Abstract
We demonstrate a composite nanomaterial, termed an aptamer nano-flare, that can directly quantify an intracellular analyte in a living cell. Aptamer nano-flares consist of a gold nanoparticle core functionalized with a dense monolayer of nucleic acid aptamers with a high affinity for adenosine triphosphate (ATP). The probes bind selectively to target molecules and release fluorescent reporters which indicate the presence of the analyte. Additionally, these nanoconjugates are readily taken up by cells where their signal intensity can be used to quantify intracellular analyte concentration. These nanoconjugates are a promising approach for the intracellular quantification of other small molecules or proteins, or as agents that use aptamer binding to elicit a biological response in living systems.
Nanomaterials hold great promise for biology and medicine due to their unique size, properties, and ability to be functionalized with biological recognition elements.1–16 Our group has developed a class of nanomaterials termed polyvalent DNA functionalized gold nanoparticles17, 18 (DNA-Au NPs) that consist of a gold nanoparticle core (2–250 nm in size) and a dense shell of synthetic oligonucleotides.19, 20 Fundamental investigations have identified several unexpected properties of these materials such as enhanced binding constants for complementary targets,21 sharp and elevated melting transitions,22 the ability to form stable complexes at room temperature with as few as two base-pairings,23 enhanced enzymatic stability,24 and the ability to readily enter living cells,25 all of which clearly distinguish DNA-Au NPs from their molecular counterparts. These properties also render DNA-Au NPs useful for a variety challenging investigations such as programmed materials assembly,26, 27 the detection and quantification of trace analytes,28–32 and intracellular genetic regulation and detection.33–37
Aptamers are nucleic acids or peptides that exhibit affinity and selectivity for a target molecule and are thus considered attractive detection and diagnostic tools.38–43 For example, Li et al. developed an important aptamer-based probe consisting of a fluorophore-labeled DNA competitor and a quencher-labeled aptamer, which can be used for small molecule detection.44 Due to their high selectivity,45–47 aptamers are also well-suited for cellular applications, including diagnostics and therapeutics. Indeed, Chang et al. have developed an aptamer-gold nanoparticle based system for sensitive and selective detection of cellular growth and proliferation using a marker for Platelet-Derived Growth Factor in homogenous solution.48 However, an aptamer-based system capable of entering a cell and remaining active in the intracellular environment has proved challenging to achieve. Indeed, it is difficult to effect the cellular uptake of nucleic acid-based agents without additional agents for improving cellular internalization. Moreover, oligonucleotides can become unstable and degraded in cellular environments, and thus their application in vivo has been challenging.49–51
Herein, we describe the development of novel aptamer-nanoparticle probes, termed aptamer nano-flares, that are designed to detect and quantify intracellular molecular analytes. These nanostructures consist of a gold nanoparticle that is functionalized with a dense monolayer (~8.4 pmol/cm2) of chemisorbed aptamer oligonucleotides hybridized to fluorophore labeled flares. Based on previous work, the dense shell of oligonucleotides is expected to provide enhanced stability24 and a mechanism for cellular entry,25 and the flare is expected to be capable of transducing a binding event into a fluorescence signal.37 In addition to synthesis and characterization, we determine the sensitivity and selectivity of the approach in the detection of adenosine triphosphate (ATP) and evaluate the ability to quantify the intracellular concentration of this target molecule.
We chose ATP as a target molecule for proof-of-concept experiments aimed at developing nanomaterials that quantify concentrations of small molecule analytes inside living cells. ATP is important for regulating cellular metabolism and biochemical pathways, and as such, the intracellular ATP level is used as an indicator of living organisms and their biological activity. Current methods of ATP detection require the lysis of cells and only measure the ATP concentration of a bulk sample. Aptamer nano-flares are designed so that the flare oligonucleotide binds to the aptamer that is attached to the surface of the Au NP (Figure 1). In this bound state, the fluorescence of the flare strand is quenched by the gold nanoparticle. In the presence of the ATP target molecules, ATP binds to the aptamer causing a conformational change and resulting in a new folded secondary structure. This folded structure disrupts the Watson-Crick base-pairing between the aptamer and the flare, which causes flares to be liberated with an increase in fluorescence due to the greater distance of the flare from the gold surface.
