Abstract
We demonstrate that novel oligonucleotide-modified gold nanoparticle probes hybridized to fluorophore-labeled complements can be used as both transfection agents and cellular “nano-flares” for detecting mRNA in living cells. Nano-flares take advantage of the highly efficient fluorescence quenching properties of gold, cellular uptake of oligonucleotide nanoparticle conjugates without the use of transfection agents, and the enzymatic stability of such conjugates, thus overcoming many of the challenges to creating sensitive and effective intracellular probes. Nano-flares exhibit high signaling, have low background fluorescence, and are sensitive to changes in the number of RNA transcripts present in cells.
Probes to visualize and detect intracellular RNA including those used for in situ staining,1 molecular beacons,2 and fluorescent resonance energy transfer (FRET) pairs3 are important tools to measure and quantify activity in living systems in response to external stimuli.4 However, these probes are often difficult to transfect, require additional agents for cellular internalization, and can be unstable in cellular environments. These factors can lead to a high background signal and the inability to detect targets. Here we show how novel oligonucleotide-modified gold nanoparticle probes hybridized to fluorophore complements can be used as both transfection agents and cellular “nano-flares” for visualizing and quantifying RNA in living cells. Nano-flares take advantage of the highly efficient fluorescence quenching properties of gold,5 cellular uptake of oligonucleotide nanoparticle conjugates without the use of transfection agents, and the enzymatic stability of such conjugates,6 thus overcoming many of the challenges to creating sensitive and effective intracellular probes. Specifically, nano-flares exhibit high signaling, have low background fluorescence, and are sensitive to changes in the number of RNA transcripts present in cells.
Approaches for visualizing RNA in cells typically utilize fluorophore-labeled synthetic oligonucleotides such as fluorescence resonance energy transfer (FRET) probes2 or molecular beacon reporters,3 which translate a specific RNA binding event into a fluorescent signal. However, the delivery of oligonucleotide-based reporters into cellular media and cells has proven to be a major challenge for intracellular detection. The cellular internalization of oligonucleotide-based probes typically requires transfection agents such as lipids7 or dendrimers8 that can be toxic or alter cellular processes. Furthermore, oligonucleotides are prone to degradation within cells,9 and in the case of fluorophore-labeled probes, this can lead to a high background signal that is indistinguishable from a true recognition event.10
The discovery and subsequent development of the oligonucleotide-nanoparticle conjugate have led to a variety of new opportunities in molecular diagnostics11 and materials design.12 Recently, it has been demonstrated that oligonucleotide-functionalized nanoparticles enter cells and can act as antisense agents to control gene expression.6 These “antisense particles” are not only delivery vehicles,13 but also single entity regulation and transfection agents that undergo facile cellular internalization, resist enzymatic degradation, and bind intracellular targets with affinity constants that are as much as two orders of magnitude greater than free oligonucleotides.14 Moreover, they can be easily modified with potent designer materials such as locked nucleic acids15 and are nontoxic under conditions required for gene regulation.
The nano-flares described herein are oligonucleotide functionalized nanoparticle conjugates designed to provide an intracellular fluorescence signal that correlates with the relative amount of a specific intracellular RNA. By utilizing nanoparticles densely functionalized with fluorophore-labeled oligonucleotides, we alleviate several difficulties commonly associated with intracellular RNA detection. These probes do not require microinjection or auxiliary transfection reagents to enter cells, are highly resistant towards enzymatic degradation like their therapeutic counterparts, and are non-toxic under the conditions studied. As a result of these properties, nano-flares can be used to quantify RNA levels and are less prone to non-specific signaling than free oligonucleotide probes.
Nano-flares take advantage of the unique optical properties of gold nanoparticles (Au NPs). Au NPs quench fluorescence with a greater efficiency5 and over greater distances16 than molecular quenchers. We designed nano-flares using 13 nm Au NPs, since this size particle is an efficient quencher, can be densely functionalized with oligonucleotides,12a and does not efficiently scatter visible light, which is important for designing optical probes with minimal interference. Au NPs were functionalized with thiolated oligonucleotides containing an 18-base recognition element to a specific RNA transcript (Figure 1c) via gold thiol bond formation.17 Oligonucleotide functionalized Au NPs were then allowed to hybridize with short cyanine (Cy5) dye-terminated reporter sequences capable of acting as “flares” when displaced by a longer target or target region (Figure 1a). In the bound state, the Cy5 fluorescence of the reporter strand is quenched due to proximity to the Au NP surface. In the presence of a target, the flare strand is displaced and liberated from the Au NP by forming the longer and more stable duplex between the target and the oligonucleotide-modified Au NP.
Figure 1.
