Abstract
A cDNA encoding a lysozyme was obtained by rapid amplification of cDNA ends-polymerase chain reaction (RACE-PCR) from females of the malaria vector Anopheles dirus A (Diptera: Culicidae). The 623 bp lysozyme (AdLys c-1) cDNA encodes the 120 amino acid mature protein with a predicted molecular mass of 13.4 kDa and theoretical pI of 8.45. Six cysteine residues and a potential calcium binding motif that are present in AdLys c-1 are highly conserved relative to those of c-type lysozymes found in other insects. RT-PCR analysis of the AdLys c-1 transcript revealed its presence at high levels in the salivary glands both in larval and adult stages and in the larval caecum. dsRNA mediated gene knockdown experiments were conducted to examine the potential role of this lysozyme during Plasmodium berghei infection. Silencing of AdLys c-1 resulted in a significant reduction in the number of oocysts as compared to control dsGFP injected mosquitoes.
Keywords: Lysozymes, muramidase activity, antimicrobial peptides, RNA interference, Plasmodium, malaria
1. Introduction
Malaria persists as one of the most serious insect-transmitted diseases in the world. Anopheles dirus is one of the main vectors of human malaria in Southeast Asia, including Thailand. The relationship between the mosquito and the parasite is complex as both reproduction and morphological development of the parasites occur within this host (Beier, 1998). Distinct stages of the parasites move between several tissue locations. Following ingestion of the infected blood meal, male and female gametocytes fuse to produce a zygote that develops to a motile ookinete that can actively traverse the midgut. The ookinete comes to rest at the interface of the midgut basal epithelium and the overlying basal lamina and develops to the oocyst stage. Within the oocyst, reproduction occurs and sporozoites begin to develop. Upon release from the oocyst, sporozoites move through the hemocoel and the dorsal vessel, and are then transported into the vicinity of the salivary glands. Sporozoites enter the glands and the canal and can then be transmitted to the next host. Understanding the molecular interactions occurring between mosquitoes and parasites during this complex life cycle might provide new avenues for malaria control (Garver et al., 2009; Li et al., 2005). In particular, the ability to manipulate the mechanisms that promote or that interfere with parasite development within the mosquito host could ultimately be used to control the transmission of this deadly disease (Hoffmann et al., 1999; Nürnberger and Brunner, 2002).
Mosquitoes can sense and counter microbial invasion via the IMD or Toll pathways (Meister et al., 2005). Inactivation of these pathways through targeted gene silencing improves the frequency of successful malaria parasite development on the midgut (Garver et al., 2009). Recently, it has been demonstrated that the natural bacterial community in the mosquito midgut plays a role in regulating the IMD and Toll pathways (Dong et al., 2009). Reduction of the midgut bacteria through antibiotic treatment resulted in lowered basal immunity (IMD and Toll activity) and subsequently, an increased susceptibility to Plasmodium infections following an infected blood meal (Dong et al., 2009). This work clearly indicates that the mechanisms of innate immunity interact with the parasites to suppress infection in a normal midgut; however many of the details of this phenomenon are unknown.
We have investigated a group of antimicrobial enzymes called lysozymes (Li et al., 2005; Li and Paskewitz, 2006; Paskewitz et al., 2008; Kajla et al., 2010). These enzymes are defined by the ability to hydrolyze the 1–4 glycosidic bonds between N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) of the peptidoglycan (PGN) present in the cell walls of bacteria. Lysozymes as antibacterial defense molecules have been well characterized in vertebrates (Jolles and Jolles, 1984; Markart et al., 2004) as well as in insects (Dunn, 1986; Hultmark, 1996; Kajla et al., 2010; Powning and Davidson, 1973; Jolles et al., 1979; Hultmark et al., 1980; Abraham et al., 1995). However, lysozymes have also recently been shown to inhibit melanization of non-biological targets in Anopheles gambiae (Li and Paskewitz, 2006) and to interact directly with the oocysts of Plasmodium berghei and P. falciparum in this mosquito (Kajla et al., 2011). These observations suggest that the functional roles of mosquito lysozymes may be broader than has been previously realized.
Because of the potential for affecting malaria infections either indirectly through regulation of the gut microbiota and basal immunity or through direct interactions with the oocysts, we initiated studies on lysozymes in a second important malaria vector. In this study, a full-length lysozyme was cloned and sequenced from An. dirus. Transcript abundance of this gene was analyzed using semiquantitative RT-PCR to compare different tissues and stages of mosquitoes. Gene silencing (RNA interference) experiments were conducted to examine the role of this lysozyme in mediating P. berghei infections in An. dirus. Our results suggest that AdLys c-1 plays a role in mediating the success of malaria parasites in this important vector.
2. Materials and methods
2.1. Mosquitoes and Plasmodium berghei parasites
Anopheles dirus A was reared under laboratory conditions in an insectary in an incubator set to 70–80% relative humidity and 25–27 °C at AFRIMS (Armed Forces Research Institute of Medical Sciences, Thailand) and at the Department of Entomology, University of Wisconsin- Madison, Wisconsin, USA according to previously described protocols (Paskewitz et al., 1999; Kajla et al., 2010 and 2011) with the modification that these mosquitoes were mated artificially.
A transgenic strain of P. berghei that was engineered to express green fluorescent protein (GFP) was obtained from MR4 (http://www.mr4.org/) and passaged in Balb/c mice as described in Kajla et al. (2011).
2.2. Isolation, cloning and sequencing of full length AdLys c-1 cDNA
For the production of cDNA, 5–10 An. dirus female mosquitoes were cold anaesthetized for five minutes and immediately processed for RNA isolation. Total RNA was extracted with TRIZOL® Reagent (Invitrogen, Carlsbad, CA) according to supplied instructions. RNA concentration was determined using spectrophotometry on a Nanodrop NT1000 (Thermo Fisher Scientific, Waltham, MA). Five hundred nanograms of total RNA were used to synthesize cDNA using SuperScript™ III Reverse Transcriptase (Invitrogen). cDNA was stored at minus 20 °C until further use.