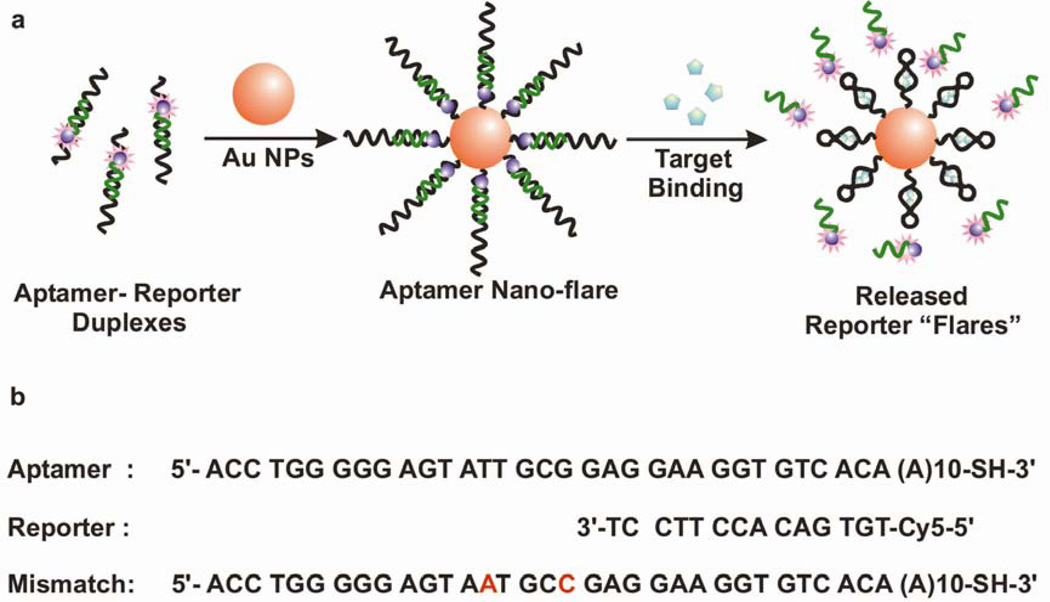
Figure 1.
(a) Aptamer nano-flares are gold nanoparticles functionalized with thiol-terminated aptamer sequences hybridized to a short complementary Cy5-labeled reporter strand. The reporter is capable of being displaced by a conformation change in the aptamer that is induced by the target molecule. (b) Oligonucleotide sequences used to prepare the aptamer nano-flares.
Aptamer nano-flares were synthesized from citrate-capped 13-nm Au NP precursors. Thiol-terminated ATP binding aptamers (5’ ACC TGG GGG AGT ATT GCG GAG GAA GGT GTC ACA (A)10 propyl thiol -3’) designed from the literature52 were allowed to hybridize with fluorophore-labeled (Cy5) reporter sequences (5’- Cy5 TGT GAC ACC TTC CT -3’) in a phosphate buffered saline solution (PBS). Following literature procedures, the duplexes were added to the Au NPs and a dense monolayer was allowed to coat the nanoparticles, exchanging the citrate-capping layer.17 Quantification of DNA surface loading by fluorescence53 reveals that each aptamer nano-flare contains approximately 27 duplexes. (see Supporting Information)
Aptamer nano-flares show a significant fluorescence increase upon target addition. For example, when 2 mM ATP is added, an immediate 3.8-fold increase in fluorescent signal is observed, which is consistent with target binding and flare release (Figure 2). Conversely, addition of the ATP analogues guanidine triphosphate, cytosine triphosphate or uridine triphosphate did not result in a significant increase in fluorescent signal. These results are consistent with a selective target binding ability. The affect of target concentration was also investigated. Aptamer nano-flares respond to the presence of ATP in a dose-dependent manner, demonstrating that the fluorescence can be used to quantify ATP concentration. The apparent Kd (~ 594 μM, defined as ATP concentration that resulted in half maximal fluorescent change) of aptamer nano-flares was found to be similar to analogous quencher-fluorophore system (~600 μM)44 but lower than the original aptamer (~6 μM)52 presumably due to partial blockage of ATP binding site by the flare strand. Importantly, aptamer nano-flares are sensitive to ATP concentrations that are typically found in cells (0.1 – 3.0 mM).54
Figure 2.
(a) Solution fluorescence spectra of aptamer nano-flares treated with ATP (2 mM) and UTP, GTP and CTP. (b) Fluorescence spectra of aptamer nano-flares in solutions with increasing concentrations of ATP (0.1 – 3.0 mM). Inset shows the peak intensity as a function of ATP concentration.
