Abstract
Arabidopsis ERD1 is a ClpC-like protein that sequence analysis suggests may interact with the chloroplast-localized ClpP protease to facilitate proteolysis. The mRNA encoded by the ERD1 gene has previously been shown to accumulate in response to senescence and to a variety of stresses and hormones. Here we show that the ERD1 protein, in contrast to the ERD1 mRNA, strongly declines in abundance with age, becoming undetectable in fully expanded leaves. Sequence analysis also suggests that ERD1 is chloroplast targeted, and we show in an in vitro system that the native protein is properly imported, processed, and present within the soluble fraction of the chloroplast, presumably the stroma. We show that ClpP protein, which is also present in the stroma, declines with age in parallel with ERD1. These results are consistent with the interaction of ERD1 and ClpP, but they suggest that it is unlikely that either plays a major role during senescence. Certain other chloroplast proteins decline with age coordinately with ERD1 and ClpP, suggesting that these declines are markers of an early age-mediated change that occurs within the chloroplast.
Arabidopsis ERD1 was first reported as an mRNA that rapidly accumulates in response to dehydration (Kiyosue et al., 1993). It was later reported, under the name SAG15, as an mRNA that accumulates during natural senescence (Lohman et al., 1994; Nakashima et al., 1997; Weaver et al., 1998). ERD1 shows strong sequence similarity to the Clp ATPases, a widely distributed family of genes characterized by two nonhomologous nucleotide-binding domains separated by a spacer region (Squires and Squires, 1992). These genes have been divided into three categories, based initially on the length of the spacer. ClpAs have short spacers and are found in bacteria. ClpBs have longer spacers and are found in bacteria, animals, plants, and fungi. ClpCs have spacers of intermediate length and are found in plants and some bacteria. Plant ClpCs have putative N-terminal transit peptides that suggest chloroplast localization (for review, see Squires and Squires, 1992).
ClpBs are heat-shock-induced chaperonins that have been shown in yeast and Escherichia coli to be necessary for survival at high temperatures (Parsell et al., 1991; Squires et al., 1991; Squires and Squires, 1992). ClpAs function as ATP-dependent regulatory subunits of the ClpP protease (Squires et al., 1991; Squires and Squires, 1992) and also have been shown to function independently as chaperonins in vitro (Wickner et al., 1994; Wawrzynow et al., 1996). The role of the ClpCs is less clear, although recent work suggests that they also may function both independently as chaperonins and as subunits of a Clp protease. An Arabidopsis ClpC (AtClpC) has been shown to interact in vitro with bacterial ClpP to facilitate ATP-dependent proteolysis (Shanklin et al., 1995), and barley ClpC and ClpP coimmunoprecipitate from chloroplast extracts, suggesting that the two may interact (Desimone et al., 1997). It also has been found that a pea (Pisum sativum) ClpC (originally called ClpA) is associated with the chloroplast-translocation apparatus (Akita et al., 1997; Nielsen et al., 1997) and may function as a molecular chaperone involved in the transport of precursor proteins into plastids.
The final stage of leaf development is often characterized by the mobilization of nutrients stored within the leaf to other parts of the plant, and this mobilization and the changes that accompany it have been referred to as the “senescence syndrome” (Bleecker and Patterson, 1997), an active process under nuclear control with a large number of genes with mRNAs, proteins, or activities that increase coordinately with it (for recent reviews, see Bleecker and Patterson, 1997; Buchanan-Wollaston, 1997; Nam, 1997; Weaver et al., 1997). An early target of the senescence process is the chloroplast, where approximately one-half of the protein in leaves is found. It is not known which proteases are involved in mobilizing the nitrogen contained within chloroplast proteins, although ATP-dependent proteolysis has been observed within chloroplasts (Liu and Jagendorf, 1984; Malek et al., 1984; Lindahl et al., 1995; Halperin and Adam, 1996). The ClpP proteolytic subunit is encoded by and present within the chloroplast, and certain nuclear-encoded ClpCs are known to be chloroplast localized (Shanklin et al., 1995). Therefore, the Clp system is a candidate for the mediation of chloroplast protein degradation during senescence. If this is the case, then the levels of its components might be expected to increase during senescence.
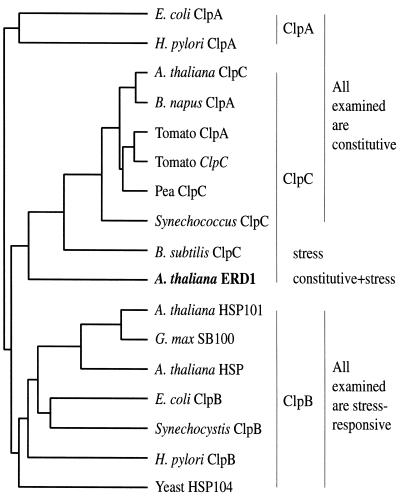
The protein or mRNA levels of ClpP and some ClpC family members have been examined in senescing leaves of several species (although rarely have mRNA and protein in the same species been studied), and none were found to be induced (Shanklin et al., 1995; Crafts-Brandner et al., 1996, 1998; Humbeck and Krupinska, 1996). An exception is the Arabidopsis ClpC family member ERD1, which, as discussed above, encodes an mRNA that accumulates during senescence (Kiyosue et al., 1993; Lohman et al., 1994; Nakashima et al., 1997; Weaver et al., 1998). The senescence-associated increase in ERD1 mRNA levels is intriguing because ERD1 is diverged from both the other Arabidopsis ClpC family member (AtClpC) and the other known plant ClpCs (Vierling, 1997; Fig. 5), which suggests that it might play a distinct role within the plant. One possibility is that ERD1 mediates Clp-dependent proteolysis during senescence (Nakashima et al., 1997; Weaver et al., 1997, 1998).