For amplification of An. dirus lysozyme, degenerate primers were designed based on conserved sequences found in mosquito c-type lysozymes. The selected primers used for amplification of the initial fragment of AdLys c-1 were as follows: Forward I 5’- GCGCGG AATTCNGAYTGGATHTGYYTN G -3’; Forward II 5’- GCGCGGAATTCNGAY TGGGTHTGYYTNG -3’; Reverse 5’ GCGCGCAAGCTTANCCRTACCANGCRT T -3’.
cDNA synthesized from the previous step was used as a template with these primer sets (Forward I and Reverse or Forward II and Reverse) in PCR with cycle condition 35 cycles of 94 °C for 1 min (denaturation), 55 °C for 1 min (annealing), and 72 °C for 1.5 min (extension). The resulting amplicons were purified and cloned into pGEM-T Easy Vector (Promega, Madison, WI) according to the manufacturer’s instructions and sequenced at the Biotechnology Center at the University of Wisconsin, Madison. The sequences were analyzed via BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm the sequence of this amplified fragment as a c-type lysozyme.
To obtain a full-length clone, the 5’ and 3’- terminal ends of lysozyme cDNA were amplified using 5’ and 3’ RLM-RACE (GeneRacer™ kit, Invitrogen), respectively. The primers for RACE were designed based on the sequence of lysozyme fragment as follows: Lys-For I 5’-TTGTTCGTCGCCGAGGTGCTGAAT-3’; Lys-Rev I 5’-CAAGTGTGCCAAGCTGATATTCAAGCGC-3’; Lys-For II 5’-TATTCC GTAGTCGGTGGACCCGTTCT-3’; Lys-Rev II 5’-GTCTAACGATTGCAAGAT AGCGTGC-3’. In the first RACE PCR, the reaction mixture (50 µl total) comprised 38.5 µl of sterile water, 5 µl of 10X PCR buffer, 1 µl of dNTPmix (10 mM of each dNTP), 3 µl of 5’or 3’ GeneRacer™ Primer (10 µM), 1 µl of gene specific Primer (10 µM), and 0.5 µl of 5 U/µl of Advance Taq polymerase. The cDNA was amplified using 35 cycles of 94 °C for 1 min (denaturation), 55 °C for 1 min (annealing), and 72 °C for 1.5 min (extension). The second PCR was prepared as the first PCR, except that GeneRacer™ 5’or 3’ Nested Primers and Lys-Rev II or Lys-For II were used for these reactions. Products were cloned and sequenced as described above.
Finally, a complete AdLys c-1 cDNA was amplified using gene-specific end primers as follows: 5’-ATTCAGTGCCTGTGATACTTGCTG-3’ and 5’-AACGAATAACGTTCATTTTGTATTG-3’. The final PCR products were separated by electrophoresis on a 1.0% agarose gel and an amplified product of about 623 bp was detected, cloned and sequenced as described above. The sequence was analyzed by using BLAST and DNASTAR software (Lasergene, Madison, WI). The sequence of the full length clone was submitted to GenBank and assigned accession number EU622903.
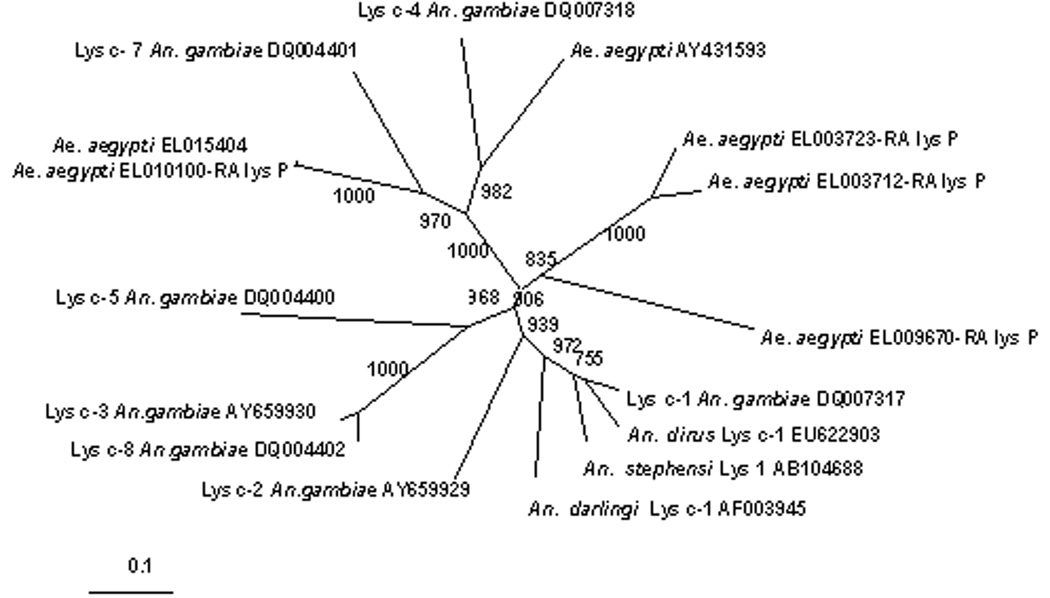
A lysozyme phylogenetic tree was constructed to investigate the molecular phylogenetic relationships among lysozymes from An. gambiae, An. stephensi, An. darlingi and Ae. aegypti. Protein sequences were aligned using Clustal W. The phylogenetic tree was generated using a neighbor-joining program in Clustal X with the ‘delete positions with gaps’ and correct for multiple substitutions’ settings on. The tree was bootstrapped 1000 times.
2.3. Plasmodium berghei infections
The protocol described in Kajla et al. (2011) was used for producing infections in An. dirus mosquitoes. The infections were monitored by visualizing GFP-expressing parasites under a fluorescent microscope.
