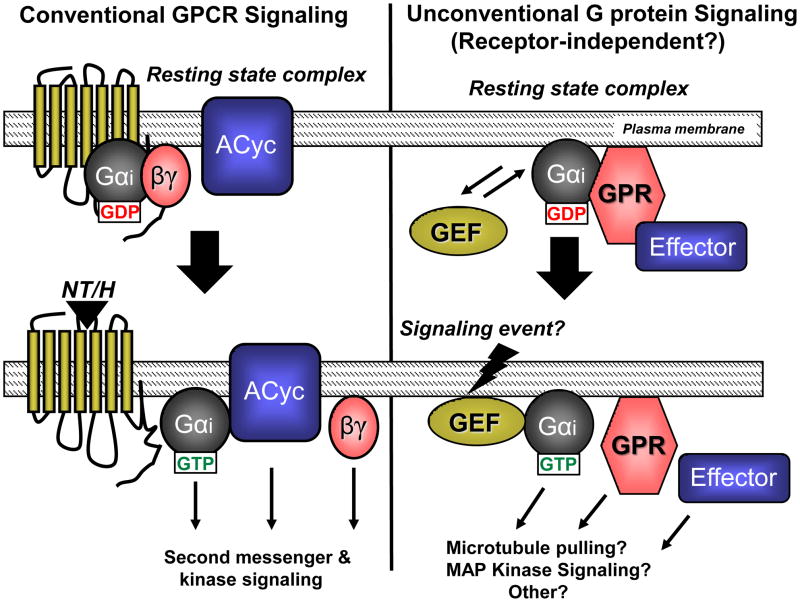
Figure 3. Conventional vs. unconventional G protein signaling.
Top: Before stimulation, conventional GPCR/G protein signaling (left) consists of a GPCR, Gαi-GDP bound to Gβγ, and a downstream effector protein (i.e. Adenylyl cyclase; ACyc). In unconventional signaling (right), a cytosolic GEF substitutes for and serves a role similar to that of the GPCR, while the GPR protein, perhaps in complex with an effector, substitutes for Gβγ. Bottom: In the presence of a stimulating neurotransmitter or hormone (NT/H), the GPCR exhibits GEF activity towards Gαi, resulting in GTP binding, heterotrimer dissociation, and subsequent Gαi-GTP and Gβγ coupling to the effector protein to regulate signaling pathways. In unconventional signaling, the cytosolic GEF catalyzes GTP exchange on the Gαi subunit, resulting in free Gαi-GTP, GPR protein, and effector that are able to regulate downstream signaling.
Early evidence for a function of GPR motifs came from studies with the Activator of G protein Signaling 3 (AGS3) protein. AGS3 was reported to activate a pheromone response pathway in Saccharomyces cerevisiae independent of a GPCR, and to selectively interact with Gα-GDP instead of Gα-GTP [44]. This finding suggested a novel mechanism of G protein activation that involved GPR domain-containing proteins functioning in the absence of GPCRs. Subsequent studies showed that the GPR domain is capable of forcing dissociation of Gαβγ heterotrimers to free Gα from Gβγ [45]. When in complex with Gα-GDP, GPR domains inhibit GDP release from Gα to prevent GTP binding [33, 34, 46, 47], thereby exhibiting guanine nucleotide dissociation inhibitor (GDI) activity. In many ways, the resulting effects of GPR association with Gα-GDP functionally resemble those of Gβγ when in complex with Gα-GDP, suggesting that Gα can exist in two distinct pools within host cells: one bound to Gβγ and one bound to GPR proteins. In this scenario, Gα either binds Gβγ at the plasma membrane or anchors GPR proteins to the plasma membrane.
The source(s) of Gα that bind to GPR proteins and how Gα comes to associate with GPR proteins is unclear, but several possibilities exist. Following activation of a GPCR and dissociation of Gαβγ, free Gα could be captured by a nearby GPR protein immediately after GTP hydrolysis, thereby swapping binding partners (GPR in place of Gβγ). Alternatively, a pool of Gα-GDP distinct from that associated with Gβγ may be sorted/targeted independently for association with GPR proteins for functions unrelated to GPCR signaling. Of note, RGS14 shares many properties with other GPR proteins in that it: 1) binds Gαi1-GDP and/or Gαi3-GDP independent of Gβγ, 2) is recruited from the cytosol to the plasma membrane by inactive Gαi1/3- GDP, and 3) can act as a GDI to inhibit nucleotide exchange [36, 43]. It remains unclear what factor(s) regulates the activation states of the RGS14:Gαi1/3-GDP complex and other GPR:Gαi- GDP complexes, though recent studies suggest a role for novel non-receptor, cytosolic GEFs [48–50].