Figure 5.
Dendrogram of selected Clp genes, including all of the sequenced full-length, higher-plant ClpCs, produced using the PileUp program from the Genetics Computer Group (Madison, WI) package. The class to which each sequence appears to belong is indicated on the right of the dendrogram (although this sometimes conflicts with the name the sequence was given), and the expression characteristics of the group are indicated to the right of that (expression patterns have not necessarily been determined for all of the individual genes). Note that ERD1, although clearly neither a ClpA nor a ClpB, appears distinct from the other plant and photosynthetic bacterial ClpCs. Accession numbers are as follows: E. coli ClpA, spP15716; Helicobacter pylori ClpA, giAE000525; Arabidopsis ClpC, giAF022909; Brassica napus ClpA, spP46523; tomato ClpA, spP31541; tomato ClpC, spP31541; pea ClpC, spP35100; Synechococcus ClpC, gi755162; B. subtilis ClpC, spP37571; Arabidopsis ERD1, spP42762; Arabidopsis HSP101, spP42730; Glycine max SB100, giL35272; Arabidopsis HSP, gnlZ97336; E. coli ClpB, spP03815; Synechocystis ClpB, gnlD90915; H. pylori ClpB, spP71404; and yeast HSP104, spP31539.
We show here in Arabidopsis that the ERD1 protein is properly imported, processed, and present within the soluble fraction of the chloroplast in an in vitro chloroplast import system, indicating that it is present within the chloroplast in the plant and is therefore potentially able to interact with ClpP. Although ERD1 mRNA levels increase during senescence, ERD1 protein levels actually decline abruptly, in parallel with ClpP, as leaves senesce.
MATERIALS AND METHODS
Plant Materials and Treatments
Arabidopsis ecotype Landsberg erecta (Ler) was originally obtained from the Arabidopsis Stock Center at The Ohio State University (Columbus). Plants were grown on germination mixture (Fafard, Agawam, MA) under cool-white fluorescent light (120 μmol m−2 s−1) in growth chambers. In most experiments, plants were grown under continuous light. In the experiments shown in Figures 1 and 3B, plants were grown under 16 h of light. Under these conditions, plants flowered after forming approximately eight rosette leaves (cotyledons were not counted). All plants used in a given experiment were taken from a single synchronously growing population and harvested in late afternoon. Only identically aged leaves were pooled.
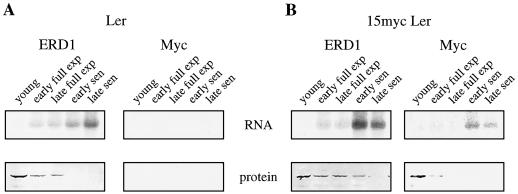
Figure 1.
ERD1 protein is down-regulated during senescence, whereas its mRNA is up-regulated. A, RNA and protein blots derived from the fifth true leaf of wild-type Arabidopsis (ecotype Ler) probed with ERD1 polyclonal antibodies or labeled cDNA (left) or with c-myc monoclonal antibodies or labeled DNA (right). B, RNA and protein blots derived from the fifth true leaf of Arabidopsis 15myc Ler, which is Ler transformed with a c-myc-tagged ERD1 genomic clone, and probed as in A. Young leaves averaged 15 mm and were harvested 16 d after germination; early fully expanded (exp) leaves averaged 27 mm and were harvested 20 d after germination; late fully expanded leaves averaged 30 mm and were harvested 23 d after germination; early senescent (sen) leaves averaged 15% yellow and were harvested 29 d after germination; late senescent leaves averaged 70% yellow and were harvested 33 d after germination. RNA and protein derived from equal volumes of leaf tissue were loaded.
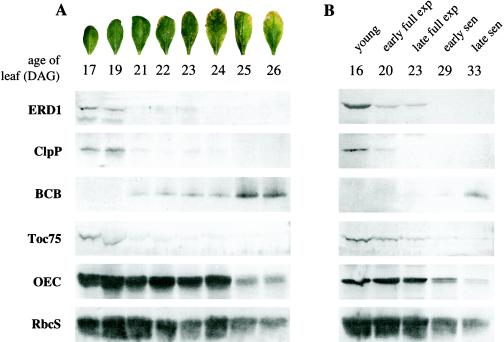
Figure 3.
Protein blots derived from two developmental series. A, Time course emphasizing the period between early full expansion and middle senescence. Arabidopsis plants from a synchronous population grown under continuous light were harvested over a period of several days, and protein from the sixth true leaf was prepared. Protein blots were examined with the indicated antibodies. The photographs indicate the developmental stages of the leaves at each time. DAG, Days after germination. B, The broader time course shown in Figure 1A, left, examined with the indicated antibodies. Plants were grown under a 16-h photoperiod, and protein and RNA from the sixth true leaf was prepared. BCB is a known senescence-associated gene, ClpP is the proteolytic subunit of the Clp protease, and Toc75 is a component of the chloroplast import apparatus. Protein derived from equal volumes of leaf tissue was loaded. exp, Expansion; sen, Senescence.
