Abstract
Two-component triblock magnetic nanorods with gold end blocks and nickel interior blocks have been synthesized and used as affinity templates for the simultaneous and efficient separation of a three component protein mixture. The gold blocks were selectively functionalized with 11-amino-1-undecanethiol, and then glutaraldehyde was used to covalently attach nitrostreptavidin to them. His-tagged proteins bind to the nickel block and biotin-tagged proteins bind to the functionalized gold ends, allowing one to separate a mixture of three proteins with a single material. Each surface bound protein can be released selectively using imidazole for the His-tagged protein and biotin for the biotinylated protein.
Keywords: Biotin tagged protein, Histidine tagged protein, Magnetic nanorod, Protein separation
Introduction
The development of efficient methods for the purification of recombinant proteins from cell extracts is very important for the field of biotechnology because of the need for purified protein in applications ranging from diagnostics to therapeutics.1-18 A number of generic affinity-based purification methods for recombinant proteins have been developed by proteomic researchers.7-18 These methods are based upon the use of specific antibodies or affinity tags that are covalently attached to the protein of interest. Usually short peptides with specific molecular recognition properties such as a maltose binding protein,8 streptag,9 cellulose binding domain,10 glutathione S-transferase,11 polyhistidine,12-14 and biotin15-20 are used as such tags. Among the affinity-based purification methods for recombinant proteins, Ni-histidine12-14 and biotin-streptavidin (or avidin)15-18 systems are particularly popular. These tags can be readily incorporated into desired proteins by the expression of the target gene in microorganism cells using commercial expression vectors,12-13,21 which typically include a gene encoding polyhistidine22 or one encoding for a specific peptide15,21 that can be biotinylated in vivo by bacterial BirA enzyme (protein-biotin ligase). Biotin tagging, which involves the in vivo biotinylation of the protein through bacterial, yeast, and mammalian cells, offers an advantage over other tagging methods including histidine for the purification of recombinant proteins in terms of reduced background binding since there are very few naturally occurring biotinylated proteins.15 When biotin tags are used in protein mixture separation schemes, nitrated streptavidin is typically used to ensure reversible binding between the streptavidin modified stationary support and the captured biotinylated proteins. 23-24 Native streptavidin binds biotin in an irreversible fashion, which prevents the release of biotin-tagged proteins in the separation procedure. The biotin-labeled proteins are released from the stationary phase by eluting them with a solution with a high concentration of biotin.16-17, 23 In the case of His-tagged proteins, Ni columns are often used as a support to selectively bind the His-tagged proteins. Protein release is effected by a change in pH or by eluting the column with a solution with a high concentration of imidazole. 12-14
Nanomaterials such as nanoparticles, nanotubes, and nanorods have been extensively used as complementary candidates for the effective separation of biomaterials 25-30 as well as in the field of biomedical diagnostics and therapeutics. 31-37 Previously we reported that two-component triblock magnetic nanorods with Au end blocks and a Ni interior block can be used as affinity templates for simple and efficient separation of His-tagged proteins from other proteins without affinity tags.25 The Ni portion of the nanorods not only provides a docking site for His-tagged proteins but it is also ferromagnetic, which enables easy isolation under a magnetic field. We hypothesized that if Au end blocks of the nanorods are modified with specific functionalities which can recognize target proteins, two different kinds of proteins with different affinity tags can be simultaneously and selectively separated from a protein mixture. Herein, we demonstrate how one can use two-component triblock magnetic nanorods with Au ends and Ni interior blocks to magnetically separate biotin-tagged proteins and His-tagged proteins from a mixed solution thereby using a single material to separate multiple proteins.