Some underlying mechanisms of GPR:Gαi signaling were elucidated following the discovery of Resistance to Inhibitors of Cholinesterase (RIC-8; also known as Synembryn) in Caenorhabditis elegans, and its mammalian counterparts Ric-8A and Ric-8B [51, 52]. Unlike GPCRs, Ric-8 proteins exist as soluble cytosolic proteins in the absence of binding partners. However, much like GPCRs, Ric-8A acts as a GEF towards Gαi1, Gαq, and Gαo subunits and binds the inactive form of these subunits in the absence of Gβγ to induce GDP release and GTP binding to the subunit [52]. Because Ric-8A only acts on inactive Gα subunits, it was thought that Ric-8A may act on GPR:Gα-GDP protein complexes. Studies designed to test this idea found that purified Ric-8A protein binds and acts on both the purified GPR protein complexes LGN:Gαi1-GDP and AGS3:Gαi1-GDP, catalyzing nucleotide binding to Gαi1 and subsequently inducing dissociation of the complexes [48, 49]. Gαi1 simultaneously binds both Ric-8A and the GPR domain of AGS3 to form a transient ternary complex [49].
With respect to RGS14, we have recently shown that the RGS14:Gαi1-GDP complex is also regulated by Ric-8A [50]. Ric-8A binds RGS14 in cells and induces dissociation of the RGS14:Gαi1-GDP complex both in cells and in vitro. Ric-8A catalyzes both GTP binding to and the steady-state GTPase activity of Gαi1 in the presence of RGS14, ultimately overcoming the GDI effects of RGS14 [50]. Of note, both RGS14 and Ric-8A co-localize within the same CA2 hippocampal neurons, supporting a functional link between these two proteins that may be involved in RGS14’s effects on learning and memory [50]. Other non-receptor cytosolic proteins besides Ric-8A and Ric-8B have been identified [53, 54] that may also serve as GEFs for GPR:Gαi complexes, illustrating an extensive (though poorly understood) network of unconventional G protein signaling pathways.
Working model for how RGS14 may integrate unconventional G protein and MAP kinase signaling pathways in hippocampal neurons
To summarize, RGS14 is a multifunctional scaffolding protein in brain that binds Ric-8A, active Gαi/o, inactive Gαi1/3, active H-Ras and Rap2, and Raf kinases. RGS14 localizes to dendritic spines and possibly the PSD of CA2 hippocampal neurons, and is important for hippocampal synaptic plasticity, learning, and memory. However, the molecular mechanisms whereby RGS14 and its binding partners integrate unconventional G protein and MAP kinase signaling to modulate synaptic plasticity remain uncertain. Even so, sufficient information is now available to propose a testable working model (Figure 4) that describes how the RGS and GPR domains of RGS14 work together to bind and modulate the functions of a soluble GEF, such as Ric-8A, Gαi, H-Ras, and Raf kinases in a coordinated signaling event.
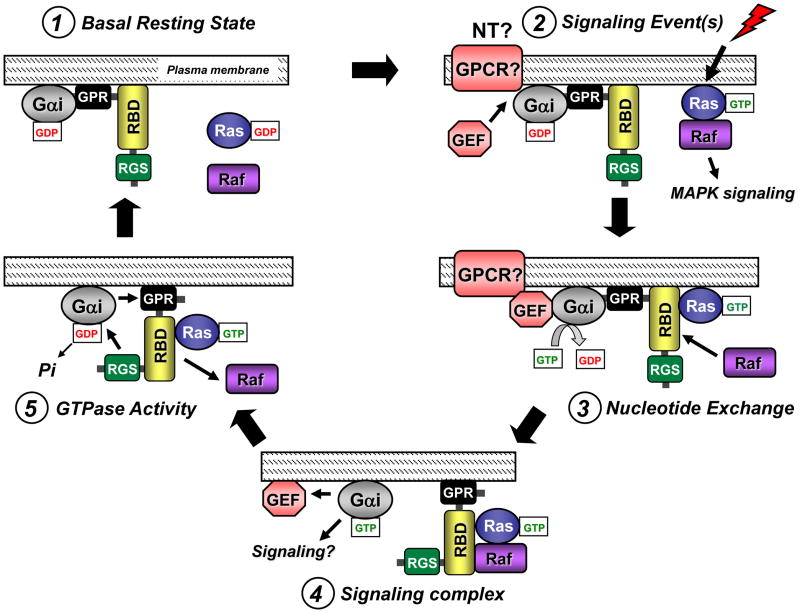
Figure 4. Proposed working model for how the RGS, RBD, and GPR domains of RGS14 may function coordinately to regulate Gαi signaling.