Protein Extraction and Immunoblotting
Leaf extracts were prepared by grinding tissue under liquid N2 and adding lysis buffer (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 100 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 0.1% Triton X-100, 0.7% 2-mercaptoethanol, and 1 mm PMSF); equal volumes of lysis buffer and frozen, ground tissue were used. Samples were vortexed and centrifuged, and the supernatant was added to an equal volume of 2× loading buffer (125 mm Tris-HCl, pH 7.5, 1% 2-mercaptoethanol, 4% SDS, 20% glycerol, and 0.01% bromphenol blue). Equal volumes of each sample (representing protein derived from equal volumes of leaf tissue) were electrophoresed on SDS-PAGE gels and electroblotted onto PVDF membranes (Bio-Rad). Immunodetection was performed as described by Shanklin et al. (1987).
RNA Extraction and Blotting
Total RNA was extracted (RNA Isolator, Genosys Biotechnologies, The Woodlands, TX). RNA was size fractionated by electrophoresis on 1% formaldehyde-agarose gels and transferred onto nylon membranes by capillary blotting. RNA derived from equal volumes of ground, frozen leaf tissue was loaded in each lane. Probes were derived from the ERD1 cDNA (Lohman et al., 1994) and the c-myc cassette of the plasmid pJR1265 (R. Hampton and J. Rine, personal communication) and 32P labeled by random priming (Prime-a-Gene kit, Promega). Hybridization was at 65°C overnight in 0.25 m NaH2PO4, pH 7.4, 7% SDS, 1% casein, and 1 mm EDTA, and membranes were washed twice for 45 min each in 0.04 m NaH2PO4, pH 7.2, 1% SDS, and 1 mm EDTA. Probe hybridization was visualized with a phosphor imager using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Antibodies
A 1.5-kb ERD1 XhoI cDNA fragment encoding a 507-amino acid residue peptide corresponding to the Arg-368 to Ile-675 region of the native ERD1 (accession no. D17582) was cloned into pET28a (Novagen, Madison, WI). The resulting plasmid, pSAG15exXho, was transformed into Escherichia coli strain BL21(DE3). Isopropylthio-β-galactoside induction resulted in the production of an insoluble protein. Induced cells were lysed by sonication and centrifuged for 20 min at 2000g, and the insoluble fraction was washed with binding buffer (5 mm imidazole, 4.5 m NaCl, and 20 mm Tris-HCl, pH 7.9). This protein was resuspended in binding buffer plus 6 m guanidine and affinity purified on a column (HisBind, Novagen) under denaturing conditions according to the manufacturer's instructions. The purified protein was dialyzed several times against 60 mm Tris-HCl, pH 6.8, and 1% SDS and used to inoculate rabbits to raise polyclonal antibodies. BCB antibody was prepared similarly (L.M. Weaver and R.M. Amasino, unpublished results). c-myc antibody was obtained from Oncogene Science (Manhasset, NY; anti-c-myc monoclonal 9E10.2, IgG1). ClpP antibody was as described by Shanklin et al. (1995), OEC was as described by Camm et al. (1987), Toc75 was as described by Nielsen et al. (1997), and RbcS was as described by Akita et al. (1997).
Construction of the myc-Tagged ERD1 Clone and Arabidopsis Transformation
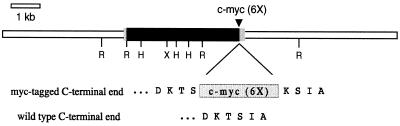
The genomic sequences used in the construction were obtained from a 13.7-kb clone derived from an Arabidopsis ecotype Columbia λGEM11 library (a gift from Ron Davis, Stanford University). The c-myc epitope cassette used was the 6-myc repeat found in pJR1265 (R. Hampton and J. Rine, personal communication). Primers were designed to PCR amplify the c-myc cassette while introducing ClaI restriction sights into its 5′ and 3′ ends. A ClaI site is also present at the 3′ end of the ERD1-coding region, at a position corresponding to amino acid 944. A 3.4-kb genomic EcoRI fragment containing this ClaI site was subcloned into the vector pGEM11Zf, and the amplified c-myc cassette was inserted into the ClaI site to yield the plasmid pSAG15genRH2bmyc. The region containing the c-myc cassette and insertion site was sequenced and found to be free of errors. The entire 13.7-kb genomic clone, with the myc-tagged EcoRI fragment from pSAG15genRH2bmyc substituted for the corresponding wild-type fragment, was cloned in a series of steps into the XbaI and SalI sites of the binary transformation vector pPZP111, to give the plasmid pSAG15myc. pSAG15myc was transformed into Arabidopsis ecotypes Ler and Columbia using vacuum infiltration (Bechtold et al., 1993; Bent and Clough, 1998) and Agrobacterium tumefaciens strain LBA4404. Two kanamycin-resistant Columbia lines and one Ler line were examined and showed identical ERD1 and myc protein-expression patterns. The Ler line was used in the work described.
Chloroplast Binding, Import, and Fractionation of ERD1
Chloroplasts were isolated from 8- to 12-d-old pea (Pisum sativum var Little Marvel) seedlings as described previously (Bruce et al., 1994). After isolation, chloroplasts were resuspended to 1 mg chlorophyll/mL import buffer (50 mm Hepes-KOH, pH 8.0, and 330 mm sorbitol). Synthesis of precursor to ERD1 was performed using a wheat-germ extract (Bruce et al., 1994) with [35S]Met (DuPont/NEN) for labeling. The template was the ERD1 cDNA clone pSAG15full, which consists of the complete ERD1-coding region, the 3′-untranslated region, and a poly(A+) tail cloned into pBluescript SK; it lacks the 5′-untranslated region. Import of ERD1 was performed as follows: Wheat-germ-translated ERD1 (approximately 500,000 dpm/150 μL import reaction) was added to pea chloroplasts in the presence of 4 mm Mg-ATP and incubated at room temperature. Aliquots were removed at given times, and import was terminated by reisolating intact chloroplasts by sedimentation through a 40% Percoll cushion. Fractions were resuspended in sample buffer and subsequently analyzed by SDS-PAGE and fluorography.
