Abstract
Atrial fibrillation (AF), the most common sustained cardiac arrhythmia, represents a major health burden to individuals and health care system within the Western world. The lifetime risk for the development of AF at age 40 years has been estimated to be approximately 1 in 4. AF is associated with substantial morbidity and a two-fold increased risk of mortality. Given its increasing prevalence with age, coupled with the aging population, the number of Americans affected with AF is expected to increase from ~ 2.3 million in the year 2000 to nearly 16 million by 2050. This AF epidemic is further complicated by the lack of highly effective therapies. One reason for the lack of effective therapies for AF stems from incomplete understanding of the complex pathophysiology of the arrhythmia. AF has often been regarded as a condition that occurs in the context of atrial electrical and structural remodeling that can result from cardiac and systemic disorders. However, up to 30% of patients have no obvious cause and are said to have idiopathic or “lone” AF. Up until recently, AF was considered to be a sporadic, non-genetic disorder, but we and others have shown that lone AF has a substantial genetic basis. Mutations in genes encoding cardiac ion channels (KCNQ1, KCNE1-5, KCNJ2, KCNA5 and SCN5A), gap junctions (GJA5), and signaling molecules (atrial natriuretic peptide [ANP], nucleoporins [NUP155]) have been reported in isolated cases and small kindreds. The advent of the human genome and HapMap projects and high-throughput genotyping has fundamentally accelerated our ability to discover the genetic contribution to common variation in human disease. In 2007, a genome-wide association study (GWAS) identified two genetic variants that associated with AF. More recently, two additional AF loci on chromosomes 16q22 and 1q21 have been identified. It is quite likely however, that the effects of alleles in many genes contribute to common complex diseases such as AF. The overall AF risk associated with common variants identified by the GWAS approach is small (odds ratios 1.1–2.5) and explains less than 10% of the heritability in lone AF. This raises the possibility that rare independent variants with large effects strong effects may account for a large fraction of the risk for lone AF.
Introduction
Atrial fibrillation (AF) is the most common arrhythmia in clinical practice and has become a disease of epidemic proportions, affecting 2–5 million Americans with increased morbidity, mortality and socioeconomic consequences of repeated hospital admissions, chronic disease management and disabilities.1 The lifetime risk for the development of AF at age 40 years has been estimated to be approximately 1 in 4. 2, 3 It is projected that the number of persons in the U.S. with AF will exceed 16 million by the year 2050.4
Despite the overall advance in the treatment of patients with cardiac arrhythmias, therapeutic options in AF have remained largely unchanged. For most patients, the goal of therapy is to restore and maintain normal sinus rhythm, control heart rate and prevent systemic embolization. New developments in surgical and ablation techniques for AF are promising, but to date are still laborious, plagued by significant complications, and have limited applicability.
One reason for the lack of effective therapies for AF stems from incomplete understanding of the complex pathophysiology of this common arrhythmia. The familial aggregation and high heritability of AF in these studies are consistent with common genetic mechanisms for the arrhythmia. Evidence of a genetic contribution in the development of AF was first provided in 1943 by Wolff,5 who documented transmission of AF in a family with an autosomal dominant pattern of inheritance. Since that time, large epidemiologic studies have documented evidence of heritability in AF. We and others have confirmed that a family history of AF is present in one-third of patients with lone AF indicating that familial AF is more common than previously recognized.6–8 To date linkage analysis, candidate gene approach and association studies have been the preferred methods to study heritability of AF. However, less than 10% of inherited variance can be explained by all AF loci.
Linkage analysis
Several genes and loci have now been described for Mendelian forms of lone AF (Table 1). Although the first AF locus was reported in 1997, the gene responsible for the arrhythmia has still not yet been identified.9 Four other AF loci have been subsequently been identified.10–13 We recently identified a novel AF locus on chromosome 5p15 and established an abnormally prolonged P-wave determined as an ‘endophenotype’ for AF.14 In 2003, the first gene (KCNQ1) responsible for an autosomal dominant form of AF was identified.15 This work was followed by subsequent studies that implicated numerous other genes encoding ion channels in the pathogenesis of AF (Table 1). Although isolated families with Mendelian AF have been described, linkage analysis has facilitated the identification of individual mutations in several familial AF kindreds. We recently identified a novel heterozygous frameshift mutation in the natriuretic peptide precursor A (NPPA) gene that encodes atrial natriuretic peptide (ANP).16 These diverse proteins are thought to predispose to AF through different electrical mechanisms, a notion that emphasizes the degree of heterogeneity that governs initiation and maintenance of AF.