2.4. dsRNA mediated silencing of AdLysc-1
The procedures for the production of dsRNA and injections in the mosquitoes were performed according to the protocol described in Kajla et al. (2010, 2011). Briefly, the template for generation of dsRNA for AdLysc-1 was produced using one step PCR. Each primer included 15 bp of T7 promoter sequence plus AdLys c-1 specific sequence: (5’-TAATACGACTCACTATAGGGTAATTTCACGCACACGCTAGCA-3’) and 5’-TAATACGACTCACTATAGGGACTTGCTGTAGTGTGCTTCAGA- 3’). The GFP sequence (Fwd: 5’-TAATACGACTCACTATAGGGCGTGATCAAGCCCGACA-3’, Rev: 5’-TAATACGACTCACTATAGGGCTTCGGCGTGCTC-3’) was used to amplify a product from phMGFP vector (Promega, Madison, Wisconsin).
A QIAquick PCR Purification Kit (Qiagen Sciences, Maryland, USA) was used to purify the PCR product. The purified PCR product was used as template for transcription by using the MEGAscript™ RNAi kit (Ambion, Austin, Texas) following the manufacturer’s instructions.
Zero to 1 d old mosquitoes were used for dsRNA injections. Female mosquitoes were injected with 1.4 µg/ 0.6 µl volume dsRNA for AdLys c-1 or unrelated control GFP. This concentration of dsRNA was chosen based on pilot experiments in which different concentrations of dsRNA were injected in the mosquitoes for varying periods of time from 24 h to 4 days. The silencing effect was observed through 4 days after injections of dsRNA.
In these experiments about 40–50 mosquitoes were injected with dsRNA for AdLys c-1 or dsGFP. Three days after dsRNA injections, mosquitoes were starved of sugar for about 2–4 h before blood feeding on a mouse infected with the GFP-expressing strain of P. berghei. This time period allows for the mosquitoes to recover from the injury caused by injections. Mosquitoes that failed to feed on the mouse were removed within 24 h. Four to five days later, midguts from each batch were dissected and the number of oocysts counted using fluorescence microscopy.
Five mosquitoes were harvested to check the efficacy of the knockdown using RT-PCR as described below. For this, the RNA isolation and cDNA was prepared as described above in section 2.2. This cDNA was used in the RT-PCR reaction for amplification of AdLys c-1. Specific primer sets for RT-PCR for AdLys c-1 were as follows: 5-GCTGAGGCCAAGAAATTCAG-3 and 5-AGCAGCTTTTTGCACGCTAT-3. All experiments were repeated three times.
2.5. Semiquantitative RT-PCR
Semiquantitative RT-PCR was performed to assess relative transcript abundance in the different developmental stages and in the adult female mosquitoes following RNAi. The different life stages and tissues tested were: Eggs, larvae (first through 4th instars), pupae, freshly emerged males and females at day zero and five day old adult males and females. In all cases 5–10 whole animals were taken for analysis. Various tissues from larvae (4th instar) and adult (2–3 d old females) mosquitoes included head, thorax, abdomen, fat bodies, midguts, malphigian tubules, and ovaries from naïve as well as from blood fed mosquitoes. From larvae, the hindguts and gastric caeca were also tested for the presence of AdLys c-1.
Total RNA was isolated from five An. dirus mosquitoes from respective samples/treatments using MasterPure™ Complete DNA and RNA Purification Kit (Epicentre Biotechnologies, Madison, WI, USA) according to the supplied instructions. Total RNA was treated with RQ1 DNAse (Promega, Madison, WI) at a concentration of 1 unit/µg RNA preparation to remove genomic DNA. The concentration of RNA was measured with micro-spectrophotometry (Nanodrop NT1000, Thermo Fisher Scientific, Waltham, MA). Five hundred nanograms of total RNA were used to synthesize cDNA using a High-Capacity cDNA Archive Kit (Applied Biosystems, Carlsbad, California) The resulting cDNA was stored at −20°C until use.
Primers based on the An. dirus cDNA sequence (GenBank: accession no. EU622903) and RPS7 (Salazar et al., 1993) were designed using Oligoperfect software (Invitrogen). The S7 ribosomal gene was used to normalize the template. Final concentrations of reagents used for RT-PCR were 1X buffer, 200 µM dNTPs, 0.5 µM each primer, 0.2 µl (4 units) Advantage Taq polymerase (Clontech Laboratories Inc., CA, USA). PCR cycle conditions were: an initial denaturation at 95°C for 3 min; then repeated cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 30 s; and a final extension step at 72°C for 10 min. For AdLys c-1, 35 cycles were used while 25 cycles were used for S7. PCR products were separated on 1% agarose gels and stained with ethidium bromide. The gels were exposed to UV-transillumination and images were captured on a gel-imaging system. For the dsRNA knockdown experiments, signal intensities of bands were quantified by densitometric analysis using the Quantity One Software (version 4.6.8) from Bio-Rad Laboratories (Bio-Rad, Hercules, CA). Fold change in expression was calculated by dividing the ratios of dsGFP/S7 control with the dsAdLys c-1/S7 ratios.
3. Results
3.1. Features of AdLys c-1 and comparison to other reported mosquito lysozymes
A full-length lysozyme was cloned and sequenced from An. dirus. The sequence information was submitted to GenBank (EU622903). The lysozyme (AdLys c-1) cDNA is 623 nt in length (Fig. 1). The SIGNAL P 3.0 program was used to predict the location of signal peptides. The PROTEAN program (DNASTAR) was used to estimate the theoretical pI and molecular mass of the predicted mature protein. For AdLys c-1, the signal peptide cleavage was shown between amino acids 20 and 21 leaving a mature protein of 120 amino acids. The predicted molecular mass of the mature protein is 13.4 kDa and the theoretical pI is 8.45.
Fig. 1.
Complementary DNA and translated amino acid sequences of An. dirus Lys c-1 (GenBank accession number EU622903). The start and stop codons and the putative signal peptide are indicated in bold. The predicted signal cleavage site is indicated by the vertical arrow.