Binding of ERD1 was performed by incubating wheat-germ-translated ERD1 with chloroplasts for 30 min in the dark at 4°C under low-ATP conditions (approximately 75 μm). The binding reaction was divided into two equal fractions. One sample was treated with thermolysin and the other sample was not. Protease digestion was allowed to continue for an additional 30 min on ice in the dark. Proteolysis was terminated by adding EDTA to a final concentration of 5 mm, and intact chloroplasts were reisolated by sedimentation through a 40% Percoll cushion with 5 mm EDTA present. Fractions were resuspended in sample buffer and subsequently analyzed by SDS-PAGE and fluorography.
Fractionation of imported proteins was performed as described previously (Bruce et al., 1994). ERD1 was imported as described above, and intact chloroplasts were recovered by sedimentation through a 40% Percoll cushion. Intact chloroplasts were lysed hypotonically in 200 μL of lysis buffer (25 mm Hepes-KOH, pH 8.0, and 4 mm MgCl2). The lysis reaction was incubated on ice for 15 min in the dark, and the supernatant and crude membrane fractions were separated by centrifugation (30 min at 100,000g; model RP 100-ATP, Sorvall). Isolated chloroplastic crude membrane and supernatant were resuspended in sample buffer and subsequently analyzed by SDS-PAGE and fluorography.
RESULTS
ERD1 Protein Levels Decline during Leaf Senescence, in Contrast to ERD1 mRNA Levels
Polyclonal antibodies raised to an E. coli-expressed ERD1 peptide detected an approximately 100-kD protein in green but not senescent leaves (Fig. 1A, bottom left). Total RNA isolated from the same tissue as the protein samples, when blotted and probed with labeled ERD1 cDNA, confirmed previous observations that ERD1 mRNA levels increase during senescence (Fig. 1A, top left). This suggested that the ERD1 protein, in contrast to the ERD1 mRNA, is not induced during senescence. One possible explanation for this unexpected result is that the polyclonal antibody recognized not ERD1 but a related Clp family member or some other cross-reacting species. To examine this possibility, an epitope-tagged ERD1 was constructed. A 13.7-kb genomic clone containing the entire ERD1-coding region and 3 to 4 kb of both upstream and downstream sequences was engineered to include six repeats of a c-myc epitope tag at the C-terminal portion of the ERD1 protein (Fig. 2). This clone was then transformed into Arabidopsis, and RNA and protein blots were prepared from transgenic lines.
Figure 2.
Construction of the c-myc-tagged ERD1 genomic clone. The 13.7-kb ERD1 genomic clone transformed into plants is diagrammed. R, H, and X, Positions of EcoRI, HindIII, and XhoI sites, respectively. The box represents the ERD1 primary transcript. The darkly shaded portion of the box indicates the area containing the coding region (introns are not distinguished). The lightly shaded portions indicate the 5′- and 3′-untranslated regions. Below the diagram (and not to scale), the C-terminal ends of the myc-tagged and wild-type proteins are shown.
In all cases, the protein detected by both the ERD1 and c-myc antibodies was observed to decline during senescence, whereas RNA detected with both ERD1 and c-myc DNA probes was observed to increase (Fig. 1B). When protein and RNA blots derived from nontransformed plants were examined with c-myc antibody or labeled c-myc DNA, no signal was detected, confirming that these reagents detect only the myc-tagged ERD1 mRNA and protein (Fig. 1A, right). This indicates that the anti-ERD1 polyclonal antibodies recognize the ERD1 protein, which declines during senescence. All further ERD1 antibody work was done using the polyclonal antibodies and wild-type plants.
To further characterize the expression pattern of the ERD1 protein in leaves, a more detailed developmental time course, emphasizing the period between early full expansion and early senescence, was examined. Detectable steady-state levels of ERD1 were again present only in expanding and young fully expanded leaves, and ERD1 became undetectable between full expansion and early senescence (Fig. 3A). As a positive control for senescence, the developmental series was examined with antibody to BCB (Van Gysel et al., 1993), also known as SAG14, a gene known to be up-regulated during senescence at both the mRNA (Lohman et al., 1994; Weaver et al., 1998) and protein levels (L.M. Weaver and R.M. Amasino, unpublished results). BCB became detectable only in late fully expanded/early senescent leaves, soon after ERD1 became undetectable. ERD1 is chloroplast localized (see below), and the series was also examined with antibodies that recognize four other chloroplast-localized proteins as controls for senescence and for the state of the chloroplast (ClpP, Toc75, OEC, and RbcS; Fig. 3). These proteins declined during senescence, as expected, two with kinetics similar to ERD1 and two more slowly (see Discussion). One of the proteins that declined in parallel with ERD1, ClpP, is the protease with which ERD1 has been hypothesized to interact.
Developmental conditions can affect the progression of the senescence syndrome. Shorter photoperiods in particular can delay the onset of senescence (Noodén et al., 1996). The plants used for the experiments shown in Figure 3A were grown in continuous light. To determine whether ERD1 was similarly regulated under different environmental conditions, with respect to both development and the other genes tested, a second developmental time course derived from plants grown in 16-h photoperiods was also examined (Fig. 3B). In both experiments, the different proteins were coordinately regulated with respect to each other, although the absolute developmental stages at which they declined or increased varied (e.g. in both cases, ERD1 became undetectable as BCB became detectable, but it first became undetectable in fully expanded leaves in the experiment shown in Fig. 3A and in early senescent leaves in the experiment shown in Fig. 3B). This suggests that, although environmental factors can influence the timing of the senescence program relative to leaf development, the program itself remains largely constant.