The proposed model for RGS14 signaling proceeds clockwise from top left. (1) RGS14 pre-exists in complex with inactive Gαi-GDP via its GPR motif at the plasma membrane in its basal resting state. (2) An unknown stimulation event, perhaps through a receptor tyrosine kinase to stimulate Ras and/or neurotransmitter (NT) activation of a GPCR, induces recruitment of a GEF to the RGS14:Gαi-GDP complex. (3) After binding the RGS14:Gαi-GDP complex, the GEF catalyzes nucleotide exchange on and GTP binding to the Gαi, thereby releasing RGS14 which is now free to bind activated Ras/Raf via its RBDs. (4) Active Gαi-GTP dissociates from RGS14, allowing it to serve as a scaffold to assemble Ras and Raf in a signaling complex. (5) In some regulated fashion, the adjacent RGS domain recognizes the active Gαi to accelerate Gα-GTP hydrolysis, resulting in signal termination. The nearby GPR domain re-binds Gαi-GDP and causes Raf and Ras to dissociate, leading to reformation of the inactive RGS14:Gαi-GDP complex and a return to the basal resting state (1).
In contrast to other RGS protein signaling models, our proposed model for RGS14 highlights the GPR domain rather than the RGS domain as the first point of contact between RGS14 and Gαi. In the basal resting state (Figure 4, Step 1), we propose that RGS14 exists in a stable complex with Gαi-GDP at the plasma membrane, or perhaps at the PSD within CA2 hippocampal neurons. We postulate that following a signaling event (as yet undefined) (Figure 4; Step 2), a soluble GEF, such as Ric-8A, recognizes and stimulates nucleotide exchange and GTP binding to Gαi, subsequently promoting dissociation of the RGS14:Gαi-GDP complex because the GPR domain cannot bind Gα-GTP. Of note, a role for a Gαi-linked GPCR or tyrosine kinase receptor in this activation step cannot be ruled out [32, 55–57]. Once released from Gαi (Figure 4; Step 3), RGS14 would be free to interact with other downstream binding partners (e.g. active H-Ras, Rap2, and Raf kinases). RGS14 may sequester H-Ras and Raf-1 in a signaling complex to passively inhibit and/or modulate MAP kinase signaling involved with LTP and synaptic plasticity (Figure 4; Step 4). We postulate that the lifetime of this RGS14 signaling complex is limited by the RGS domain (Figure 4; Step 5), which would act on the nearby Gαi-GTP to restore Gαi-GDP and promote reformation of the RGS14:Gαi-GDP complex via the GPR domain. We speculate that Gαi-GDP binding to RGS14 is coupled with dissociation of H-Ras and Raf-1 and a return to the basal resting state (Figure 4; Step 1).
Although speculative, this proposed activation/deactivation cycle is entirely consistent with reported findings, though many steps remain to be tested. One attractive feature of this model is that it reconciles the need for RGS and GPR domains within RGS14, and also highlights the possibility that other GPR proteins and RGS proteins can work together in specific cellular contexts. This model also accounts for the idea that the RGS and GPR domains are functioning together to limit the presence of activated Gαi subunits, favoring the accumulation of Gαi-GDP. Furthermore, having the GPR and RGS domains built into the same protein could serve to spatially restrict the RGS domain GAP activity towards the pre-bound Gα, thus the RGS domain would exhibit GAP activity towards the activated Gα that is released from the GPR domain. This would be a logical point for tight regulation, for example, by a reversible phosphorylation step, as RGS14 is a target of phosphorylation by both cAMP-PKA and ERK [58, 59]. Future studies will examine this idea and other untested steps in this model. Although our model addresses the mechanics of how RGS14 might integrate G protein and MAP kinase signaling pathways, it does not address how RGS14 integrates these signaling steps at the PSD of dendritic spines to modulate synaptic plasticity.