C-type lysozymes are characterized by the presence of conserved amino acids at the catalytic cleft as well as eight cysteine residues. These features were examined in AdLys c-1 sequence. Two critical amino acids, E32 and D49 that are necessary for muramidase enzymatic activity were identified. These two amino acids also were identified in Lys c-1 and c-2 from An. gambiae as well as in lysozymes from An. stephensi and An. darlingi (Fig. 2) and define a group of catalytically active mosquito enzymes. Mammals and birds contain a group of calcium binding c-type lysozymes (Prager and Jolles, 1996). In a previous study, both Lys c-3 and c-8 of An. gambiae displayed similarity to the binding site in the vertebrate calcium-binding lysozymes (Prager and Jolles, 1996; Li et al., 2005). In AdLys c-1 sequence, a similar site (DDDITDD) was identified.
Fig. 2.
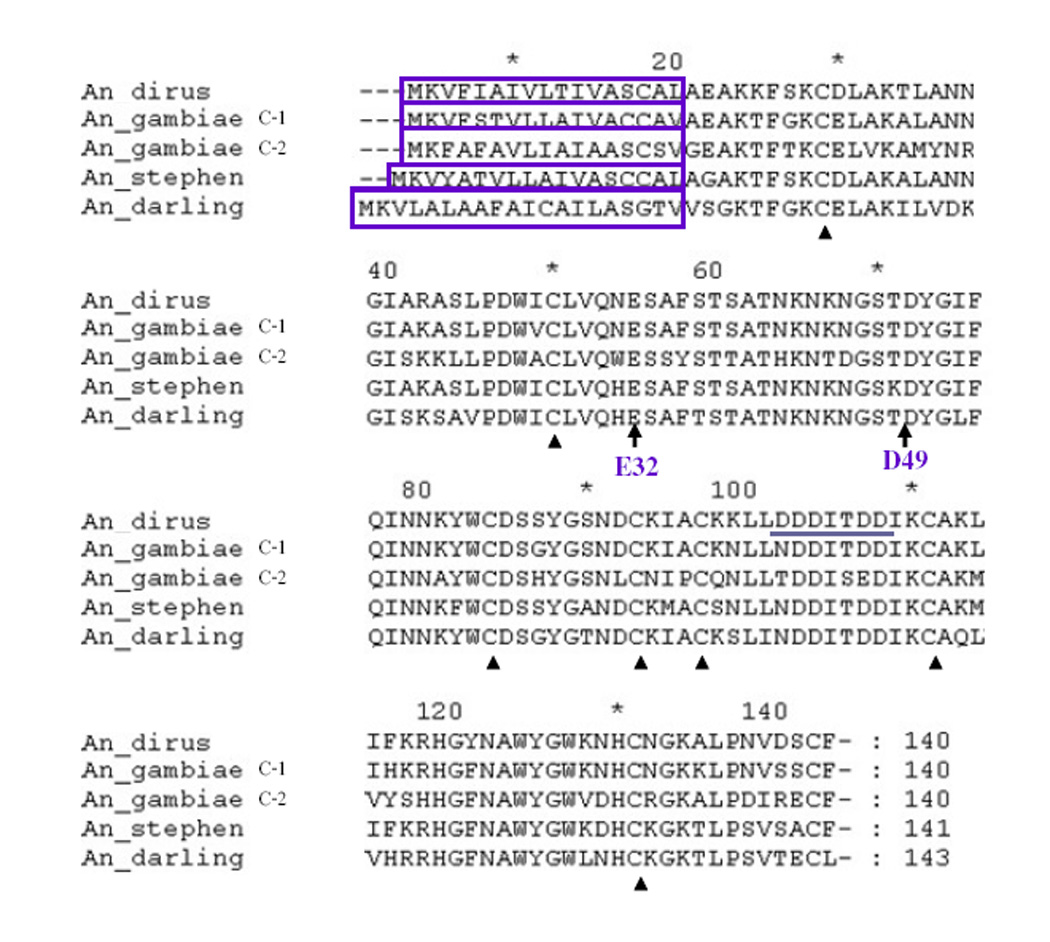
Alignment of deduced amino acid sequences of AdLys c-1 from An. dirus and other Anopheles mosquitoes. The predicted signal peptides are boxed. Conserved cysteines are marked with black triangles. E32 and D49, the two amino acids that are essential for muramidase function are denoted by black arrows. The potential calcium binding site (DDDITDD) from An. dirus Lys c-is underlined.
A phylogenetic tree was constructed to investigate the molecular phylogenetic relationships among lysozymes from An. dirus, An. gambiae, and additional members that were identified in An. stephensi, An. darlingi, and Ae. aegypti (Fig. 3). A neighbor joining analysis in ClustalX was used to determine the closest associations among mosquito lysozymes. AdLys c-1 grouped into a clade of lysozymes from anopheline mosquitoes. This clade includes Lys c-1 and Lys c-2 from An. gambiae as well as enzymes from An. stephensi and An. darlingi (Fig. 3). All of these enzymes share the two critical active site amino acids, while other mosquito lysozymes have lost one or both of these.
Fig. 3.
Phylogenetic analysis of the c-type lysozyme proteins from An. dirus, An. gambiae, Ae. aegypti, An. stephensi, and An. darlingi. Numbers at the nodes shows bootstrap proportions (BP) on 1000 replicates. Only BPs over 75% are shown. The GenBank accession numbers for each sequence are provided.