Experimental Section
Synthesis of multicomponent magnetic nanorods
A thin layer of silver (200 nm) was evaporated on one side of an alumina filter (Whatman International Ltd, d = 13 mm, pore size = 20 nm; the pore diameter in the central region of the filter is substantially larger than the quoted 20 nm) and served as a cathode in a three electrode electrochemical cell after making physical contact with aluminum foil. Platinum wire was used as a counter electrode, and a commercial Ag/AgCl reference electrode (Bio Analytical System, Inc) was used in all experiments. The nano-pores were filled with Ag (Technic ACR silver RTU solution from Technic, Inc.) at a constant potential, −0.9 V vs Ag/AgCl, by passing 1.1 C/cm2 for 30 min. An Au block was then electroplated from Orotemp 24 RTU solution (Technic, Inc.) at −0.95 V vs Ag/AgCl followed by a Ni block from nickel sulfamate RTU solution (Tecnnic Inc.) at −0.9 V vs Ag/AgCl. The procedure involving Au was repeated to form the final capping block. Each segment length was controlled by monitoring the charge passed through the membrane. The first 2 μm (± 0.2) long block of Au was generated by passing 1.6 C. The 6 μm (± 0.4) block of Ni required 10.8 C, and the final 2 μm (± 0.2) Au block involved 1.6 C (the exposed membrane surface area is ~1 cm2). The Ag backing and alumina membrane were dissolved with concentrated nitric acid and 3 M sodium hydroxide solutions, respectively.
Preparation of nitrostreptavidin
Nitrostreptavidin was prepared according to published procedures.23 A sample of streptavidin (2.5 mg in 1 ml of 50 mM Tris buffer, pH 8) was treated with tetranitromethane (50 mM) for 50 min at room temperature. The sample was dialyzed overnight: once against 4 liters of 1 M NaCl and twice against deionized distilled water.
Preparation of the nanorods modified with nitrostreptavidin for bioseparation
Nanorods were repeatedly washed with alcohol to remove contaminants from their surfaces. The Au ends of triblock nanorod structures (~109) were modified with 11-amino-1-undecanethiol in methanol (1 mM) overnight at room temperature. They were subsequently washed with absolute ethanol, distilled water, and phosphate buffered saline (0.15 M PBS, pH 7.4), respectively. The rinsing solutions were sonicated for 30 sec every 1 hr to keep the rods dispersed. The primary amine groups on the surfaces of the Au end blocks were activated with 8 % glutaraldehyde in 0.15 M PBS (pH 7.4). Nitrostreptavidin was attached to the Au end blocks of the nanorods by the incubation of the activated nanorods in a nitrostreptavidin solution (1 μM in 0.15 M PBS, pH 7.4) overnight at room temperature, followed by washing the nanorods with 0.1 % Tween 20 in 0.15 M PBS (pH 7.4). The solution was sonicated for 30 sec every 1 hr. The residual amine groups of the 11-amino-1-undecanethiol modified rods were inactivated with ethanolamine (0.2 M). The Schiff bases (formed by the reaction between glutaraldehyde and the primary amines of ethanolamine and nitrostreptavidin) were reduced to secondary amines using sodium borohydride (0.1 M).
Selective blocking of unmodified recognition sites of nitrostreptavidin
Although streptavidin was modified with tetranitromethane to form nitrospretavidin, the unmodified sites originating from incomplete modification can potentially pose problems in the subsequent separation steps. Therefore, the unmodified recognition sites of the nitrostreptavidin immobilized on the nanorods were selectively blocked with excess free biotin (5 mM in 0.15 M PBS, pH 7.4). Biotin that occupied binding sites of nitrostreptavidin could be selectively released using 50 mM sodium carbonate buffer, pH 10. Biotin molecules, which occupy the unmodified biotin-binding sites, are retained under these conditions.23-24
Selective binding and release of tagged proteins
Nanorods with Au end blocks functionalized with nitrostreptavidin and Ni interiors were incubated in a mixture of tagged proteins (0.15 M PBS at pH 7.4, 0.1 % Tween 20, 0.1 mM imidazole) for 1hr at room temperature. Then, the nanorods were rinsed with PBS buffer. During each rinsing step, the rods were magnetically separated from the supernatant with a BioMag® microcentrifuge tube separator (Polysciences, Inc.). His-tagged proteins bound to the Ni interior blocks were released by copious rinsing with a concentrated solution of imidazole (500 mM in 0.15 M PBS, pH 7.4). The nanorods were magnetically separated from supernatant, and the supernatant containing the released proteins was collected. Biotin-tagged proteins were released from the Au blocks by rinsing with a solution of biotin (5 mM in 0.15 M PBS, pH 7.4). The nanorods were separated with a bar magnet, and the supernatant was collected. As synthesized Au-Ni-Au rods are very stable at room temperature in PBS solution. We also investigated the stability of blocks modified with nitrostreptavidin and found that they are stable over at least a one week time span when stored at 4°C.