Possible mechanistic roles for RGS14 and its binding partners in synaptic plasticity
RGS14 appears to localize to post-synaptic densities of dendritic spines [9, 11], well-established focal points for the induction and expression of synaptic plasticity. Furthermore, as outlined above, RGS14 binds specific G proteins and key components of the MAP kinase pathway that are important for synaptic plasticity. Based on these findings, we postulate that RGS14 serves as a regulatory brake to reduce LTP and synaptic plasticity initiated by presynaptic input. CA2 neurons express a unique profile of signaling genes and proteins [60, 61], RGS14 among them, that may contribute to the region’s unusual regulation of LTP. How RGS14 may interact with these or other signaling proteins to integrate G protein and MAP kinase signaling pathways and modulate synaptic plasticity is unclear. Here we will entertain various possibilities based on what is currently known about CA2 synaptic transmission (Figure 2C).
The G protein-regulated second messengers calcium and cAMP each play crucial roles in learning, memory, and synaptic plasticity by regulating gene expression and modulating postsynaptic signaling events. Calcium in particular is important in mediating MAP kinase signaling in neurons [24, 62, 63]. Within CA2 neurons, there exist both calcium-dependent and calcium-independent mechanisms that regulate LTP. RGS14 may therefore regulate calcium signaling directly or indirectly by one or more mechanisms. A particularly robust calcium extrusion system normally suppresses LTP in proximal and middle regions of CA2 dendrites, yet stimulation of Gq/11-linked vasopressin (Avp1b) receptors potentiates postsynaptic transmission that normally requires calcium and calmodulin kinase II (CamKII) [16, 17, 64]. Potentiation of postsynaptic transmission in CA2 neurons is also induced by calcium independent-mechanisms requiring cAMP/PKA following inhibition of Gi/o-linked adenosine A1 receptors [65]. Loss of RGS14 and the capacity of its RGS domain to limit Gαi/o signaling may alter postsynaptic cAMP levels to affect LTP and learning. Since GPR motifs can, in some cases, compete with Gβγ for Gαi binding [45], then loss of RGS14 may allow activated Gβγ to bind free Gαi to form an inactive complex, thus terminating any Gβγ-mediated effects on calcium channels.
Alternatively, RGS14 actions on synaptic plasticity in CA2 neurons may be linked to its capacity to bind Rap2, H-Ras, and Raf kinase and regulate MAP kinase signaling. Both H-Ras and Rap2 are implicated in different forms of synaptic plasticity in the hippocampus. Expression of active Rap2A in hippocampal neurons of mice results in spatial learning deficits that are coupled with increases in Janus kinase (JNK) activity, decreases in synaptic AMPA receptor activity and dendritic branch complexity, and an enhancement of long-term depression (LTD) of neurotransmission [20–22]. By contrast, H-Ras enhances AMPA receptor-mediated synaptic transmission and induces up-regulation of AMPA receptors on spine surfaces [66]. However, other studies also show that Ras regulates LTP in hippocampal neurons by facilitating NMDA receptor phosphorylation [67]. Furthermore, transgenic mice that express active H-Ras exhibit enhanced spatial learning and LTP [23], a phenotype similar to the RGS14-KO mice [11]. Our observation that the nascent LTP caused by the loss of RGS14 is blocked by MEK inhibitors [11] suggests that RGS14 modulates synaptic plasticity through the inhibition of Ras/Raf/MAP kinase signaling pathways in hippocampal neurons (Figure 2C). Of note, CA2 neurons express high levels of the EGF receptor (EGFR) and basic-FGF receptor, FGF1R [60, 61], both tyrosine kinase receptors that activate H-Ras and ERK signaling. However, H-Ras-mediated ERK signaling is also directly activated by G proteins, calcium, and/or cAMP regulated pathways [24], so RGS14 may serve as a brake on ERK signaling downstream of any of these pathways
Despite this wealth of information, much remains unknown about negative regulation of synaptic plasticity and the role of RGS14 in this process. Although evidence suggests a role for H-Ras/ERK/MEK signaling, we cannot rule out the possibility that RGS14 suppresses LTP in CA2 neurons through modulation of conventional or unconventional Gαi/o signaling, Rap2 signaling, or some combination of these and H-Ras/ERK/MEK signaling. Binding of RGS14 to these proteins may either inhibit and/or redirect their signaling to alter regulation of synaptic plasticity. Studies are ongoing to determine which of these pathways underlie RGS14-mediated suppression of synaptic plasticity in CA2 neurons.