3.2. Temporal and tissue expression profiles of AdLys c-1 transcript levels
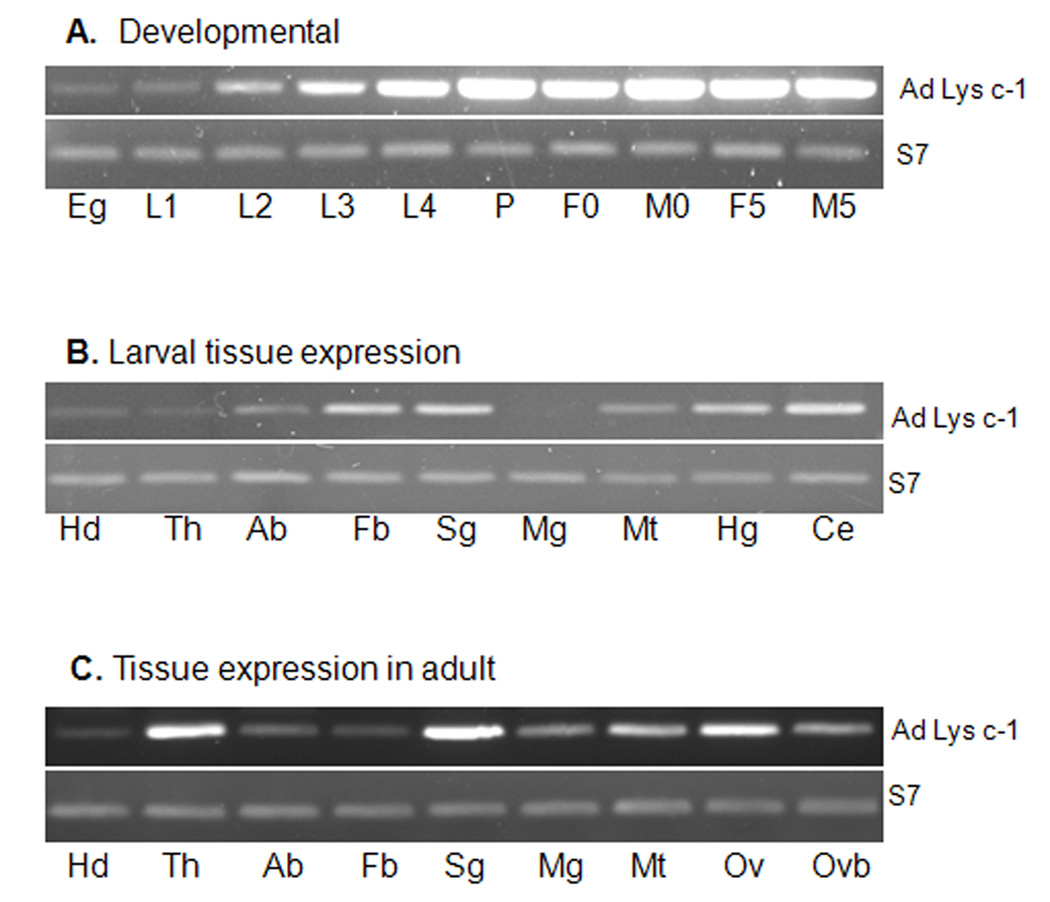
AdLys c-1 expression was examined using RT-PCR at various developmental stages and in different tissues from both larval and adult An. dirus mosquitoes (Fig. 4). The expression of AdLys c-1 in these RT-PCR assays was normalized against the ribosomal S7 gene. AdLys c-1 was expressed in all developmental stages. It appeared to be highly expressed in later stages of larval development as compared to eggs and earlier stages (Fig. 4A). In larval tissues, AdLys c-1 appeared high in salivary gland, fat body, and hindgut but not in the midgut (Fig. 4B). Interestingly, AdLys c-1 transcripts were very abundant in the larval caecum, a potential site of residence for bacteria. In the adults, AdLys c-1 transcripts were detected in most of the tested tissues with highest expression in the thorax, salivary glands and ovaries (Fig. 4C).
Fig. 4.
RT-PCR expression analyses of AdLys c-1 in developmental and tissue samples from An. dirus. The ribosomal protein S7 was used to normalize the samples. (A) Developmental: Eg=eggs, L1, L2, L3, L4= 1st, 2nd, 3rd, and 4th instar larvae respectively, P=pupae, F0=adult females on day of emergence, M0=adult males on day of emergence, F5=adult females 5 days after emergence, M5=adult males 5 days after emergence. (B) Larval tissue expression: Hd=head, Th=thorax, Ab=abdomen, Mg=midgut, Sg=salivary gland, Fb=fat body (abdominal body wall), Mt=malpighian tubules, Hg=Hindgut, Ce=Caecum. (C) Tissue expression in adult: Hd=head, Th=thorax, Ab=abdomen, Mg=midgut, Sg=salivary gland, Fb=fat body (abdomal body wall), Mt=malpighian tubules, Ov=ovary OvB=ovary post blood feeding.