ERD1 Is Localized within the Soluble Fraction of the Chloroplast
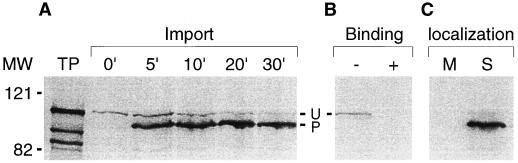
Because ERD1 appears to have an N-terminal transit peptide and is similar in sequence to the chloroplast-localized AtClpC, we wanted to determine whether the ERD1 protein is also imported into the plastid. An expression construct was made using the full ERD1-coding sequence, and radiolabeled ERD1 protein was produced in a wheat-germ transcription/translation system. This protein was then introduced into an in vitro plastid-uptake system (Bruce et al., 1994). In the presence of ATP, ERD1 was imported into the chloroplast and processed in a manner consistent with the N-terminal transit peptide being removed (Fig. 4A). Under low-ATP conditions, ERD1 also bound the chloroplast but was not imported or processed (Fig. 4B). These results are consistent with the behavior of known chloroplast-imported proteins in this system. After import, chloroplasts were lysed and crude membrane and soluble fractions prepared, and ERD1 was observed to be present within the soluble fraction (Fig. 4C). These results indicate that in the cell ERD1 is present within the chloroplast and likely within the stroma as well.
Figure 4.
ERD1 import, binding, and fractionation using pea chloroplasts. A, Import of ERD1 into pea chloroplasts was performed as described in Methods. Aliquots were removed at given times, and import was terminated by reisolating intact chloroplasts by sedimentation through a 40% Percoll cushion. Fractions were resuspended in sample buffer and subsequently analyzed by SDS-PAGE and fluorography. B, Binding of ERD1 to pea chloroplasts under low-ATP conditions. The binding reaction was equally divided into two fractions. One sample was treated with thermolysin (+) and the other sample was not (−). Protease digestion was allowed to continue for an additional 30 min on ice in the dark. Proteolysis was terminated by adding EDTA to a final concentration of 5 mm, and intact chloroplasts were reisolated by sedimentation through a 40% Percoll cushion with 5 mm EDTA present. C, ERD1 was imported into pea chloroplasts and fractionated as described in Methods. All fractions were analyzed by SDS-PAGE and fluorography. TP, Ten percent of translation product added to the import reaction; M, crude membrane pellet; S, supernatant fraction; U, unprocessed form of ERD1; P, processed form of ERD1; MW, Mr standards.
DISCUSSION
Recent studies indicate that many, and probably most, genes that are induced during age-associated senescence (i.e. senescence occurring in leaves of nonstressed plants progressing through normal stages of development) are also induced by one or more stresses or stress-related hormones (Azumi and Watanabe, 1991; Hsieh et al., 1995; Stirpe et al., 1996; Buchanan-Wollaston and Ainsworth, 1997; Park et al., 1998; Weaver et al., 1998). It seems likely that in at least some cases these genes in fact function directly in both senescence and stress responses, and one role they may be playing is to protect the cell from damage that may be common to both. It is important to remember in this context that the senescence syndrome is an active, ordered developmental process, not simply passive cell death, so cells undergoing senescence must remain sufficiently healthy to allow the program to progress. The induction of stress-response systems during senescence may thus function to help maintain the necessary cellular integrity. Most of the work describing senescence- and stress-associated gene induction has been done at the level of steady-state mRNA, under the assumption that mRNA levels will reflect protein levels and ultimately biological function. At the mRNA level, ERD1 is one such senescence-associated gene that is also induced by a variety of stresses and hormones (Kiyosue et al., 1993; Nakashima et al., 1997; Weaver et al., 1998).
ERD1 is a particularly interesting example of the class of senescence- and stress-induced mRNAs because its sequence suggests that it is localized in the chloroplast, which is a major target of the senescence syndrome. As a member of the ClpC family, its sequence also suggests what it might do there, i.e. mediate proteolysis via the Clp system. The possibility has been proposed that ERD1 might interact with the plastid-localized ClpP to promote senescence-induced chloroplast protein degradation or stress-necessitated chloroplast restoration (Kiyosue et al., 1993; Nakashima et al., 1997; Weaver et al., 1997). It has been reported that the N-terminal region of ERD1, when fused to GFP (green fluorescent protein), targets it to the chloroplast (Nakashima et al., 1997). Here we demonstrate in an in vitro chloroplast import system that the native protein is imported, processed, and localized within the nonmembranous fraction of the chloroplast, presumably the stroma. ClpP is also present within the stroma (Shanklin et al., 1995), and the two are thus potentially able to interact.
We also observed similar developmental regulation of the two proteins, which likewise is consistent with their interaction. Our data, however, do not show ERD1 to be in the senescing chloroplast; ERD1 protein, in contrast to ERD1 mRNA, is present at high levels in expanding and green fully expanded leaves, but it decreases below the level of detection at or before the onset of visible senescence. ERD1 mRNA is present at low levels in young leaves, and the high protein abundance at that time could be explained if the mRNA present is translated very actively, if the protein produced is very stable, or both. In senescing leaves, in which mRNA levels are high but protein levels are below detection, translation may not occur, and any protein that is produced may be very unstable (perhaps because another factor, such as ClpP, is not present).