Concluding Remarks
In summary, compelling evidence now indicates that RGS14 is a multifunctional scaffold that integrates G protein and MAP kinase signaling pathways important for synaptic plasticity in CA2 hippocampal neurons. Although much is known about RGS14 binding partners and how they interact, more studies are needed to examine how these proteins and RGS14 may work together to suppress hippocampal synaptic plasticity in CA2 neurons. RGS14 can be added to a growing list of genes/proteins that have been linked to enhanced cognition [68]. The challenge going forward will be to determine how RGS14 fits into these key pathways to suppress LTP, and how this process is regulated. Besides these signaling proteins involved with enhanced cognition, other GPR proteins that share similarities with RGS14 are also important for brain function. The mammalian partner of inscutable (mPins, aka LGN) and AGS3 both contain GPR domains that bind Gαi/o-GDP to stabilize their association with membranes, are regulated by Ric-8A, and are enriched in brain. AGS3 is localized within neurons throughout most of the CNS, including the hippocampus [69], and in the prefrontal cortex and nucleus accumbens AGS3 is reported to be important for cocaine-seeking and ethanol-seeking relapse behavior, respectively [70, 71]. mPins/LGN is enriched in synaptic membranes of CA1 hippocampal neurons, where it associates with PSD-95 and MAGUK scaffolding proteins in a Gαi1-dependent manner to influence membrane trafficking, NMDA receptor surface expression, and dendritic remodeling [42]. RGS14 and its binding partners in CA2 neurons likely serve roles mechanistically similar to, though functionally distinct from those of mPins/LGN and AGS3 in brain physiology. Together, these proteins and RGS14 represent a newly appreciated class of G protein binding partners important for brain physiology/disease that could serve as future therapeutic targets for a range of CNS pathologies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.De Vries L, et al. The Regulator of G protein Signaling family. Annu Rev Pharmacol Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 2.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 3.Ross EM, Wilkie TM. GTPase-Activating proteins for heterotrimeric G proteins: Regulators of G protein Signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 4.Willars GB. Mammalian RGS proteins: multifunctional regulators of cellular signalling. Semin Cell Devel Biol. 2006;17:363–376. doi: 10.1016/j.semcdb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 6.Neitzel KL, Hepler JR. Cellular mechanisms that determine selective RGS protein regulation of G protein-coupled receptor signaling. Semin Cell Devel Biol. 2006;17:383–389. doi: 10.1016/j.semcdb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Snow BE, et al. Molecular cloning and expression analysis of rat Rgs12 and Rgs14. Biochem Biophys Res Comm. 1997;233:770–777. doi: 10.1006/bbrc.1997.6537. [DOI] [PubMed] [Google Scholar]
- 8.Traver S, et al. RGS14 is a novel Rap effector that preferentially regulates the GTPase activity of Gαo. Biochem J. 2000;350:19–29. [PMC free article] [PubMed] [Google Scholar]
- 9.Hollinger S, et al. RGS14 is a bifunctional regulator of Galphai/o activity that exists in multiple populations in brain. J Neurochem. 2001;79:941–949. doi: 10.1046/j.1471-4159.2001.00629.x. [DOI] [PubMed] [Google Scholar]
- 10.Cho H, et al. RGS14, a GTPase-activating protein for Giα, attenuates Giα- and G13α-mediated signaling pathways. Mol Pharmacol. 2000;58:569–576. doi: 10.1124/mol.58.3.569. [DOI] [PubMed] [Google Scholar]
- 11.Lee SE, et al. RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc Natl Acad Sci USA. 2010;107:16994–16998. doi: 10.1073/pnas.1005362107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neves G, et al. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat Rev Neurosci. 2008;9:65–75. doi: 10.1038/nrn2303. [DOI] [PubMed] [Google Scholar]