Having demonstrated the signaling ability of the aptamer nano-flares, we next tested their ability to enter cells and detect intracellular ATP using a HeLa cell (human cervical cancer) model. For control experiments, a second probe containing a mismatched aptamer sequence (two base substitutions) was prepared. The mismatch probe was designed and determined to have similar background fluorescence, melting properties, and total number of duplex ligands as the aptamer nano-flare (see Supporting Information). Cells were cultured on slide chambers, incubated with aptamer nano-flares and imaged using scanning confocal microscopy. HeLa cells treated with aptamer nano-flares were highly fluorescent as compared to those treated with the mismatch controls, with fluorescence seen primarily in the cytoplasm, consistent with the localization of the DNA-Au NPs (Figure 3).
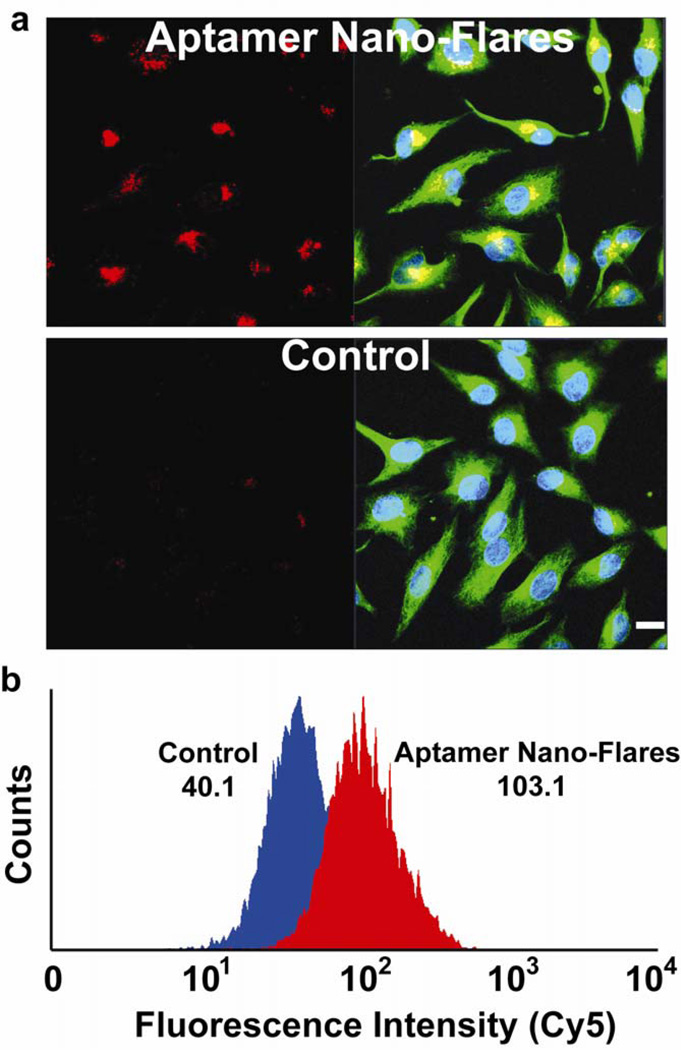
Figure 3.
(a) Fluorescence microscopy images of HeLa cells incubated with aptamer nano-flares and control particles for 2 hrs. The red channel is Cy5 fluorescence associated with activated aptamer nano-flares, the green channel is fluorescence associated taxol-Alexa 488 and specific for tubulin, and the blue channel is fluorescence associated with Hoechst 3342 and specific for the nucleus. Scale bar is 20μm. (b) Cell-associated fluorescence (Cy5) of populations treated with aptamer nano-flares and control particles as determined by flow cytometry. Inset numbers indicate the mean fluorescence value of the population.
In order to quantify the intracellular signaling of the aptamer nano-flares, we examined cells treated with probes using analytical flow cytometry. Flow cytometry reveals that the HeLa cell population treated with aptamer nano-flares is highly fluorescent and 2.6-times more fluorescent than the population treated with mismatch control particles (Figure 3). These flow cytometry experiments are in excellent agreement with confocal imaging and demonstrate the uniform cellular internalization and intracellular signaling of the aptamer nano-flares.