Spectroscopy and microscopy
Scanning electron microscopy (SEM) images were obtained using a Hitachi S-4500 cFEG SEM (Electron Probe Instruments Center (EPIC), NUANCE, Northwestern University). Optical and fluorescence images were obtained using a Zeiss Axiovert 100A inverted optical/fluorescence microscope (Thomwood, NY) equipped with a Penguin 600CL digital camera and StreamPix software. Fluorescence emission spectra were obtained on a Jobin Yvon SPEX Fluorolog fluorometer using quartz cells (10 × 4 mm light path). UV-vis absorption measurements were performed with a Varian Cary 5000 UV-Vis-NIR spectrophotometer.
Results and Discussion
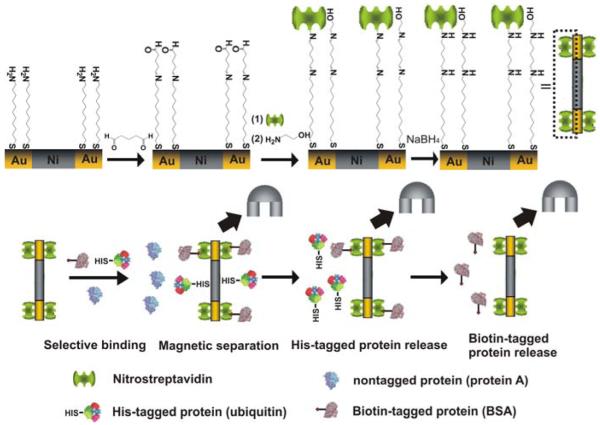
As proof-of-concept for our novel protein separation scheme, we evaluated its effectiveness with a mixture consisting of fluorophore labeled protein A (1.4 μM, Alexa 350, blue), ubiquitin (3.2 μM, Alexa 488, green) and bovine serum albumin (1.4 μM, BSA, Alexa 594, red), Scheme 1. Ubiquitin and BSA were tagged with histidine and biotin, respectively, and protein A was unmodified. In our scheme, the His-tagged ubiquitin and the biotin-tagged BSA should selectively bind to the nitrostreptavidin modified Au-Ni-Au nanorods. The mixture of proteins fluoresces strongly in all three spectral regions associated with the three dyes, Figure 1A (I).
Scheme 1.
Procedure for the simultaneous and selective separation of His-tagged proteins and biotin-tagged proteins from nontagged proteins.
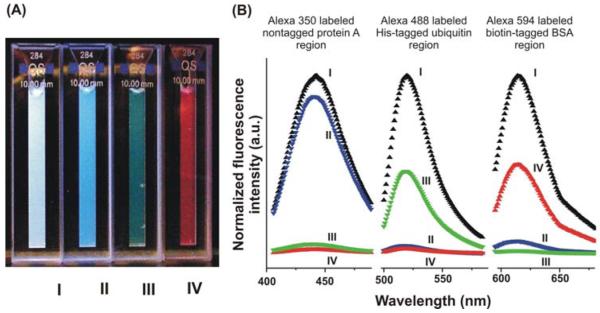
Figure 1.
(A) A photograph of four protein solutions in cuvettes I: A solution mixture containing Alexa 488 labeled His-tagged ubiquitin, Alexa 594 labeled biotin-tagged BSA, and Alexa 350 labeled nontagged protein A before separation, II: After separation of Alexa 350 labeled non-tagged protein A from two other proteins with nanorods, III: The green solution formed after release of Alexa 488 labeled His-tagged ubiquitin in the elution buffer from nanorods, IV: The red solution formed after release of Alexa 594 labeled biotin-tagged BSA in the elution buffer from nanorods. (B) Fluorescence spectra corresponding to Figure 1A. The fluorescence spectrum of solution (I) is in black. The fluorescence spectrum of solution (II) is in blue. The fluorescence spectrum of solution (III) is in green. The fluorescence spectrum of solution (IV) is in red. (Note: each spectrum is normalized to the fluorescence intensity of the initial solution containing all proteins before separation).