This disparity between ERD1 protein and mRNA levels may indicate that ERD1 does not play a role in senescence. If so, there are several possible explanations for the increase of ERD1 mRNA during senescence. One is that it is an evolutionary relic of some ancestral gene that was involved in the senescence syndrome. Another is that ERD1 is functionally a stress-response gene (in addition to any constitutive functions it might have) and senescence indirectly activates its transcription by stressing the cell. The other known plant ClpCs (Shanklin et al., 1995) and photosynthetic bacterial ClpCs (Clarke and Eriksson, 1996) are constitutively present and are not stress responsive. The ClpC of the nonphotosynthetic bacterium Bacillus subtilis, in contrast, is known to accumulate as part of a general stress response (Kruger et al., 1994). Photosynthesis exposes an organism to oxidative stress, and it has been proposed that in photosynthetic organisms ClpC may have evolved away from stress responsiveness and toward constitutive expression to defend against these constitutive stresses (Clarke, 1997). As discussed above, ERD1 mRNA, unlike the other ClpC mRNAs that have been examined, is induced by a variety of stresses and stress-related hormones.
It is intriguing that the ERD1 sequence is as similar to the sequence of the stress-responsive B. subtilis ClpC as it is to that of the stress-insensitive Arabidopsis ClpC (approximately 48% identity to both), whereas all other known plant ClpCs are quite similar to each other (e.g. 88% identity between Arabidopsis and pea ClpC; Fig. 5). In fact, it was recently suggested based on sequence alone that ERD1 should be described not as a ClpC but as the founding member of a ClpD family (Vierling, 1997). It seems likely that in the future related genes will be identified in other plants, making the existence of a separate ERD1/ClpD subfamily more evident.
Finally, it is also possible that ERD1 mRNA accumulation during senescence has little significance at all, whether evolutionary or stress related. All activity that occurs during senescence need not be functionally related to it, and it has been pointed out that the “molecular ramblings of cells going senile” might not be so easy to distinguish from more biologically relevant processes (Bleecker, 1998). It should be noted that at present very few senescence-associated genes have been examined at the level of protein expression, so it cannot be predicted just how unusual this disparity actually is between protein and mRNA levels.
A second model to explain the disparity between ERD1 protein and mRNA levels is that the ERD1 protein is functionally involved in senescence but is highly unstable in senescent tissue. This would presuppose that the protein is biologically active at low levels and that in senescent tissue high levels of mRNA are required to maintain even this low level of protein. If ERD1 stimulates general proteolysis in senescing chloroplasts, it is perhaps not unreasonable that it would also be a target of the process, and there is evidence that E. coli ClpA mediates its own degradation (Gottesman et al., 1990; Maurizi et al., 1990). Our results do not allow us to distinguish between these two models.
ERD1 is present within the chloroplast, which is an early target of senescence-induced degradation, so it is possible that the developmental decline of ERD1 simply parallels a general decline in chloroplasts. ERD1 protein levels, however, decline well before the onset of leaf yellowing (Fig. 3), i.e. before changes in the chloroplast are apparent (Thomas, 1977). To explore the significance of the timing of ERD1 decline, we examined the levels of four other chloroplast proteins that have different functions and are present in various compartments. Toc75 is part of the chloroplast import apparatus and is present within the outer chloroplast membrane (Nielsen et al., 1997). ClpP is the proteolytic subunit of the Clp protease and is present within the stroma (Shanklin et al., 1995). RbcS is also present within the stroma (Akita et al., 1997). OEC is present within the thylakoid lumen (Camm et al., 1987). Toc75 and ClpP declined in parallel with ERD1, before the onset of visible senescence. The CAB (chlorophyll a/b-binding) protein behaved similarly (data not shown). RbcS and OEC, in contrast, both remained readily detectable throughout the course of the experiment, although by the later time they had clearly declined (Fig. 3).
Senescence is known to begin at approximately the time of full leaf expansion in many species (Mae et al., 1987; Hensel et al., 1993; Crafts-Brandner et al., 1996), and gradual declines in RbcS protein, correlated with a gradual decline in photosynthesis, have been observed in other plants (Gepstein, 1988; Jiang et al., 1993; Jiang and Rodermel, 1995). The coordinated, abrupt declines in the other molecules is more unusual, however. One interpretation of these results is that before the onset of visible senescence the decision is made to no longer maintain the chloroplast, so proteins involved in long-term maintenance, as opposed to short-term function, are not replaced. This model suggests that both ERD1 and ClpP are involved in chloroplast maintenance and that their simultaneous declines before the onset of visible senescence are markers of a more general, senescence-related change that occurs in the chloroplast at that time.
Based on its similarity to ClpAs and ClpCs, ERD1 could function as a chaperonin, repairing denatured proteins; as a protease regulatory subunit, presenting denatured proteins to the ClpP protease for degradation; or as both, repairing or degrading as interactions with other factors dictate (Squires and Squires, 1992; Wickner et al., 1994). In plants, ClpP is encoded by and localized within the chloroplast, although nuclear-encoded ClpPs with putative chloroplast transit peptides are present in the genome (Schaller and Ryan, 1995; L.M. Weaver, J.E. Froehlich, and R.M. Amasino, unpublished observations). Little is known of the role of ClpP in plants, although it appears to be essential, because it is present even within the minimal plastid of the nonphotosynthetic flowering plant Epifagus virginiana, which contains only 42 genes, 38 of which specify components of the plastid gene-expression apparatus (Wolfe et al., 1992). In bacteria, it is known that ClpP can interact with several different catalytic subunits, which are thought to determine its specificity. We have shown here that ClpP protein levels, like those of ERD1, decline during senescence. These results are consistent with studies in which ClpP mRNA was shown to decline during senescence in other species (Crafts-Brandner et al., 1996; Humbeck and Krupinska, 1996). It appears unlikely, however (although it cannot be ruled out), that either ClpP or ERD1 is involved in proteolysis during senescence. It seems most likely that ERD1 is involved in chloroplast assembly or maintenance during normal presenescent growth and, possibly, like certain of its bacterial ClpC cousins, during stresses.