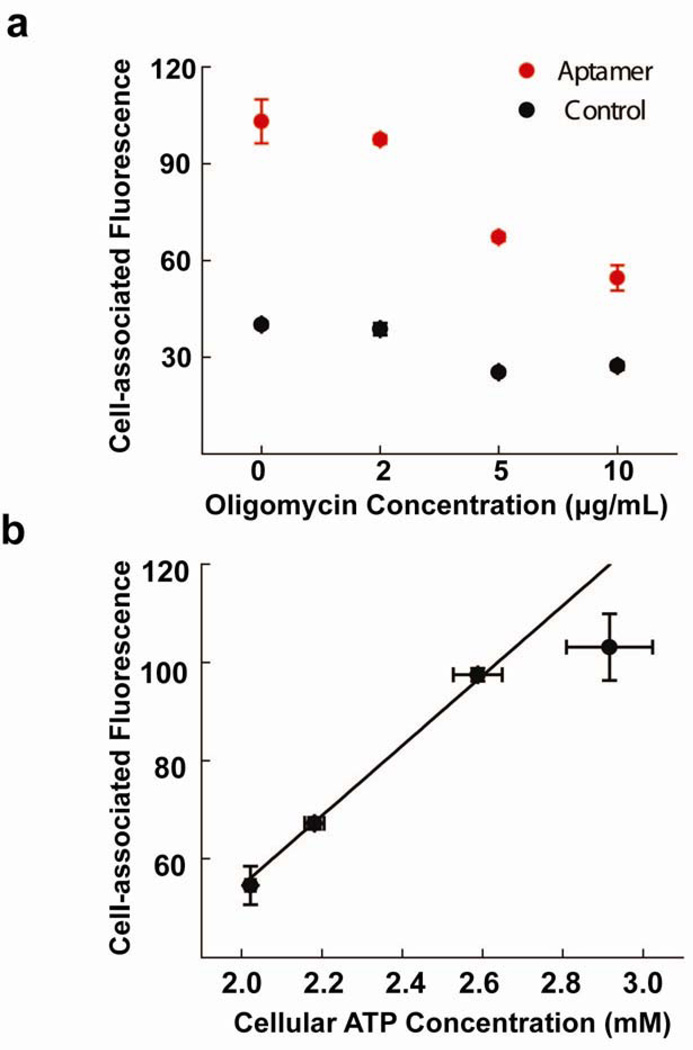
To demonstrate that aptamer nano-flares can be used to quantify intracellular amounts of ATP, we conducted ATP depletion experiments to reduce the levels of intracellular ATP in the HeLa cell models. Cells were treated with doses of oligomycin and 2-deoxy-D-glucose (a drug combination that is known to reduce ATP)55 and aptamer nano-flares (see Supporting Information for experimental details). Analytical flow cytometry was used to quantify cell-associated fluorescence. In these experiments we observe an oligomycin concentration-dependent decrease in the cell-associated fluorescence, which is consistent with drug induced ATP depletion (Figure 4a). The decrease in cell-associated fluorescence is not observed in the control population nor is it a result of fewer particles entering the treated cells (Mass spectrometry measurements confirm similar levels of Au NP internalization for all samples; see Supporting Information). To quantify intracellular ATP concentration we compared cell-associated fluorescence to ATP levels (determined for the bulk sample using a commercial ATP measuring kit normalized for the total number of cells). Accordingly, we found that cell-associated fluorescence corresponds to cellular ATP concentration in a nearly linear manner (Figure 4b). Deviations from linearity at high ATP concentrations are likely due to probe saturation at the highest intracellular ATP concentrations. The overall set of observations shows that aptamer nano-flares can be used to directly quantify intracellular ATP levels in live samples.
Figure 4.
(a) Fluorescence of cell populations treated with doses of oligomycin followed by aptamer nano-flares (red) or control particles (black). The error bars represent standard deviation from at least 3 independent replicates. (b) Cell-associated fluorescence of living cells plotted as a function of cellular ATP concentration as bulk measurement by a commercial ATP detection kit. (see Supporting Information for experimental details) The error bars along the X- and Y-axis represent standard deviations from at least 3 replicates in cellular ATP concentrations and cell-associated fluorescence as measured by flow cytometry, respectively.