3.3. Presence of AdLys c-1 was found to promote the development of Plasmodium berghei
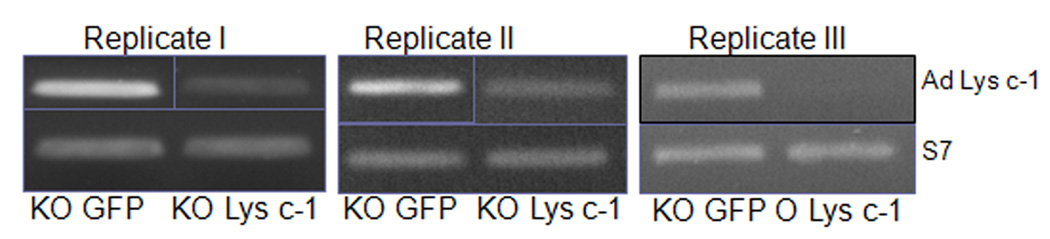
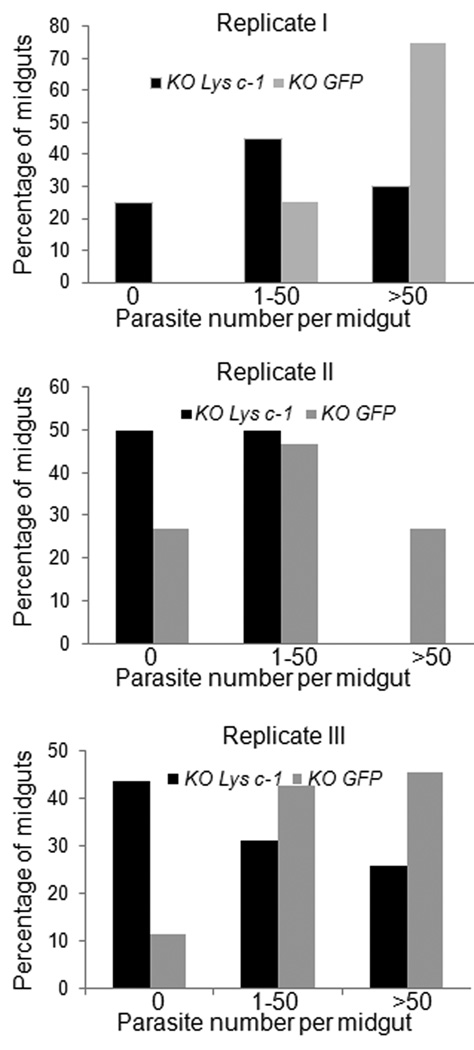
Transient depletion using dsRNA-mediated gene silencing is an effective and widely used approach in the mosquito for assessing the potential role of a target gene of interest and its effect on the development of Plasmodium parasites (Blandin et al., 2002; Kajla et al., 2011). The injection of dsLys c-1 successfully suppressed the systemic expression of AdLys c-1 in three replicate experiments (2.8–5.3 fold; Fig. 5). dsRNA mediated silencing of AdLys c-1 also resulted in a significant reduction (p<0.05, Kruskal-Wallis test) in the number of developing oocysts four days after the feeding of an infectious blood meal as compared with dsGFP-injected mosquitoes (Fig. 6 and 7). The morphology of parasites did not differ between AdLys c-1 knockdown and control dsGFP injected mosquitoes. Moreover, melanization of parasites did not occur in any of the three experiments (data not shown). Thus, knockdown of AdLys c-1 did not affect this immune process. The reduced parasite number was most evident in the 3rd replicate (Fig. 7) which correlated with the strongest reduction in the AdLys c-1 transcript.
Fig. 5.
RT-PCR verification of RNA interference-mediated gene silencing. A ribosomal S7 primer set was used as reference gene for comparisons. Inhibition of expression of AdLys c-1 was most efficient in the 3rd replicate. KO GFP = knock down of GFP, KO Lys c-1 = knock down of AdLys c-
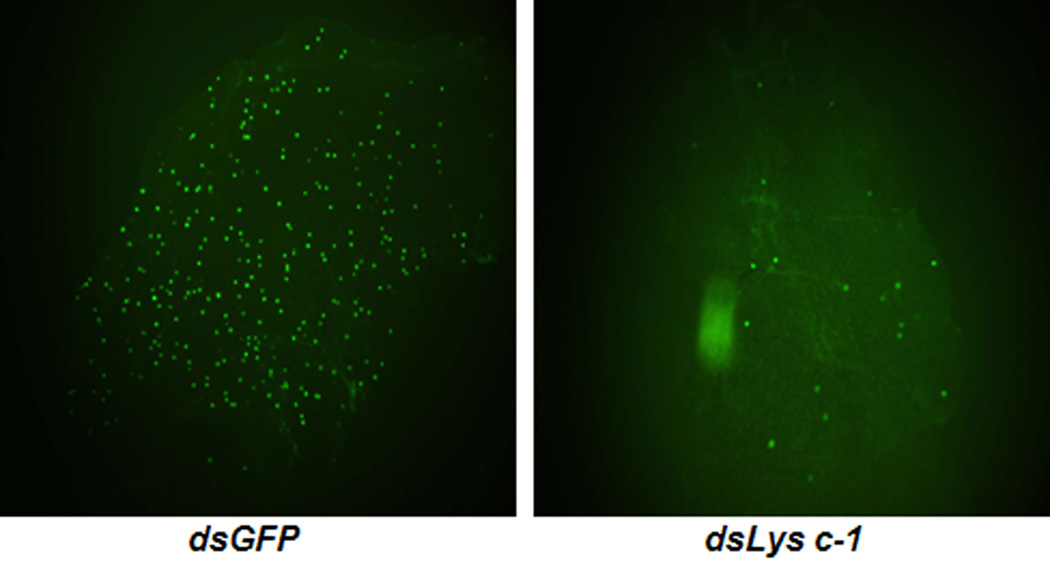
Fig. 6.
Comparison of An. dirus midguts infected with a GFP-expressing strain of P. berghei after the mosquitoes were injected with dsGFP or dsAdLys c-1. At 4 days after infection, midguts from each batch were dissected and the number of oocysts was counted using fluorescence microscopy. The number of parasites in dsAdLys c-1 KO mosquitoes was dramatically reduced when compared with control dsGFP mosquitoes.
Fig. 7.
The effect of AdLys c-1 knockdown on number of P. berghei.
At 4 days after infection, midguts from each batch were dissected and the number of oocysts counted using fluorescence microscopy. Results of three independent replicates are presented. (KO GFP = knock down of GFP, KO Lys c-1 = knock down of AdLys c-1, Parasite number per midguts 1= 0, 2= 1–50, 3= >50).
4. Discussion
Our results support an agonistic role of AdLys c-1 in the response of mosquitoes during P. berghei infection. Knockdown of this gene resulted in a significant reduction in the number of oocysts. This suggests that AdLys c-1 may play roles in mosquito biology that assist the development of the Plasmodium parasites.