ACKNOWLEDGMENTS
We thank John Shanklin for generously providing the ClpP antibody and Janet Meehl and Andrew Staehelin for providing the OEC antibodies.
Abbreviations:
- OEC
oxygen-evolving complex
- RbcS
small subunit of Rubisco
Footnotes
This work was supported through the Consortium for Plant Biotechnology Research (grant no. DE-FG02-97ER20280) and the Graduate School of the University of Wisconsin. L.M.W. was partially supported by National Institutes of Health training grant no. T32 GM07215.
LITERATURE CITED
- Akita M, Nielsen E, Keegstra K. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J Cell Biol. 1997;136:983–994. doi: 10.1083/jcb.136.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azumi Y, Watanabe A. Evidence for a senescence-associated gene induced by darkness. Plant Physiol. 1991;95:577–583. doi: 10.1104/pp.95.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thalianaplants. CR Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Bent AF, Clough SJ. Agrobacteriumgerm-line transformation: transformation of Arabidopsis without tissue culture. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual, Vol B7. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–14. [Google Scholar]
- Bleecker A, Patterson S. Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell. 1997;9:1169–1179. doi: 10.1105/tpc.9.7.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB. The evolutionary basis of leaf senescence: method to the madness? Curr Opin Plant Biol. 1998;1:73–78. doi: 10.1016/s1369-5266(98)80131-3. [DOI] [PubMed] [Google Scholar]
- Bruce BD, Perry S, Froehlich J, Keegstra K. In vitroimport of proteins into chloroplasts. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual, Vol J1. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–15. [Google Scholar]
- Buchanan-Wollaston V. The molecular biology of leaf senescence. J Exp Bot. 1997;48:181–199. [Google Scholar]
- Buchanan-Wollaston V, Ainsworth C. Leaf senescence in Brassica napus: cloning of senescence related genes by subtractive hybridisation. Plant Mol Biol. 1997;33:821–834. doi: 10.1023/a:1005774212410. [DOI] [PubMed] [Google Scholar]
- Camm EL, Green BR, Allred DR, Staehelin LA. Association of the 33 kDa extrinsic polypeptide (water-splitting) with PS II particles: immunochemical quantification of residual polypeptide after membrane extraction. Photosynth Res. 1987;13:69–80. doi: 10.1007/BF00032266. [DOI] [PubMed] [Google Scholar]
- Clarke AK. Synechococcussp. PCC 7942 ClpC. In: Gething M, editor. Guidebook to Molecular Chaperones and Protein-Folding Catalysts. Oxford, UK: Oxford University Press; 1997. pp. 247–249. [Google Scholar]
- Clarke AK, Eriksson MJ. The cyanobacterium Synechococcussp. PCC 7942 possesses a close homologue to the chloroplast ClpC protein of higher plants. Plant Mol Biol. 1996;31:721–730. doi: 10.1007/BF00019460. [DOI] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, Holzer R, Feller U. Influence of nitrogen deficiency on senescence and the amounts of RNA and proteins in wheat leaves. Physiol Plant. 1998;102:192–200. [Google Scholar]
- Crafts-Brandner SJ, Klein RR, Klein P, Holzer R, Feller U. Coordination of protein and mRNA abundances of stromal enzymes and mRNA abundances of the Clp protease subunits during senescence of Phaseolus vulgaris(L.) leaves. Planta. 1996;200:312–318. doi: 10.1007/BF00200298. [DOI] [PubMed] [Google Scholar]
- Desimone M, Weisswichert C, Wagner E, Altenfeld U, Johanningmeier U. Immunochemical studies on the Clp-protease in chloroplasts: evidence for the formation of a ClpC/P complex. Bot Acta. 1997;110:234–239. [Google Scholar]
- Gepstein S. Photosynthesis. In: Noodén L, Leopold A, editors. Senescence and Aging in Plants. San Diego, CA: Academic Press; 1988. pp. 85–109. [Google Scholar]
- Gottesman S, Clark WP, Maurizi MR. The ATP-dependent Clp protease of Escherichia coli: sequence of ClpA and identification of a Clp-specific substrate. J Biol Chem. 1990;265:7886–7893. [PubMed] [Google Scholar]
- Halperin T, Adam Z. Degradation of mistargeted OEE33 in the chloroplast stroma. Plant Mol Biol. 1996;30:925–933. doi: 10.1007/BF00020804. [DOI] [PubMed] [Google Scholar]
- Hensel LL, Grbic V, Baumgarten DA, Bleecker AB. Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell. 1993;5:553–564. doi: 10.1105/tpc.5.5.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh HM, Liu WK, Huang PC. A novel stress-inducible metallothionein-like gene from rice. Plant Mol Biol. 1995;28:381–389. doi: 10.1007/BF00020388. [DOI] [PubMed] [Google Scholar]
- Humbeck K, Krupinska K. Does the Clp protease play a role during senescence-associated protein degradation in barley leaves? J Photochem Photobiol. 1996;36:321–326. [Google Scholar]
- Jiang CZ, Rodermel SR. Regulation of photosynthesis during leaf development in RbcS antisense DNA mutants of tobacco. Plant Physiol. 1995;107:215–224. doi: 10.1104/pp.107.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CZ, Rodermel SR, Shibles RM. Photosynthesis, Rubisco activity and amount, and their regulation by transcription in senescing soybean leaves. Plant Physiol. 1993;101:105–112. doi: 10.1104/pp.101.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of cDNA for a dehydration-inducible gene that encodes a CLP A,B-like protein in Arabidopsis thalianaL. Biochem Biophys Res Commun. 1993;196:1214–1220. doi: 10.1006/bbrc.1993.2381. [DOI] [PubMed] [Google Scholar]