Table 1.
Genes and loci implicated in familial AF.
| Chromosome | Culprit Gene |
Functional Effect | Inheritance | Reference |
|---|---|---|---|---|
| 11p15.5 | KCNQ1 | Enhanced slow component of the delayed rectifier potassium current (Iks) | AD | 15 |
| 21q22.1 | KCNE2 | Enhanced KCNQ1-KCNE2 potassium current | AD | 26 |
| 17q23.1 | KCNJ2 | Enhanced inward rectifier current (IK1) | AD | 27 |
| 12p13 | KCNA5 | Decreased ultrarapid component of the delayed rectifier potassium current (IKur) | AD | 28 |
| 3p22.2 | SCN5A | Hyperpolarizing/depolarizing shift in Nav1.5 inactivation | AD | 24, 29, 30 |
| 1p35-p36 | NPPA | Increased circulating levels of mutant atrial natriuretic peptide | AD | 16 |
| 5p13 | NUP155 | Affects transport of hsp70 | AR | 31 |
| 1q21 | GJA5 | Decreased gap junction conductance | Somatic mutation | 18 |
| 14q11 | MYH6 | Unknown | AD | 32 |
| Genetic loci | ||||
| 10q22-24 | Unknown | Unknown; overlaps with locus for dilated cardiomyopathy | AD | 9 |
| 6q14-16 | Unknown | Unknown; overlaps with locus for dilated cardiomyopathy | AD | 10 |
| 10p11-q21 | Unknown | Unknown | AD | 13 |
| 5p15 | Unknown | Unknown; associated with ↑ P-wave duration | AD | 14 |
AD, autosomal dominant; AF, atrial fibrillation; AR, autosomal recessive; hsp70, heat hock protein.
Candidate gene studies
A candidate gene can be any gene that is hypothesized to cause a disease. Based on the work relating KCNQ1 to AF, investigators have considered other potassium (and ion) channels as potential candidate genes and screened for mutations in these genes in cohorts of subjects with AF. The genes that encode connexins, gap-junction proteins that mediate the spread of action potentials (APs) between cardiac myocytes, have also been examined as potential candidates for AF. Prior work has shown that mice with null alleles of GJA5, the gene for connexin-40, exhibit atrial reentrant arrhythmias.17 From this work, Gollob et al.18 considered this gene as a potential candidate in individuals with lone AF who underwent pulmonary vein isolation surgery. An analysis of DNA isolated from their cardiac tissue showed that 4 of the 15 subjects had mutations in GJA5 that markedly interfered with the electrical coupling between cells. In three of the patients, DNA isolated from their lymphocytes lacked the same mutation in GJA5 indicating that the connexin-40 mutation had been acquired after fertilization or was a somatic mutation. One of the four individuals carried the mutation in both cardiac tissue and lymphocytes consistent with a germ line rather than somatic mutation.
Association studies
Most patients with AF have one or more identifiable risk factors, but many patients with these same risk factors do not develop AF. Studies comparing cases of non-familial AF to age-related and gender-matched controls (association studies) have provided some insight into the genetic basis of acquired AF. These studies have typically tested a small number of variants and have been directed at candidate genes previously believed to be involved in AF. Examples include genes encoding the renin-angiotensin-aldosterone system (RAAS), ion channels, neurohormonal and signaling pathways. Major limitations of such studies are relatively small sample sizes and a lack of replication in distinct populations, as well as phenotypic and genetic heterogeneity.