(a) Nanoparticles functionalized with a recognition sequence are hybridized with a short complementary Cy5 labeled reporter strand, which is capable of being displaced by the target. (b) Fluorescence spectra of 1 nM nano-flares alone (green), in the presence of 1 mM target (red), and in the presence of 1 mM non-complementary sequence (blue). (c) Oligonucleotide sequences.
Testing the nano-flare design using synthetic complementary targets demonstrates that the probes respond with a 3.8 fold increase in fluorescence signal upon target recognition and binding (Figure 1b). In contrast, the signal does not change in the presence of a non-complementary target, and is of comparable magnitude to background fluorescence (Figure 1b). These results thus demonstrate that nano-flares are efficient at signaling the presence of a specific target.
Having established the signaling ability of nano-flare probes with synthetic targets, their ability to enter cells, and detect RNA targets was investigated. Nano-flares were designed to incorporate a complementary region for the survivin transcript, a target that has received significant attention due to its potential use in cancer therapeutics and diagnostics.18 The SKBR3 cell line (human breast cancer), which expresses a high number of surviving transcripts,2c was used as a model to test survivin-targeting nano-flares. As a control, a second probe containing a non-complementary sequence was prepared. The non-complementary probe was designed and determined to have similar background fluorescence, melting properties, and signaling ability as the survivin probe (Supporting Information).
Cells were cultured on glass microscope cover slips, incubated with nano-flares, and imaged using scanning confocal microscopy. SKBR3 cells treated with survivin nano-flares were highly fluorescent as compared to those treated with the non-complementary controls (Figure 2). To further confirm that this signaling is consistent with the presence of survivin, a C166 cell-line (mouse endothelial) was used as a control since it does not contain the human survivin transcript. C166 cells were treated with both the survivin and control probes. In this case, no distinguishable difference in the fluorescence of the cells was observed after treatment. These imaging results were consistent with reverse transcriptase PCR (RT-PCR) measurements (vide infra).
Figure 2.
Intracellular testing of nano-flares. Differential contrast and fluorescence image of survivin-expressing SKBR3 cells treated with survivin nano-flares (top left panel) and non-complementary nano-flares (top right panel). Analogously treated non survivin-expressing C166 cells (bottom panels). Scale bar is 20 mm. Flow cytometry data is shown below each image. The bold numbers to the right of the histogram are the total mean fluorescence of the cell populations. The background fluorescence (untreated cells) was 3.4, and 4.7 for the C166 and SKBR3 cells, respectively.
In order to quantify the intracellular signaling of the nano-flares, we examined cells treated with probes using analytical flow cytometry. Additionally, flow cytometry allows one to collect fluorescence data for a large population of cells. This eliminates variations and experimental artifacts that can be observed using techniques such as fluorescence imaging which only permit the examination of a small sample of cells. Cell-lines transfected with nano-flares showed uniform single populations of fluorescent cells, consistent with the greater than 99% cell penetration that we observe when transfecting antisense particles.6 Flow-cytometry revealed that SKBR3 cells treated with surviving nano-flares were highly fluorescent and 2.5 times more fluorescent than the population treated with non-complementary controls (Figure 2). For comparison, in C166 cell models, both probes resulted in a similarly low fluorescent signal. These flow cytometry experiments are in excellent agreement with confocal imaging and demonstrate the uniform cellular internalization and intracellular signaling of the nano-flares.
We next designed experiments to understand the unique properties of these probes in the context of intracellular detection experiments. We first compared the intracellular performance of nano-flares with a molecular beacon reporter delivered using Lipofectamine, a commercial transfection agent. Molecular beacons and nano-flares were introduced to SKBR3 cells (Supporting Information). A higher cell-associated fluorescence was observed with complementary probes using both nano-flares and molecular beacons. We then compared the background fluorescence contributed by the non-complementary probes (both molecular beacon and nano-flare). The fluorescence of the non-complementary molecular beacon probe is 1.9 times greater than that of the non-complementary nano-flare (Supporting Information). Since the difference between the background and signal is critical for accurate target detection, the lower background of nano-flares provides an important advantage when detecting intracellular targets.
We hypothesized that the greater signaling ability and low background of the nano-flares was both the result of efficient gold quenching and enhanced nuclease resistance, since it has been previously shown that immobilization of oligonucleotides on a nanoparticle surface provides stability in the presence of enzymes.6 To probe how enzymatic degradation leads to non-specific signaling, we incubated nano-flares with the endonuclease DNAse I (0.38 mg/L, significantly greater than what would be found in a cellular environment), and measured the rate of degradation by monitoring the increase in fluorescence signal as a function of time. The results of the assay reveal that the nano-flare is degraded at a normalized rate of 0.275 nmol min−1 under these conditions. In comparison, a molecular beacon is degraded at a normalized rate of 1.25 nmol min−1, approximately 4.5 times more rapidly than the nano-flare (Supporting Information). Since nuclease activity increases background fluorescence with a conventional probe, the reduced nuclease activity of the nano-flares leads to a system with lower background signal.