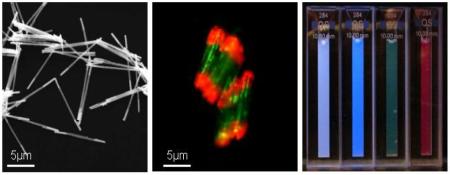
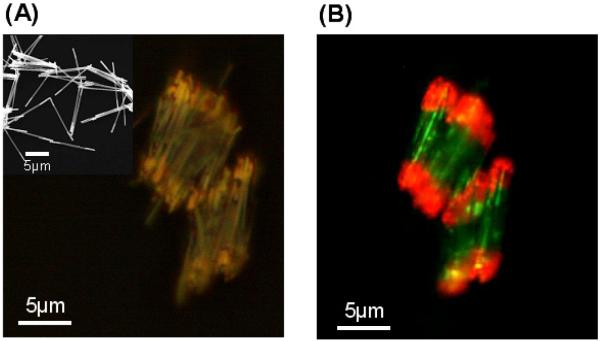
Prior to evaluating the protein mixture separation scheme, we characterized the rods through scanning electron microscopy (SEM), optical microscopy, and fluorophore labeling experiments, Figure 2A and B. The SEM and optical microscopy images allow one to clearly visualize the Au and Ni segments of the rods, Figure 2A. Rods modified with nitrostreptavidin on their Au ends were treated with green fluorescein-tagged poly-histidine (His×6) and red atto 590-tagged biotin and characterized by fluorescence microscopy, Figure 2B. Fluorescence microscopy images show that the red dye-labeled biotin cleanly binds to the nitrostreptavidin modified Au segments, and the green-dye labeled poly-histidine binds to the interior Ni segments. Rods that have not been modified with nitrostreptavidin show no evidence for biotin binding to the Au ends.
Figure 2.
(A) Optical microscopy image of Au-Ni-Au rods: inset shows FESEM image of Au-Ni-Au rods. (B) Fluorescence microscopy image of Au-Ni-Au after modification of Ni interior and Au end blocks with fluorescein-tagged poly-His and atto 590-tagged biotin, respectively.
In a typical protein mixture separation experiment, the three segment Au-Ni-Au rods (yield = 109-1010 rods), determined by SEM pore density measurements) were selectively functionalized on the Au ends with 11-amino-1-undecanethiol, Scheme 1. The amine-ends were capped with glutaraldehyde and subsequently functionalized with nitrostreptavidin. Unreacted aldehydes were converted to alcohols with ethanolamine. These rods were then added to a PBS solution (pH 7.4) containing the mixture of proteins. The spectrum of the mixture before the addition of nanorods exhibits three bands at λmax = 442, 519, and 617 nm that correspond to the three dye labels, Figure 1B. After addition of the nanorods, the solution is allowed to stand for 60 min, and a magnet is then used to attract the rods to the reaction vessel sidewall. Fluorescence spectroscopy of the supernatant shows that there is a 95 % (λmax = 519 nm) and 93 % (λmax = 617 nm) decrease in the signals for the dye labels associated with the His-tagged proteins and biotin-tagged proteins, respectively, and only 10 % for the protein without the affinity tag (λmax = 442 nm), Figure 1B. The supernatant containing predominantly (almost exclusively) protein A is decanted and characterized by fluorescence spectroscopy, Figure 1A (II) and 1B. The supernatant fluoresces strongly in the blue as one would expect for the blue-dye labeled protein A but not in the red or green. This demonstrates clean separation of the His-tagged ubiquitin and biotin-tagged BSA from protein A.
The rods can be moved to a second vial and suspended in PBS. Addition of imidazole results in the displacement of the His-tagged ubiquitin from the Ni segment over 15 min. A bar magnet can be used again to collect the rods and separate them from the solution containing the ubiquitin. The supernatant fluoresces strongly in the green portion of the spectrum but not in the red or blue, consistent with clean separation and isolation of the ubiquitin from the other two proteins, Figure 1A (III) and 1B. From fluorescence spectroscopy, we estimate that the solution consists of 94.1 % ubiquitin, 1.5 % BSA and 4.4 % protein A. The ubiquitin isolated in this procedure is 47.4 % of the total ubiquitin in the original mixture.