- Kruger E, Volker U, Hecker M. Stress induction of ClpC in Bacillus subtilisand its involvement in stress tolerance. J Bacteriol. 1994;176:3360–3367. doi: 10.1128/jb.176.11.3360-3367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Yang DH, Andersson B. Regulatory proteolysis of the major light-harvesting chlorophyll a/b protein of photosystem II by a light-induced membrane-associated enzymic system. Eur J Biochem. 1995;231:503–509. doi: 10.1111/j.1432-1033.1995.tb20725.x. [DOI] [PubMed] [Google Scholar]
- Liu X-Q, Jagendorf AT. ATP-dependent proteolysis in pea chloroplasts. FEBS Lett. 1984;166:248–252. [Google Scholar]
- Lohman K, Gan S, John M, Amasino RM. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol Plant. 1994;92:322–328. [Google Scholar]
- Mae T, Makino A, Ohira K (1987) Carbon fixation and changes with senescence in rice leaves. In W Thomson, E Nothnegel, R Huffaker, eds, Plant Senescence: Its Biochemistry and Physiology. American Society of Plant Physiologists, Rockville, MD, pp 123–131
- Malek L, Bogorad L, Ayers AR, Goldberg AL. Newly synthesized proteins are degraded by an ATP-stimulated proteolytic process in isolated pea chloroplasts. FEBS Lett. 1984;166:253–257. [Google Scholar]
- Maurizi MR, Clark WP, Katayama Y, Rudikoff S, Pumphrey J, Bowers B, Gottesman S. Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1990;265:12536–12545. [PubMed] [Google Scholar]
- Nakashima K, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. A nuclear gene, erd1, encoding a chloroplast-targeted Clp protease regulatory subunit homolog is not only induced by water stress but also developmentally up-regulated during senescence in Arabidopsis thaliana. Plant J. 1997;12:851–861. doi: 10.1046/j.1365-313x.1997.12040851.x. [DOI] [PubMed] [Google Scholar]
- Nam HG. The molecular genetic analysis of leaf senescence. Curr Opin Biotechnol. 1997;8:200–207. doi: 10.1016/s0958-1669(97)80103-6. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noodén LD, Hillsberg JW, Schneider MJ. Induction of leaf senescence in Arabidopsis thalianaby long days through a light-dosage effect. Physiol Plant. 1996;96:491–495. [Google Scholar]
- Park J-H, Oh SA, Kim YH, Woo HR, Nam HG. Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Mol Biol. 1998;37:445–454. doi: 10.1023/a:1005958300951. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Sanchez Y, Stitzel JD, Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- Schaller A, Ryan CA. Cloning of a tomato cDNA (GenBank L38581) encoding the proteolytic subunit of a Clp-like energy dependent protease (PGR 95-001) Plant Physiol. 1995;108:1341. [Google Scholar]
- Shanklin J, DeWitt ND, Flanagan JM. The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coliClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell. 1995;7:1713–1722. doi: 10.1105/tpc.7.10.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, Jabben M, Vierstra RD. Red light-induced formation of ubiquitin-phytochrome conjugates: identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci USA. 1987;84:359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C, Squires CL. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol. 1992;174:1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires CL, Pedersen S, Ross BM, Squires C. ClpB is the Escherichia coliheat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirpe F, Barbieri L, Gorini P, Valbonesi P, Bolognesi A, Polito L. Activities associated with the presence of ribosome-inactivating proteins increase in senescent and stressed leaves. FEBS Lett. 1996;382:309–312. doi: 10.1016/0014-5793(96)00188-3. [DOI] [PubMed] [Google Scholar]
- Thomas H. Ultrastructure, polypeptide composition and photochemical activity of chloroplasts during foliar senescence of a non-yellowing mutant genotype of Festuca pratensisHuds. Planta. 1977;137:53–60. doi: 10.1007/BF00394435. [DOI] [PubMed] [Google Scholar]
- Van Gysel A, Van Montagu M, Inze D. A negatively light-regulated gene from Arabidopsis thalianaencodes a protein showing high similarity to blue copper-binding proteins. Gene. 1993;136:79–85. doi: 10.1016/0378-1119(93)90450-h. [DOI] [PubMed] [Google Scholar]
- Vierling E. Chloroplast-localized Clp proteins. In: Gething M, editor. Guidebook to Molecular Chaperones and Protein-Folding Catalysts. Oxford, UK: Oxford University Press; 1997. pp. 255–258. [Google Scholar]
- Wawrzynow A, Banecki B, Zylicz M. The Clp ATPases define a novel class of molecular chaperones. Mol Microbiol. 1996;21:895–899. doi: 10.1046/j.1365-2958.1996.421404.x. [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatments. Plant Mol Biol. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Weaver LM, Himelblau E, Amasino RM (1997) Leaf senescence: gene expression and regulation. In JK Setlow, ed, Genetic Engineering, Vol 19: Principles and Methods. Plenum Press, New York, pp 215–234
- Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi MR. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]