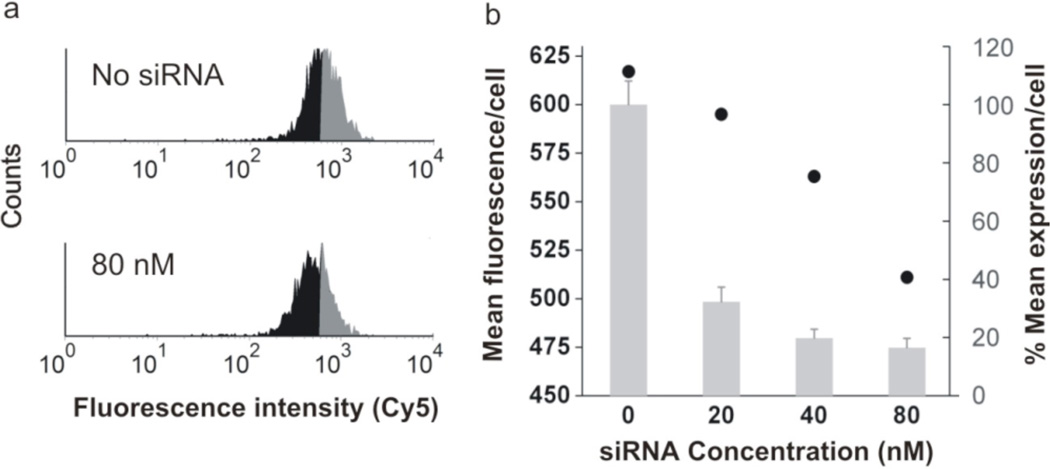
To demonstrate an application where the cellular entry, elevated signaling, and low background of the nano-flare translate into a high sensitivity for changes in intracellular amounts of RNA, we conducted siRNA knockdown experiments to reduce the levels of survivin RNA transcripts in the SKBR3 cell models. Cells were initially treated with siRNA against survivin, and the intracellular RNA levels were quantified using nano-flares and flow cytometry. We observe a siRNA concentration dependent shift in the fluorescence of the cell population as a function of the concentration of siRNA added to the cell culture (decreasing grey, increasing black, Figure 3a). To confirm that this shift was commensurate with a decrease in the number of surviving transcripts, RT-PCR measurements were conducted on samples treated with the same concentrations of siRNA. The decrease in the fluorescence signal within the population of cells is in agreement with the decrease in the number of survivin RNA copies as determined by RT-PCR measurements (Figure 3b). Differences in magnitude of decrease between nano-flares and RT-PCR measurements may be due to smaller pieces of mRNA that are still present when siRNA leads to degradation of long transcripts. Short mRNAs may be detected by nano-flares, but not amplified by RT-PCR. Taken together however, these results indicate that the nano-flares are sensitive to changes in the number of intracellular transcripts.
Figure 3.
Quantification of survivin knockdown using nano-flares. (a) Flow cytometry data collected on siRNA treated SKBR3 cells. The siRNA concentration is given in the graph to the left of the histogram. In the untreated sample, half of the population exhibiting equal or greater fluorescence than the mean is shaded grey. Treated samples show a smaller fraction of the cell population exhibiting the mean fluorescence (declining grey, increasing black). (b) Plot of mean fluorescence (black circles) and survivin expression (grey bar-graph) as a function of siRNA concentration. The RT-PCR reactions were conducted in triplicate, and the error bars shown above are the standard deviations of those measurements.
In summary, we have described a new class of intracellular probe termed “nano-flares.” Nano-flares are novel and potentially very useful since they are the only probe that combines cellular transfection, enzymatic protection, and RNA detection and quantification. In addition to their demonstrated use in the context of siRNA knockdown experiments, nano-flares will also be of utility in other areas such as cell sorting, gene profiling, and real-time drug validation studies. Finally, given the ability of these materials to also act as gene regulation agents,6,15 these probes may be easily adapted to simultaneously transfect, control and visualize gene expression in real time.
Supplementary Material
Acknowledgement
C.A.M. acknowledges a Cancer Center for Nanotechnology Excellence (CCNE) award for support of this research. C.A.M. is also grateful for a NIH Director’s Pioneer Award. D. S. S. was supported by the LUNGevity Foundation – American Cancer Society Postdoctoral Fellowship in Lung Cancer. H.D.H. acknowledges the U.S. Department of Homeland Security (DHS) for a Graduate Fellowship under the DHS Scholarship and Fellowship Program. Technical assistance was provided by James Marvin of The Flow Cytometry Core Facility of the Robert H. Lurie Comprehensive Cancer Center.