The rods are then redispersed in PBS buffer and presumably have only biotin-tagged BSA attached to their surfaces. Addition of biotin to the solution effects release of the BSA, and the rods are separated from the supernatant with the bar magnet. Consistent with this conclusion, the supernatant fluoresces strongly in the red but not the blue and the green, Figure 1A (IV) and 1B. This solution consists of 85.6 % BSA, 10.5 % ubiquitin and 3.9 % protein A. The total BSA isolated in this procedure is 50.0 % of the BSA in the original mixture.
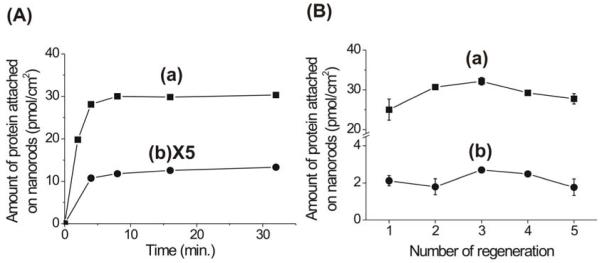
The binding kinetics of His-tagged proteins and biotin-tagged proteins to the nanorods were studied by UV-vis spectroscopy by monitoring the change in absorbance at 280 nm as a function of exposure time to the nanorods. The band at 280 nm is an electronic transition associated with the aromatic rings that are part of the amino acids (e.g. phenylalanine, tryptophan, histidine, and tyrosine) that make up the proteins.38 In a typical experiment, triblock Au-Ni-Au nanorods are added to His-tagged ubiquitin (100 nmol/mL) and biotin-tagged BSA (10 nmol/mL), respectively. For the biotin-tagged proteins, the Ni surface of the nanorods was chemically passivated with imidazole in order to minimize nonspecific binding of the biotin-tagged proteins to the Ni surface of the nanorods. The nanorods were saturated with His-tagged ubiquitin and biotin-tagged BSA within 8 and 16 min, respectively, as evidenced by the decrease in absorbance at 280 nm, Figure 3A. Since in affinity chromatography adsorption can be slow because of the low flux of sample passing through a packed bed of porous particles, it is remarkable that the surface of the nanorods in this experiment can be saturated with each protein within minutes.
Figure 3.
(A) A plot of surface concentration of His-tagged (a) and biotin tagged (b) proteins as a function of exposure time to nanorods. (B) A plot of surface concentration of His-tagged (a) and biotin tagged (b) proteins as a function of nanorod use.
The recyclability of the nanorods was also examined by repeated application of His-tagged proteins and biotin-tagged proteins to a single batch of triblock nanorods. In a typical experiment, a batch of nanorods was added to a His-tagged ubiquitin solution (60 nmol/mL), and after 60 min the nanorods were moved to the side walls with a magnetic field. The resulting supernatant was studied by UV-vis spectroscopy, and the amount of His-tagged ubiquitin adsorbed to the Ni segments was determined to be 28.9 ± 2.7 pmol/cm2 based on the decreased peak intensity at 280 nm and the physical dimensions of the nanorods (7.5 μm2 surface area for each Ni block). The nanorods were then rinsed with buffer solution (0.1 M glycine·HCl, pH 2.8)25, 39 in order to release the protein and regenerate a clean Ni surface. The same experiment was repeated 5 times with the same batch of rods. The plot of surface adsorbed protein as a function of nanorod use shows that the amount of His-tagged ubiquitin attached on the nanorods is fairly constant for each cycle (Figure 3B, (a)).
We performed the similar experiments with biotin-tagged BSA, and obtained a similar trend, Figure 3B, (b). In this case, the Ni surface of the nanorods was chemically passivated with imidazole in order to minimize nonspecific binding of the biotin-tagged proteins to the Ni surface of the nanorods. A batch of nanorods was added to biotin-tagged BSA solution (4.5 nmol/mL), and the nanorods were moved to the side walls by a magnetic field. The resulting supernatant was studied by UV-vis spectroscopy, and the amount of biotin-tagged BSA adsorbed to Au segments was determined to be 2.1 ± 0.4 pmol/cm2 based on the decreased peak intensity at 280 nm and the physical dimensions of the nanorods (6.5 μm2 surface area for each Au block). The identical batch of nanorods was rinsed with buffer solution (50 mM of sodium carbonate buffer, pH 10)23 to regenerate the nitrostreptavidin on the Au segments. The experiment was repeated 5 times with similar results (Figure 3B, (b)).