It is possible that this agonistic role relates to regulation of the gut bacterial community. A number of studies have shown an increased lysozyme expression in the mosquitoes after challenge with bacteria (Fujimoto et al., 2001; Lee and Brey, 1995; Sun et al., 1991; Gao and Fallon, 2000; Hernandez et al., 2003; Bedoya et al., 2005; Kajla et al., 2010). In blood fed mosquitoes the bacterial population increases significantly within the midgut (Beier, 1998, DeMaio et al., 1996; Pumpuni et al., 1993, 1996; Bedoya et al., 2005). Recently Dong et al. (2009) showed that the presence of gut microbiota indirectly impacted the susceptibility of the mosquitoes to become infected with malaria parasites apparently via modulation of the basal immune response. It is conceivable that in the absence of AdLys c-1, certain types of gut bacteria might grow at a higher rate and could upregulate basal immunity and/or directly injure parasites, preventing their development. However, silencing of lysozyme c-1 in An. gambiae did not result in a significant increase in the total number of culturable midgut bacteria (Kajla et al., 2011). Blandin et al. (2002) reported that knockdown of another antibacterial protein, defensin, in An. gambiae, did not cause significant differences in oocyst morphology or the frequency distribution of oocyst numbers although an altered microbial community would be predicted as a result of this manipulation. Further work will be needed to determine the interaction between AdLys c-1 and other antibacterial proteins, the midgut microbial community and the malaria parasites within mosquitoes.
Lysozyme c-1 has also been shown to inhibit melanization of abiotic foreign targets in An. gambiae (Li and Paskewitz, 2006). Recent work showed that An. gambiae lysozyme c-1 physically associates with P. berghei oocysts (Kajla et al., 2011). This suggested the possibility that the presence of mosquito lysozyme c-1 on the surface of the oocyst could protect it from melanization. However, we did not observe melanization of parasites following silencing of the AdLys c-1 gene.
Because mosquito lysozymes belong to an expanded gene family (Li et al., 2005; Bedoya et al., 2005), we are also interested in investigating the biological functions of each member of the group. As a first step towards that goal, we analyzed the expression profiles for AdLys c-1 across stages and tissues of An. dirus. AdLys c-1 was expressed in all developmental stages. Surprisingly, the larval gastric caecae exhibited very high levels of transcript abundance. The function of the mosquito gastric caecum is not known but it may harbor microorganisms that promote digestion or protect the gut from colonization by other, more pathogenic bacteria. The presence of AdLys c-1 in this tissue suggests that the protein may be present, perhaps to regulate the bacterial community either in the caecum or in the midgut itself. Alternatively, the gastric caecum may secrete lysozyme c-1 into the midgut where it might participate in regulation of the microbiota. How larval immunity differs from adults and whether larval exposure to bacteria can affect immune status of the subsequent adult are relevant questions that should be examined in more detail.
The patterns of AdLys c-1 transcription are similar to the temporal and tissue expression profiles of Lysc-1 from An. gambiae. AgLys c-1 and Lys c-2 were inducible when bacteria were introduced into the mosquito (Li et al., 2005). Because the phylogenetic analysis indicated that AdLys c-1 is closely related to these proteins, we expect that AdLys c-1 will be similarly involved in responses to bacteria.
In conclusion, we found that the presence of An. dirus Lys c-1 was important to the development of Plasmodium parasites as has been reported for An. gambiae Lys c-1 (Kajla et al., 2011). Future investigations on the relationships among mosquito lysozymes, midgut bacteria and malaria parasites will be required to understand the intricacies of these complex relationships.
Acknowledgements
We gratefully acknowledge financial support from the Development and Promotion of Science and Technology Talents Projects (DPST) grants, Thailand. We would like to thank Olga Andreeva for sharing her expertise in dsRNA injections; Aditya Singh for statistical analysis of oocyst count data and Eric J. Shelley for help with P. berghei/mouse experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham EG, Nagaraju J, Salunke D, Gupta HM, Dalta RK. Purification and partial characterization of an induced antibacterial protein in the silkworm, Bombyx mori. J. Invertebr. Pathol. 1995;65:17–24. doi: 10.1006/jipa.1995.1003. [DOI] [PubMed] [Google Scholar]
- Bedoya RJU, Mitzey AM, Obraztsova M, Lowenberger C. Molecular cloning and transcriptional activation of lysozyme-encoding cDNAs in the mosquito Aedes aegypti (Diptera: Culicidae) Insect Mol. Bio. 2005;14:89–94. doi: 10.1111/j.1365-2583.2004.00534.x. [DOI] [PubMed] [Google Scholar]
- Beier JC. Malaria parasite development in mosquitoes. Annu. Rev. Entomol. 1998;43:519–543. doi: 10.1146/annurev.ento.43.1.519. [DOI] [PubMed] [Google Scholar]
- Blandin S, Moita LF, Köcher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae (Diptera: Culicidae): targeted disruption of the Defensin gene. EMBO Rep. 2002;3:852–856. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaio J, Pumpuni CB, Kent M, Beier JC. The midgut bacterial flora of wild Aedes triseriatus (Diptera: Culicidae), Culex pipiens (Diptera: Culicidae), and Psorophora columbiae (Diptera: Culicidae) mosquitoes. Am. J. Trop. Med. Hyg. 1996;54:219–223. doi: 10.4269/ajtmh.1996.54.219. [DOI] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. Implication of the Mosquito Midgut Microbiota in the Defense against Malaria Parasites. PLOS Pathog. 2009;5(5) doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn PE. Biochemical aspects of insect immunology. Ann. Rev. Entomol. 1986;31:321–339. [Google Scholar]