- 13.Nakazawa K, et al. Hippocampal CA3 NMDA receptors are crucial for memory acquisition of one-time experience. Neuron. 2003;38:305–315. doi: 10.1016/s0896-6273(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 14.Rolls ET, Kesner RP. A computational theory of hippocampal function, and empirical tests of the theory. Prog Neurobiol. 2006;79:1–48. doi: 10.1016/j.pneurobio.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Lorente de No R. Studies on the structure of the cerebral cortex II. Continuation of the study of the ammonic system. J Psychol Neurol. 1934;46:113–117. [Google Scholar]
- 16.Simons SB, et al. Regional differences in hippocampal calcium handling provide a cellular mechanism for limiting plasticity. Proc Natl Acad Sci USA. 2009;106:14080–14084. doi: 10.1073/pnas.0904775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao M, et al. Synaptic plasticity (and the lack thereof) in hippocampal CA2 neurons. J Neurosci. 2007;27:12025–12032. doi: 10.1523/JNEUROSCI.4094-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevaleyre V, Siegelbaum SA. Strong CA2 pyramidal neuron synapses define a powerful disynaptic cortico-hippocampal loop. Neuron. 2010;66:560–72. doi: 10.1016/j.neuron.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pineda VV, et al. Removal of G(ialpha1) constraints on adenylyl cyclase in the hippocampus enhances LTP and impairs memory formation. Neuron. 2004;41:153–163. doi: 10.1016/s0896-6273(03)00813-4. [DOI] [PubMed] [Google Scholar]
- 20.Fu Z, et al. Differential roles of Rap1 and Rap2 small GTPases in neurite retraction and synapse elimination in hippocampal spiny neurons. J Neurochem. 2007;100:118–131. doi: 10.1111/j.1471-4159.2006.04195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu J, et al. Constitutively active Rap2 transgenic mice display fewer dendritic spines, reduced extracellular signal-regulated kinase signaling, enhanced long-term depression, and impaired spatial learning and fear extinction. J Neurosci. 2008;28:8178–8188. doi: 10.1523/JNEUROSCI.1944-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, et al. Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron. 2005;46:905–916. doi: 10.1016/j.neuron.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 23.Kushner SA, et al. Modulation of presynaptic plasticity and learning by the H-ras/Extracellular Signal-Regulated Kinase/Synapsin I signaling pathway. J Neurosci. 2005;25:9721–9734. doi: 10.1523/JNEUROSCI.2836-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy MB, et al. Integration of biochemical signalling in spines. Nat Rev Neurosci. 2005;6:423–434. doi: 10.1038/nrn1685. [DOI] [PubMed] [Google Scholar]
- 25.Kiel C, et al. Recognizing and defining true Ras binding domains II: in silico prediction based on homology modelling and energy calculations. J Mol Biol. 2005;348:759–775. doi: 10.1016/j.jmb.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 26.Shu F-j, et al. RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways. Cell Signal. 2010;22:366–376. doi: 10.1016/j.cellsig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willard FS, et al. Regulator of G-protein signaling 14 (RGS14) is a selective H-Ras effector. PLoS One. 2009:4. doi: 10.1371/journal.pone.0004884. ( http://www.plosone.org) [DOI] [PMC free article] [PubMed]
- 28.Marty C, Ye RD. Heterotrimeric G protein signaling outside the realm of seven transmembrane domain receptors. Mol Pharmacol. 2010;78:12–18. doi: 10.1124/mol.110.063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conway AM, et al. Platelet-derived-growth-factor stimulation of the p42/p44 mitogen-activated protein kinase pathway in airway smooth muscle: role of pertussis-toxin-sensitive G-proteins, c-Src tyrosine kinases and phosphoinositide 3-kinase. Biochem J. 1999;337:171–177. [PMC free article] [PubMed] [Google Scholar]
- 30.Woodard GE, et al. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol Cell Biol. 2010;30:3519–3530. doi: 10.1128/MCB.00394-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alderton F, et al. Tethering of the platelet-derived growth factor β receptor to G-protein-coupled receptors. A novel platform for integrative signaling by these receptor classes in mammalian cells. J Biol Chem. 2001;276:28578–28585. doi: 10.1074/jbc.M102771200. [DOI] [PubMed] [Google Scholar]