Conclusion
We have developed chemistry for modifying the Au ends of two-component triblock (Au-Ni-Au) magnetic nanorods with nitrostreptavidin and shown that these can be used for separation of three component mixtures, two of which can be selectively removed based upon chemical affinity for different portions of the nanorod structure. The ferromagnetic Ni portion of these multiblock nanostructures serves two roles by providing a docking site for His-tagged proteins and imparting magnetic properties for easy separation in a magnetic field. Our novel method demonstrates the potential for chemically tailored nanorod modification in protein mixture separation protocols and provides an alternative to the currently used affinity chromatography protocols. This method of using multiblock nanostructures as separation agents could be very useful in terms of separating small amounts of proteins. The concentration of the multiblock nanorods can be easily adjusted, depending on protein concentration, in order to maximize separation yield.
The easy recovery of the nanorods with a magnetic field allows for rapid and effective protein separation because it provides pseudo-homogeneous mixing between the nanostructure and sample solutions. The approach is limited at present in terms of the quantity of proteins that can be separated due to the capacity of the currently used solid-templates. This problem will be overcome as advances are made in the solution phase synthesis of nanorod structures.
Acknowledgement
C.A.M. acknowledges NSF, AFOSR, and NIH for generous financial support. C.A.M is also grateful for a NIH Director’s Pioneer Award. B.-K. Oh and S. W. Lee acknowledge the Korea Research Foundation Grant funded by the Korean Government for post-doctoral fellowships (grant no. M01-2003-000-20168-0, M01-2003-000-20278-0).
References
- (1).Dyr JE, Suttnar J. J. Chromatogr. B. 1997;699:383–401. doi: 10.1016/s0378-4347(97)00201-6. [DOI] [PubMed] [Google Scholar]
- (2).Linder MB, Qiao M, Laumen F, Selber K, Hyytiä T, Nakari-Setälä T, Penttilä ME. Biochemistry. 2004;43:11873–11882. doi: 10.1021/bi0488202. [DOI] [PubMed] [Google Scholar]
- (3).Fexby S, Bülow L. Trends Biotechnol. 2004;22:511–516. doi: 10.1016/j.tibtech.2004.08.005. [DOI] [PubMed] [Google Scholar]
- (4).Meyer DE. Nat. Biotechnol. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- (5).van Reis R, Zydney AL. Curr. Opin. Biotechnol. 2001;12:208–211. doi: 10.1016/s0958-1669(00)00201-9. [DOI] [PubMed] [Google Scholar]
- (6).Peng CC, Chen JCF, Shyu DJH, Chen MJ, Tzen JTC. J. Biotechnol. 2004;111:51–57. doi: 10.1016/j.jbiotec.2004.03.013. [DOI] [PubMed] [Google Scholar]
- (7).Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- (8).Maina CV, Riggs PD, Grandea AG, III, Slatko BE, Moran LS. Gene. 1988;74:365–373. doi: 10.1016/0378-1119(88)90170-9. [DOI] [PubMed] [Google Scholar]
- (9).Skerra A, Schmidt TMG. Biomol. Eng. 1999;16:79–86. doi: 10.1016/s1050-3862(99)00033-9. [DOI] [PubMed] [Google Scholar]
- (10).Ong E, Greenwood JM, Gilkes NR, Kilburn DG, Miller RC, Warren RA. Trends Biotechnol. 1989;7:239–243. [Google Scholar]
- (11).Smith DB, Johnson KS. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- (12).Stiborova H, Kostal J, Mulchandani A, Chen W. Biotechnol. Bioeng. 2003;82:605–611. doi: 10.1002/bit.10609. [DOI] [PubMed] [Google Scholar]