In summary, we present a new nanoparticle composite probe, termed an aptamer nanoflare, that can be used to detect and quantify a small molecule analyte in a live cell. Aptamer nano-flares are sensitive to physiologically relevant changes in ATP concentrations (0.1 – 3 mM) and show a high selectivity for ATP when compared to other nucleoside-triphosphate analogs. Aptamer nano-flares readily enter cells where they can be used to directly quantify intracellular ATP levels, which makes them well-suited for cell imaging and sorting studies. Our results show that aptamer ligands remain functional through cellular internalization and acts as a nanoparticle-based intracellular small-molecule detector. In contrast to molecular beacon based systems, the particles are naturally taken up by the cells, and resist nuclease degradation,24 which in the case of molecular beacons leads to high background fluorescence.37 Given that similar nanoconjugates can be prepared with a variety of nucleic acid aptamers, the concept of the aptamer nano-flare can be extended for detecting a range of other analytes based on aptamer binding in a living sample.
Supplementary Material
Acknowledgements
C.A.M. acknowledges a Cancer Center for Nanotechnology Excellence (NCI-CCNE) award for support of this research. C.A.M. is also grateful for an NIH Director’s Pioneer Award. D.S.S. was supported by the LUNGevity Foundation−American Cancer Society Postdoctoral Fellowship in Lung Cancer. P.C.P. was supported by a Ryan Fellowship.
References
- 1.Xiao Y, Patolsky F, Katz E, Hainfeld JF, Willner I. Science. 2003;299(5614):1877–1881. doi: 10.1126/science.1080664. [DOI] [PubMed] [Google Scholar]
- 2.Tkachenko AG, Xie H, Coleman D, Glomm W, Ryan J, Anderson MF, Franzen S, Feldheim DL. J. Am. Chem. Soc. 2003;125(16):4700–4701. doi: 10.1021/ja0296935. [DOI] [PubMed] [Google Scholar]
- 3.Sandhu KK, McIntosh CM, Simard JM, Smith SW, Rotello VM. Bioconj. Chem. 2002;13(1):3–6. doi: 10.1021/bc015545c. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Lu Y. J. Am. Chem. Soc. 2003;125(22):6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Liu GD, Merkoci A. J. Am. Chem. Soc. 2003;125(11):3214–3215. doi: 10.1021/ja029668z. [DOI] [PubMed] [Google Scholar]
- 6.Nativo P, Prior IA, Brust M. Acs Nano. 2008;2(8):1639–1644. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]
- 7.Chithrani BD, Ghazani AA, Chan WCW. Nano Letters. 2006;6(4):662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 8.Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Jr, Schultz PG. Nature. 1996;382(6592):609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 9.Wu YR, Phillips JA, Liu HP, Yang RH, Tan WH. Acs Nano. 2008;2(10):2023–2028. doi: 10.1021/nn800325a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Sayed IH, Huang XH, El-Sayed MA. Nano Lett. 2005;5(5):829–834. doi: 10.1021/nl050074e. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Sun XM, Nakayama-Ratchford N, Dai HJ. Acs Nano. 2007;1(1):50–56. doi: 10.1021/nn700040t. [DOI] [PubMed] [Google Scholar]
- 12.Medintz IL, Berti L, Pons T, Grimes AF, English DS, Alessandrini A, Facci P, Mattoussi H. Nano Letters. 2007;7(6):1741–1748. doi: 10.1021/nl070782v. [DOI] [PubMed] [Google Scholar]
- 13.Dillenback LM, Goodrich GP, Keating CD. Nano Letters. 2006;6(1):16–23. doi: 10.1021/nl0508873. [DOI] [PubMed] [Google Scholar]
- 14.Graham D, Thompson DG, Smith WE, Faulds K. Nature Nanotechnology. 2008;3(9):548–551. doi: 10.1038/nnano.2008.189. [DOI] [PubMed] [Google Scholar]
- 15.Chan WCW, Nie SM. Science. 1998;281(5385):2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 16.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Nano Letters. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 17.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382(6592):607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 18.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277(5329):1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 19.Lee J-S, Seferos DS, Giljohann DA, Mirkin CA. J. Am. Chem. Soc. 2008;130(16):5430–5431. doi: 10.1021/ja800797h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Anal. Chem. 2006;78(24):8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lytton-Jean AKR, Mirkin CA. J. Am. Chem. Soc. 2005;127(37):12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 22.Jin RC, Wu GS, Li Z, Mirkin CA, Schatz GC. J. Am. Chem. Soc. 2003;125(6):1643–1654. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- 23.Hurst SJ, Hill HD, Mirkin CA. J. Am. Chem. Soc. 2008;130(36):12192–12200. doi: 10.1021/ja804266j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Nano Lett. 2009;9(1):308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Nano Lett. 2007;7(12):3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA. Nature. 2008;451(7178):553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 27.Hill HD, Macfarlane RJ, Senesi AJ, Lee B, Park SY, Mirkin CA. Nano Lett. 2008;8(8):2341–2344. doi: 10.1021/nl8011787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289(5485):1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 29.Cao YWC, Jin RC, Mirkin CA. Science. 2002;297(5586):1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 30.Park SJ, Taton TA, Mirkin CA. Science. 2002;295(5559):1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 31.Nam J-M, Thaxton CS, Mirkin CA. Science. 2003;301(5641):1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 32.Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. J. Am. Chem. Soc. 2006;128(26):8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 33.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312(5776):1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 34.Seferos DS, Giljohann DA, Rosi NL, Mirkin CA. Chembiochem. 2007;8(11):1230–1232. doi: 10.1002/cbic.200700262. [DOI] [PubMed] [Google Scholar]
- 35.Patel PC, Giljohann DA, Seferos DS, Mirkin CA. Proc Natl Acad Sci U S A. 2008;105(45):17222–17226. doi: 10.1073/pnas.0801609105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. J Am Chem Soc. 2009;131(6):2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. J. Am. Chem. Soc. 2007;129(50):15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellington AD, Szostak JW. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 39.Ellington AD, Szostak JW. Nature. 1992;355(6363):850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 40.Cox JC, Rudolph P, Ellington AD. Biotechnol Progr. 1998;14(6):845–850. doi: 10.1021/bp980097h. [DOI] [PubMed] [Google Scholar]
- 41.Breaker RR. Nature Biotechnology. 1997;15(5):427–431. doi: 10.1038/nbt0597-427. [DOI] [PubMed] [Google Scholar]
- 42.Chen JW, Liu XP, Feng KJ, Liang Y, Jiang JH, Shen GL, Yu RQ. Biosens. Bioelectron. 2008;24(1):66–71. doi: 10.1016/j.bios.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Li W, Nie Z, Xu XH, Shen QP, Deng CY, Chen JH, Yao SZ. Talanta. 2009;78(3):954–958. doi: 10.1016/j.talanta.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Nutiu R, Li YF. J. Am. Chem. Soc. 2003;125(16):4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 45.Drake TJ, Tan WH. Appl Spectrosc. 2004;58(9):269a–280a. doi: 10.1366/0003702041959406. [DOI] [PubMed] [Google Scholar]
- 46.Nutiu R, Li YF. Angew Chem Int Edit. 2005;44(7):1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- 47.Yang LT, Ellington AD. Anal Biochem. 2008;380(2):164–173. doi: 10.1016/j.ab.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang CC, Chiu SH, Huang YF, Chang HT. Analytical Chemistry. 2007;79(13):4798–4804. doi: 10.1021/ac0707075. [DOI] [PubMed] [Google Scholar]
- 49.Barton GM, Medzhitov R. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(23):14943–14945. doi: 10.1073/pnas.242594499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell P. Nat Biotechnol. 2001;19(11):1013–1017. doi: 10.1038/nbt1101-1013. [DOI] [PubMed] [Google Scholar]
- 51.Cheung CY, Murthy N, Stayton PS, Hoffman AS. Bioconjugate Chem. 2001;12(6):906–910. doi: 10.1021/bc0100408. [DOI] [PubMed] [Google Scholar]
- 52.Huizenga DE, Szostak JW. Biochemistry. 1995;34(2):656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 53.Demers LM, Mirkin CA, Mucic RC, Reynolds RA, III, Letsinger RL, Elghanian R, Viswanadham G. Anal. Chem. 2000;72(22):5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 54.Traut TW. Molecular and Cellular Biochemistry. 1994;140(1):1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 55.Izyumov DS, Avetisyan AV, Pletjushkina OY, Sakharov DV, Wirtz KW, Chernyak BV, Skulachev VP. Biochimica Et Biophysica Acta-Bioenergetics. 2004;1658(1–2):141–147. doi: 10.1016/j.bbabio.2004.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.