- Fujimoto S, Toshimori-Tsuda I, Kishimoto K, Yamano Y, Morishima I. Protein purification, cDNA cloning and gene expression of lysozyme from eti-silkworm, Samia Cynthia ricini (Lepidoptera: Saturniidae) Comp. Biochem. Physiol. 2001;128:709–718. doi: 10.1016/s1096-4959(00)00368-7. [DOI] [PubMed] [Google Scholar]
- Gao Y, Fallon AM. Immune activation upregulates lysozyme gene expression in Aedes aegypti (Diptera: Culicidae) mosquito cell culture. Insect Mol. Biol. 2000;9:553–558. doi: 10.1046/j.1365-2583.2000.00216.x. [DOI] [PubMed] [Google Scholar]
- Garver LS, Dong Y, Dimopoulos G. Caspar controls resistance to Plasmodium falciparum (Haemospororida: Plasmodiidae) in diverse anopheline species. PLoS Pathog. 2009;5(3) doi: 10.1371/journal.ppat.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez VP, Higgins L, Fallon AM. Characterization and cDNA cloning of an immune-induced lysozyme from cultured Aedes albopictus (Diptera: Culicidae) mosquito cells. Dev. Comp. Immunol. 2003;27:11–20. doi: 10.1016/s0145-305x(02)00065-4. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- Hultmark D, Steiner H, Rasmuson T, Boman HG. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur. J. Biochem. 1980;106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x. [DOI] [PubMed] [Google Scholar]
- Hultmark D. Insect lysozymes. In: Jollès P, editor. Lysozymes: Model Enzymes in Biochemistry and Molecular Biology. Basel: Birkhäuser Verlag; 1996. pp. 87–102. [Google Scholar]
- Jollès J, Schoentgen F, Croizier G, Croizier L, Jollès P. Insect lysozymes from three species of Lepidoptera: their structural relatedness to the C (chicken) type lysozyme. J. Mol. Evol. 1979;14:267–721. doi: 10.1007/BF01732494. [DOI] [PubMed] [Google Scholar]
- Jolles P, Jolles J. What is new in lysozyme research. Always a model system, today as yesterday. Mol. Cell. Biochem. 1984;63:165–189. doi: 10.1007/BF00285225. [DOI] [PubMed] [Google Scholar]
- Kajla MK, Shi L, Li B, Luckhart S, Li J, Paskewitz SM. A New Role for an Old Antimicrobial: Lysozyme c-1 Can Function to Protect Malaria Parasites in Anopheles Mosquitoes. PLoS One. 2011;6:e19649. doi: 10.1371/journal.pone.0019649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajla MK, Andreeva O, Gilbreath TM, 3rd., Paskewitz SM. Characterization of expression, activity and role in antibacterial immunity of Anopheles gambiae lysozyme c-1. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010;155:201–209. doi: 10.1016/j.cbpb.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Brey PT. Isolation and characterization of the lysozyme-encoding gene from the silkworm Bombyx mori (Lepidoptera: Bombycidae) Gene. 1995;161:199–203. doi: 10.1016/0378-1119(95)00199-g. [DOI] [PubMed] [Google Scholar]
- Li B, Calvo E, Marinotti O, James AA, Paskewitz SM. Characterization of the c-type lysozyme gene family in Anopheles gambiae (Diptera: Culicidae) Gene. 2005;360:131–139. doi: 10.1016/j.gene.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Li B, Paskewitz SM. A role for lysozyme in melanization of Sephadex beads in Anopheles gambiae (Diptera: Culicidae) J. Insect. Physiol. 2006;52:936–942. doi: 10.1016/j.jinsphys.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Markart P, Korfhagen TR, Weaver TE, Akinbi HT. Mouse lysozyme M is important in pulmonary host defense against Klebsiella pneumoniae infection. Am. J. Respir. Crit. Care Med. 2004;169:454–458. doi: 10.1164/rccm.200305-669OC. [DOI] [PubMed] [Google Scholar]
- Meister S, Kanzok SM, Zheng X, Luna C, Li T, Hoa NT, Clayton JR, White KP, Kafatos FC, Christophides GK, Zheng L. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. PNAS. 2005;102:11420–11425. doi: 10.1073/pnas.0504950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T, Brunner F. Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant. Biol. 2002;5:318–324. doi: 10.1016/s1369-5266(02)00265-0. [DOI] [PubMed] [Google Scholar]
- Paskewitz SM, Reese-Stardy S, Gorman MJ. An easter-like serine protease from Anopheles gambiae exhibits changes in transcript abundance following immune challenge. Insect Mol Biol. 1999;8:329–337. doi: 10.1046/j.1365-2583.1999.83124.x. [DOI] [PubMed] [Google Scholar]
- Paskewitz SM, Li B, Kajla MK. Cloning and molecular characterization of two invertebrate-type lysozymes from Anopheles gambiae (Diptera: Culicidae) Insect. Mol. Bio. 2008;17:217–225. doi: 10.1111/j.1365-2583.2008.00797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powning RF, Davison WJ. Studies on insect bacteriolytic enzymes. I. Lysozyme in haemolymph of Galleria mellonella and Bombyx mori. Comp. Biochem. Physiol. 1973;45B:669–686. [PubMed] [Google Scholar]
- Prager EM, Jollès P. Animal lysozymes c and g: an overview. In: Jollès P, editor. Lysozymes: Model Enzymes in Biochemistry and Molecular Biology. Basel: Birkhäuser Verlag; 1996. pp. 9–31. [Google Scholar]
- Pumpuni CB, Beier MS, Nataro JP, Guers LD, Davis JR. Plasmodium falciparum: Inhibition of sporogonic development in Anopheles stephensi (Diptera: Culicidae) by Gram-negative bacteria. Exp. Parasitol. 1993;77:195–199. doi: 10.1006/expr.1993.1076. [DOI] [PubMed] [Google Scholar]
- Pumpuni CB, Demaio J, Kent M, Davis JR, Beier JC. Bacterial population dynamicsin three anopheline species: the impact on Plasmodium (Haemospororida: Plasmodiidae) sporogonic development. Am. J. Trop. Med. Hyg. 1996;54:214–218. doi: 10.4269/ajtmh.1996.54.214. [DOI] [PubMed] [Google Scholar]
- Salazar CE, Mills-Hamm D, Kumar V, Collins FH. Sequence of a cDNA from the mosquito Anopheles gambiae encoding a homologue of human ribosomal protein S7. Nucleic Acids Res. 1993;21:4147. doi: 10.1093/nar/21.17.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Asling B, Faye I. Organization and expression of the immunoresponsive lysozyme gene in the giant silk moth, Hyalophora cecropia (Lepidoptera: Saturniidae) J. Biol. Chem. 1991;266:6644–6649. [PubMed] [Google Scholar]