- (13).Noubhani AM, Dieryck W, Bakalara N, Latxague L, Santarelli X. J. Chromatogr. B. 2003;790:153–159. doi: 10.1016/s1570-0232(03)00088-6. [DOI] [PubMed] [Google Scholar]
- (14).Clemmit RH, Chase HA. Biotechnol. Bioeng. 2000;67:206–216. doi: 10.1002/(sici)1097-0290(20000120)67:2<206::aid-bit10>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- (15).de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Stolz J, Demar BD, Sauer N. FEBS Lett. 1995;377:167–171. doi: 10.1016/0014-5793(95)01333-4. [DOI] [PubMed] [Google Scholar]
- (17).Tucker J, Grisshammer R. Biochem. J. 1996;317:891–899. doi: 10.1042/bj3170891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mascagni P, Ball HL, Bertolini G. Analytica Chimica Acta. 1997;352:375–385. [Google Scholar]
- (19).Tatsumi H, Fukuda S, Kikuchi M, Koyama Y. Anal. Biochem. 1996;243:176–180. doi: 10.1006/abio.1996.0498. [DOI] [PubMed] [Google Scholar]
- (20).Smith PA, Tripp BC, DiBlasio-Smith EA, Lu Z, LaVallie ER, McCoy JM. Nucleic Acids Res. 1998;26:1414–1420. doi: 10.1093/nar/26.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Cress D, Shultz J, Breitlow S. Promega Notes Magazine. 1993;42:02. [Google Scholar]
- (22).pET system manual. Novagen, Inc.; WI, USA: 2003. [Google Scholar]
- (23).Morag E, Bayer EA, Wilchek M. Biochem. J. 1996;316:193–199. doi: 10.1042/bj3160193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Morag E, Bayer EA, Wilchek M. Anal. Biochem. 1996;243:257–263. doi: 10.1006/abio.1996.0514. [DOI] [PubMed] [Google Scholar]
- (25).Lee K-B, Park S, Mirkin CA. Angew. Chem., Int. Ed. 2004;43:3048–3050. doi: 10.1002/anie.200454088. [DOI] [PubMed] [Google Scholar]
- (26).Kuhara M, Takeyama H, Tanaka T, Matsunaga T. Anal. Chem. 2004;76:6207–6213. doi: 10.1021/ac0493727. [DOI] [PubMed] [Google Scholar]
- (27).Bucak S, Jones DA, Laibinis PE, Hatton TA. Biotechnol. Prog. 2003;19:477–484. doi: 10.1021/bp0200853. [DOI] [PubMed] [Google Scholar]
- (28).Khng HP, Cunliffe D, Davies S, Turner N, Vulfson EN. Biotechnol. Bioeng. 1998;60:419–424. [PubMed] [Google Scholar]
- (29).Son SJ, Reichel J, He B, Schuchman M, Lee SB. J. Am. Chem. Soc. 2005;127:7316–7317. doi: 10.1021/ja0517365. [DOI] [PubMed] [Google Scholar]
- (30).Mitchell DT, Lee SB, Trofin L, Li N, Nevanen TK, Söderlund H, Martin CR. J. Am. Chem. Soc. 2002;124:11864–11865. doi: 10.1021/ja027247b. [DOI] [PubMed] [Google Scholar]
- (31).Nam J-M, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- (32).Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- (33).Cui Y, Wei Q, Park H, Lieber CM. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- (34).Zhao X, Tapec-Dytioco R, Tan W. J. Am. Chem. Soc. 2003;125:11474–11475. doi: 10.1021/ja0358854. [DOI] [PubMed] [Google Scholar]
- (35).Niemeyer CM, Ceyhan B. Angew. Chem. Int. Ed. 2001;40:3685–3688. doi: 10.1002/1521-3773(20011001)40:19<3685::aid-anie3685>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- (36).Penn SG, He L, Natan MJ. Curr. Opin. Chem. Biol. 2003;7:609–615. doi: 10.1016/j.cbpa.2003.08.013. and references therein. [DOI] [PubMed] [Google Scholar]
- (37).Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- (38).Stoscheck CM. Methods in Enzymology. 1990;182:50–69. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- (39).Godat B, Orr L, Simpson D, Johnson T, Kobs G. MagneHis™ protein purification system technical manual. Promega, corp.; WI, USA: 2003. [Google Scholar]