In recent years, genome-wide association (GWA) studies have been made possible by advances in genotyping technology that allow investigators to assay hundreds of thousands of SNPs spread over the entire human genome. Recently, a locus on the long arm of chromosome 4 (4q25) was identified in a GWA study to have a highly significant association with AF with a relative risk ranging from 1.39 to 1.72, depending on the population tested.19 Although the mechanism for this observed association remains unknown, the locus is adjacent to the paired-like homeodomain transcription factor 2 (PITX2) gene which is critical for cardiac development.20 More recently, the CHARGE-AF/AFGen Consortium21 and Decode investigators22 identified SNPs within the transcription factor ZFXH3 that associated with AF. A novel locus associated with lone AF at 1q21 was identified in 2010.23
Challenges in AF genetics
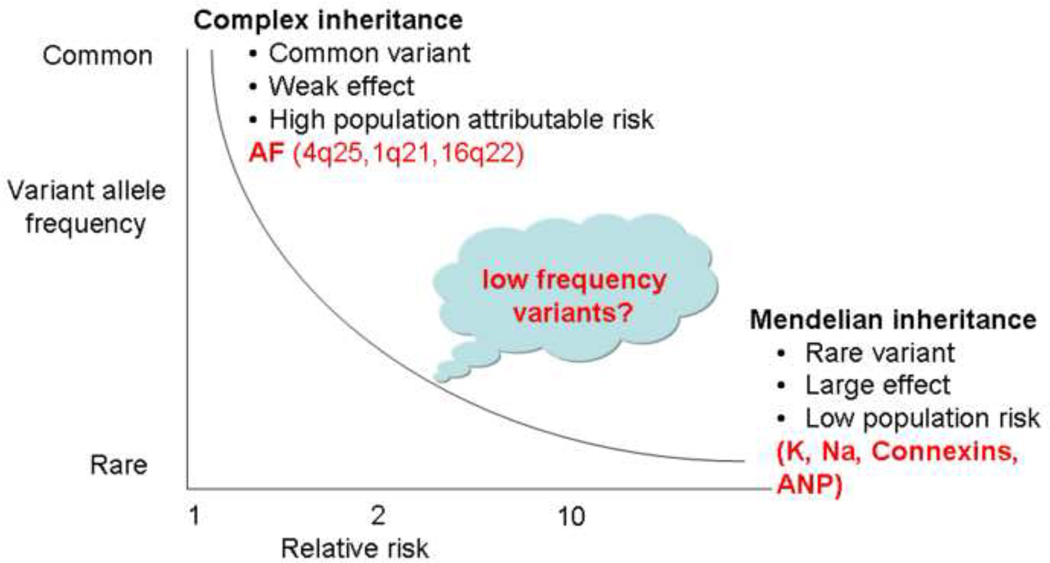
It is now widely accepted that up to one-third of all lone AF patients have a genetic basis for their disease and that a large fraction of AF cases are multifactorial. However, the common variants identified by GWA studies and the AF genes discovered by linkage analysis and candidate gene approaches, explain only a small fraction of the genetic heritability of the arrhythmia. The ‘missing’ heritability of AF may be explained by low frequency variants with an intermediate penetrance (Figure 1). An important role for rare variants in inherited multifactorial susceptibility to AF was first suggested by the effects of rare missense variants in SCN5A, the α-subunit of the cardiac sodium channel.24 This raises the possibility that rare independent variants with intermediate penetrance may account for a large fraction of the risk of developing AF.
Figure 1. Missing link in atrial fibrillation (AF) heritability.
Rare independent variants with intermediate effects may explain the missing heritability of AF.
A major focus in genetics recently has been identification of disease pathways, genes, and mutations in rare monogenic diseases. Discovery in these settings, in turn, provides the starting point for testing the role of pathway variants in commoner forms of disease. Ji et al.25 resequenced the coding regions of SLC12A3, SLC12A1, and KCNJ1 (each associated with a rare disease causing abnormal blood pressure via altered renal salt handling) in 2492 subjects from the Framingham Heart Study; they identified 92 non-synonymous variants and inferred a relationship between blood pressure and rare variant status. While most AF is secondary to other conditions, 10–30% of patients have lone AF. Although GWA studies have uncovered common genetic variants associated with AF, the overall risk for the arrhythmia with these variants is small (OR 1.1–2.5) and explains less than 10% of the heritability in lone AF (Figure 1).
Until recently, it has been difficult to systematically identify rare variants. However, recent advances in next generation sequencing, has now made it feasible to rapidly sequence whole exomes or protein-coding regions of the genome. The combination of exome sequencing with bioinformatic selection now permits the systematic evaluation of rare variants associated with AF. Defining the genotype-phenotype relationships in AF kindreds carrying these rare genetic variants will add greatly to our understanding of the pathogenesis and genetic risk of AF.