- 32.Vellano CP, et al. GPCRs and Ric-8A both regulate the RGS14:Gαi1 complex. FASEB J. 2011;25:804.803. [Google Scholar]
- 33.Kimple RJ, et al. RGS12 and RGS14 GoLoco motifs are Gαi interaction sites with guanine nucleotide dissociation inhibitor activity. J Biol Chem. 2001;276:29275–29281. doi: 10.1074/jbc.M103208200. [DOI] [PubMed] [Google Scholar]
- 34.Mittal V, Linder ME. The RGS14 GoLoco domain discriminates among Gαi isoforms. J Biol Chem. 2004;279:46772–46778. doi: 10.1074/jbc.M407409200. [DOI] [PubMed] [Google Scholar]
- 35.Shu F-j, et al. Selective interactions between Giα1 and Giα3 and the GoLoco/GPR domain of RGS14 influence its dynamic subcellular localization. Cell Signal. 2007;19:163–176. doi: 10.1016/j.cellsig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Sato M, et al. Accessory proteins for G proteins: partners in signaling. Annu Rev Pharmacol Toxicol. 2006;46:151–187. doi: 10.1146/annurev.pharmtox.46.120604.141115. [DOI] [PubMed] [Google Scholar]
- 37.Willard FS, et al. Return of the GDI: the GoLoco motif in cell division. Annu Rev Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- 38.Colombo K, et al. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- 39.Groves B, et al. A specific role of AGS3 in the surface expression of plasma membrane proteins. Proc Natl Acad Sci USA. 2007;104:18103–18108. doi: 10.1073/pnas.0709282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hampoelz B, Knoblich JA. Heterotrimeric G proteins: new tricks for an old dog. Cell. 2004;119:453–456. doi: 10.1016/j.cell.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds, et al. Convergent, RIC-8-dependent Galpha signaling pathways in the Caenorhabditis elegans synaptic signaling network. Genetics. 2005;169:651–670. doi: 10.1534/genetics.104.031286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sans N, et al. mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surface expression. Nat Cell Biol. 2005;7:1179–1190. doi: 10.1038/ncb1325. [DOI] [PubMed] [Google Scholar]
- 43.Siderovski DP, et al. The GoLoco motif: a Galphai/o binding motif and potential guanine-nucleotide exchange factor. Trends Biochem Sci. 1999;24:340–341. doi: 10.1016/s0968-0004(99)01441-3. [DOI] [PubMed] [Google Scholar]
- 44.Takesono A, et al. Receptor-independent activators of heterotrimeric G-protein signaling pathways. J Biol Chem. 1999;274:33202–33205. doi: 10.1074/jbc.274.47.33202. [DOI] [PubMed] [Google Scholar]
- 45.Ghosh M, et al. Receptor- and nucleotide exchange-independent mechanisms for promoting G protein subunit dissociation. J Biol Chem. 2003;278:34747–34750. doi: 10.1074/jbc.C300271200. [DOI] [PubMed] [Google Scholar]
- 46.Natochin M, et al. Inhibition of GDP/GTP exchange on G alpha subunits by proteins containing G-protein regulatory motifs. Biochemistry. 2001;40:5322–5328. doi: 10.1021/bi015505w. [DOI] [PubMed] [Google Scholar]
- 47.Peterson YK, et al. Stabilization of the GDP-bound conformation of Gialpha by a peptide derived from the G-protein regulatory motif of AGS3. J Biol Chem. 2000;275:33193–33196. doi: 10.1074/jbc.C000509200. [DOI] [PubMed] [Google Scholar]
- 48.Tall GG, Gilman AG. Resistance to inhibitors of cholinesterase 8A catalyzes release of Gαi-GTP and nuclear mitotic apparatus protein (NuMA) from NuMA/LGN/Gαi-GDP complexes. Proc Natl Acad Sci USA. 2005;102:16584–16589. doi: 10.1073/pnas.0508306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas CJ, et al. Ric-8A catalyzes guanine nucleotide exchange on Gαi1 bound to the GPR/GoLoco exchange inhibitor AGS3. J Biol Chem. 2008;283:23150–23160. doi: 10.1074/jbc.M802422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vellano CP, et al. Activation of the Regulator of G protein Signaling 14-Galphai1-GDP signaling complex is regulated by Resistance to Inhibitors of Cholinesterase-8A. Biochemistry. 2011;50:752–762. doi: 10.1021/bi101910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller KG, et al. RIC-8 (Synembryn): a novel conserved protein that is required for G(q)alpha signaling in the C. elegans nervous system. Neuron. 2000;27:289–299. doi: 10.1016/s0896-6273(00)00037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tall GG, et al. Mammalian Ric-8A (synembryn) is a heterotrimeric Gα protein guanine nucleotide exchange factor. J Biol Chem. 278:8356–8362. doi: 10.1074/jbc.M211862200. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Marcos, et al. GIV is a nonreceptor GEF for Gαi with a unique motif that regulates Akt signaling. Proc Natl Acad Sci USA. 2009;106:3178–3183. doi: 10.1073/pnas.0900294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le-Niculescu H, et al. Identification and characterization of GIV, a novel Gαi/s -interacting protein found on COPI, endoplasmic reticulum-Golgi transport vesicles. J Biol Chem. 2005;280:22012–22020. doi: 10.1074/jbc.M501833200. [DOI] [PubMed] [Google Scholar]
- 55.Cao C, et al. Gαi1 and Gαi3 are required for epidermal growth factor-mediated activation of the Akt-mTORC1 pathway. Sci Signal. 2009:2. doi: 10.1126/scisignal.2000118. ( http://stke.sciencemag.org) [DOI] [PMC free article] [PubMed]
- 56.Oner SS, et al. Regulation of the AGS3·Gαi signaling complex by a seven-transmembrane span receptor. J Biol Chem. 2010;285:33949–33958. doi: 10.1074/jbc.M110.138073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oner SS, et al. Receptor-regulated interaction of Activator of G-protein Signaling-4 and Galphai. J Biol Chem. 2010;285:20588–20594. doi: 10.1074/jbc.C109.088070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hollinger S, Hepler JR. Methods for measuring RGS protein phosphorylation by G protein-regulated kinases. Methods Mol Biol. 2003;237:205–219. doi: 10.1385/1-59259-430-1:205. [DOI] [PubMed] [Google Scholar]
- 59.Hollinger S, et al. Phosphorylation of RGS14 by Protein Kinase A potentiates its activity toward G alpha i. Biochemistry. 2003;42:811–819. doi: 10.1021/bi026664y. [DOI] [PubMed] [Google Scholar]
- 60.Lein ES, et al. Redefining the boundaries of the hippocampal CA2 subfield in the mouse using gene expression and 3-dimensional reconstruction. J Comp Neurol. 2005;485:1–10. doi: 10.1002/cne.20426. [DOI] [PubMed] [Google Scholar]
- 61.Lein ES, et al. Defining a Molecular Atlas of the Hippocampus Using DNA Microarrays and High-Throughput In Situ Hybridization. J Neurosci. 2004;24:3879–3889. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem. 2007;76:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- 63.Sweatt JD. Mitogen-activated protein kinases in synaptic plasticity and memory. Curr Opin Neurobiol. 2004;14:311–317. doi: 10.1016/j.conb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 64.Zhao M, Dudek SM. Neuroscience Meeting Planner. San Diego, CA: Society for Neuroscience; 2010. Vasopressin induces synpaptic potentiation in hippocampal CA2 neurons. Program Number 550.7. Online. [Google Scholar]
- 65.Dudek SM, Simons SB. Neuroscience Meeting Planner. San Diego, CA: Society for Neuroscience; 2010. A novel mechanism for caffeine induced cognitive enahncement. Program Number 550.8. Online. [Google Scholar]
- 66.Zhu JJ, et al. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell. 2002;110:443–455. doi: 10.1016/s0092-8674(02)00897-8. [DOI] [PubMed] [Google Scholar]
- 67.Manabe T, et al. Regulation of long-term potentiation by H-Ras through NMDA receptor phosphorylation. J Neurosci. 2000;20:2504–2511. doi: 10.1523/JNEUROSCI.20-07-02504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee YS, Silva AJ. The molecular and cellular biology of enhanced cognition. Nat Rev Neurosci. 2009;10:126–140. doi: 10.1038/nrn2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blumer JB, et al. Expression analysis and subcellular distribution of the two G-protein regulators AGS3 and LGN indicate distinct functionality. Localization of LGN to the midbody during cytokinesis. J Biol Chem. 2002;277:15897–15903. doi: 10.1074/jbc.M112185200. [DOI] [PubMed] [Google Scholar]
- 70.Bowers MS, et al. Nucleus accumbens AGS3 expression drives ethanol seeking through G betagamma. Proc Natl Acad Sci USA. 2008;105:12533–12538. doi: 10.1073/pnas.0706999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bowers MS, et al. Activator of G protein signaling 3: a gatekeeper of cocaine sensitization and drug seeking. Neuron. 2004;42:269–281. doi: 10.1016/s0896-6273(04)00159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]