Footnotes
Supporting Information Available: Experimental details and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.(a) Femino A, Fay FS, Fogarty K, Singer RH. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]; (b) Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RHA. Nat. Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 2.(a) Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]; (b) Sokol DL, Zhang XL, Lu PZ, Gewitz AM. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Peng X-H, Cao Z-H, Xia J-T, Carlson GW, Lewis MM, Wood WC, Yang L. Cancer Res. 2005;65:1909–1917. doi: 10.1158/0008-5472.CAN-04-3196. [DOI] [PubMed] [Google Scholar]; (d) Perlette J, Tan WH. Anal. Chem. 2001;73:5544–5550. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]; (e) Nitin N, Santangelo PJ, Kim G, Nie SM, Bao G. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Santangelo PJ, Nix B, Tsourkas A, Bao G. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santangelo P, Nitin N, Bao G. Annals of Biomedical Engineering. 2006;34:39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 5.Dubertret B, Calame M, Libchaber A. J. Nat. Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 6.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 7.Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. J. Bio. Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 8.Kukowska-Latallo JF, Bielinska AU, Johnson J, Spindler R, Tomalia DA, Baker JR., Jr. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4897–4902. doi: 10.1073/pnas.93.10.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Opalinska JB, Gewirtz AM. Nat. Rev. Drug Disc. 2002;1:503–514. doi: 10.1038/nrd837. [DOI] [PubMed] [Google Scholar]
- 10.(a) Li JJ, Geyer R, Tan W. Nucleic Acids Res. 2004;28:e52. doi: 10.1093/nar/28.11.e52. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rizzo J, Gifford LK, Zhang X, Gewirtz AM, Lu P. Molecular and Cellular Probes. 2002;16:277–283. doi: 10.1006/mcpr.2002.0423. [DOI] [PubMed] [Google Scholar]
- 11.(a) Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]; (b) Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. J. Am. Chem. Soc. 1998;120:1959–1964. [Google Scholar]; (c) Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]; (d) He L, Musick MD, Nicewarner SR, Salinas FG, Benkovic SJ, Natan MJ, Keating CD. J. Am. Chem. Soc. 2000;122:9071–9077. [Google Scholar]; (e) Wang J, Xu D, Kawde A-N, Polsky R. Anal. Chem. 2001;73:5576–5581. doi: 10.1021/ac0107148. [DOI] [PubMed] [Google Scholar]; (f) Liu JW, Lu Y. J. Am. Chem. Soc. 2004;124:12298–12305. doi: 10.1021/ja046628h. [DOI] [PubMed] [Google Scholar]; (g) Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. J. Am. Chem. Soc. 2006;128:8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]; (h) Han MS, Lytton-Jean AKR, Oh BK, Heo J, Mirkin CA. Angew. Chem. Int. Ed. 2006;45:1807–1810. doi: 10.1002/anie.200504277. [DOI] [PubMed] [Google Scholar]
- 12.(a) Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]; (b) Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Jr, Shultz PG. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]; (c) Demers LM, Park SJ, Taton TA, Li Z, Mirkin CA. Angew. Chem. Int. Ed. 2003;40:3071–3073. doi: 10.1002/1521-3773(20010817)40:16<3071::AID-ANIE3071>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]; (d) Liu J, Lu Y. J. Am. Chem. Soc. 2005;127:12677–12683. doi: 10.1021/ja053567u. [DOI] [PubMed] [Google Scholar]; (e) Lee JS, Lytton-Jean AKR, Hurst SJ, Mirkin CA. Nano Lett. 2007;7:2112–2115. doi: 10.1021/nl071108g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Sandhu KK, McIntosh CM, Simard JM, Smith SW, Rotello VM. Bioconjugate Chem. 2002;13:3–6. doi: 10.1021/bc015545c. [DOI] [PubMed] [Google Scholar]; (b) Tkachenko AG, Xie H, Colman D, Glomm W, Ryan J, Anderson MF, Franzen S, Feldhein DL. J. Am. Chem. Soc. 2003;125:4700–4701. doi: 10.1021/ja0296935. [DOI] [PubMed] [Google Scholar]
- 14.Lytton-Jean AKR, Mirkin CA. J. Am. Chem. Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 15.Seferos DS, Giljohann DA, Rosi NL, Mirkin CA. ChemBioChem. 2007;8:1230–1232. doi: 10.1002/cbic.200700262. [DOI] [PubMed] [Google Scholar]
- 16.Dulkeith E, Ringler M, Klar TA, Feldman J, Javier AM, Parack WJ. Nano Lett. 2005;5:585–589. doi: 10.1021/nl0480969. [DOI] [PubMed] [Google Scholar]
- 17.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Chem. Rev. 2005;105:1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 18.Altieri DC. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.