Acknowledgments
This work was supported by U19 HL65962, HL075266, and an AHA Established Investigator Award (0940116N).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feinberg WM, Blackshear JL, Laupacis A, Kronmal R, Hart RG. Prevalence, age distribution, and gender of patients with atrial fibrillation. Analysis and implications. Arch Intern Med. 1995;155:469–473. [PubMed] [Google Scholar]
- 2.Lloyd-Jones DM, Wang TJ, Leip EP, Larson MG, Levy D, Vasan RS, et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation. 2004;110:1042–1046. doi: 10.1161/01.CIR.0000140263.20897.42. [DOI] [PubMed] [Google Scholar]
- 3.Wolf PA, Abbott RD, Kannel WB. Atrial fibrillation as an independent risk factor for stroke: the Framingham Study. Stroke. 1991;22:983–988. doi: 10.1161/01.str.22.8.983. [DOI] [PubMed] [Google Scholar]
- 4.Miyasaka Y, Barnes ME, Gersh BJ, Cha SS, Bailey KR, Abhayaratna WP, et al. Secular trends in incidence of atrial fibrillation in Olmsted County, Minnesota, 1980 to 2000, and implications on the projections for future prevalence. Circulation. 2006;114:119–125. doi: 10.1161/CIRCULATIONAHA.105.595140. [DOI] [PubMed] [Google Scholar]
- 5.Wolff L. Familial auricular fibrillation. N Engl J Med. 1943:396–397. [Google Scholar]
- 6.Fox CS, Parise H, D'Agostino RB, Sr., Lloyd-Jones DM, Vasan RS, Wang TJ, et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA. 2004;291:2851–2855. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 7.Darbar D, Herron KJ, Ballew JD, Jahangir A, Gersh BJ, Shen WK, et al. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol. 2003;41:2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 8.Ellinor PT, Yoerger DM, Ruskin JN, Macrae CA. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;118:179–184. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 9.Brugada R, Tapscott T, Czernuszewicz GZ, Marian AJ, Iglesias A, Mont L, et al. Identification of a genetic locus for familial atrial fibrillation. N Engl J Med. 1997;336:905–911. doi: 10.1056/NEJM199703273361302. [DOI] [PubMed] [Google Scholar]
- 10.Ellinor PT, Shin JT, Moore RK, Yoerger DM, MacRae CA. Locus for atrial fibrillation maps to chromosome 6q14-16. Circulation. 2003;107:2880–2883. doi: 10.1161/01.CIR.0000077910.80718.49. [DOI] [PubMed] [Google Scholar]
- 11.Schott JJ, Probst V, Mabo P. A new locus for atrial fibrillation maps to chromosome 20q12-13. Circulation. 2004;110 (Suppl III (abstract)):1245. [Google Scholar]
- 12.Oberti C, Wang L, Li L, Dong J, Rao S, Du W, et al. Genome-Wide Linkage Scan Identifies a Novel Genetic Locus on Chromosome 5p13 for Neonatal Atrial Fibrillation Associated With Sudden Death and Variable Cardiomyopathy. Circulation. 2004;110:3753–3759. doi: 10.1161/01.CIR.0000150333.87176.C7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volders PG, Zhu Q, Timmermans C, Eurlings PM, Su X, Arens YH, et al. Mapping a novel locus for familial atrial fibrillation on chromosome 10p11-q21. Heart Rhythm. 2007;4:469–475. doi: 10.1016/j.hrthm.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 14.Darbar D, Hardy A, Haines JL, Roden DM. Prolonged signal-averaged P-wave duration as an intermediate phenotype for familial atrial fibrillation. J Am Coll Cardiol. 2008;51:1083–1089. doi: 10.1016/j.jacc.2007.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 16.Hodgson-Zingman DM, Karst ML, Zingman LV, Heublein DM, Darbar D, Herron KJ, et al. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. N Engl J Med. 2008;359:158–165. doi: 10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon AM, Goodenough DA, Paul DL. Mice lacking connexin40 have cardiac conduction abnormalities characteristic of atrioventricular block and bundle branch block. Curr Biol. 1998;8:295–298. doi: 10.1016/s0960-9822(98)70113-7. [DOI] [PubMed] [Google Scholar]
- 18.Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, Shao Q, et al. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med. 2006;354:2677–2688. doi: 10.1056/NEJMoa052800. [DOI] [PubMed] [Google Scholar]
- 19.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;488:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A. 107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;8:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, et al. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–1935. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji w, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Xia M, Jin Q, Bendahhou S, Shi J, Chen Y, et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am J Hum Genet. 2004;75:899–905. doi: 10.1086/425342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia M, Jin Q, Bendahhou S, He Y, Larroque MM, Chen Y, et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012–1019. doi: 10.1016/j.bbrc.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 28.Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–2191. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- 29.Ellinor PT, Nam EG, Shea MA, Milan DJ, Ruskin JN, MacRae CA. Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm. 2008;5:99–105. doi: 10.1016/j.hrthm.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 30.Makiyama T, Akao M, Shizuta S, Doi T, Nishiyama K, Oka Y, et al. A novel SCN5A gain-of-function mutation M1875T associated with familial atrial fibrillation. J Am Coll Cardiol. 2008;52:1326–1334. doi: 10.1016/j.jacc.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135:1017–1027. doi: 10.1016/j.cell.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 32.Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet. 43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]