Abstract
KIBRA has recently been identified as a gene associated with human memory performance. Despite the elucidation of the role of KIBRA in several diverse processes in non-neuronal cells, the molecular function of KIBRA in neurons is unknown. We found that KIBRA directly binds to the protein interacting with C-kinase 1 (PICK1) and forms a complex with α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate receptors (AMPARs), the major excitatory neurotransmitter receptors in the brain. KIBRA knockdown accelerates the rate of AMPAR recycling following N-methyl-D-aspartate receptor induced internalization. Genetic deletion of KIBRA in mice impairs both long-term depression and long-term potentiation at hippocampal Schaffer collateral-CA1 synapses. Moreover, KIBRA knockout mice have severe deficits in contextual fear learning and memory. These results indicate that KIBRA regulates higher brain function by regulating AMPAR trafficking and synaptic plasticity.
Introduction
α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate receptors (AMPARs) mediate the majority of fast excitatory synaptic transmission in the brain. The modulation of AMPAR membrane trafficking and synaptic targeting is critical for several forms of synaptic plasticity thought to be cellular mechanisms underlying learning and memory (Malinow and Malenka, 2002; Shepherd and Huganir, 2007). AMPARs are heterotetrameric assemblies of four highly related subunits, GluA1-4 (Shepherd and Huganir, 2007). AMPAR trafficking into and out of the synapse is highly dynamic and is modulated by subunit specific AMPAR-interacting proteins that link neuronal signaling pathways to the insertion and retrieval of AMPARs from synaptic sites (Shepherd and Huganir, 2007). The synaptic PDZ domain-containing protein, protein interacting with C-kinase 1 (PICK1), directly interacts with the C-terminus of GluA2/3 AMPAR subunits and is required for hippocampal long-term potentiation (LTP) and long-term depression (LTD), cerebellar LTD, Ca2+-permeable AMPAR plasticity, and mGluR LTD in the perirhinal cortex (Clem et al., 2010; Gardner et al., 2005; Jo et al., 2008; Liu and Cull-Candy, 2005; Steinberg et al., 2006; Terashima et al., 2008; Volk et al., 2010; Xia et al., 1999). Genetic deletion of PICK1 has revealed its crucial role in hippocampal synaptic plasticity (Terashima et al., 2008; Volk et al., 2010) and inhibitory avoidance learning (Volk et al., 2010). Recent studies have shown that PICK1 regulates AMPAR membrane trafficking by retaining GluA2-containing AMPARs in intracellular pools and inhibiting their recycling to the plasma membrane (Citri et al., 2010; Lin and Huganir, 2007); however, the mechanisms by which PICK1 regulates the dynamic bidirectional trafficking of AMPARs are complex and remain unclear.
Advances in genome-wide screening methods have enabled searches for genes associated with higher brain function. A recent study identified KIBRA as a gene linked with human memory performance (Papassotiropoulos et al., 2006). Carriers of a T to C single nucleotide polymorphism in the ninth intron of KIBRA were found to perform better on several episodic memory tasks (Papassotiropoulos et al., 2006). Importantly, links between this gene and human memory have been highly reproducible by other groups using different subject populations (Almeida et al., 2008; Bates et al., 2009; Schaper et al., 2008; Schneider et al., 2010). The T allele of KIBRA is associated with superior memory in healthy subjects and is also protective against Alzheimer's disease (Corneveaux et al., 2008). While these reports are very compelling, they raise the important question of how KIBRA controls higher brain function at the molecular level.
KIBRA is highly expressed in kidney and in memory-related brain regions (Johannsen et al., 2008; Kremerskothen et al., 2003; Papassotiropoulos et al., 2006). In podocytes, KIBRA interacts with the polarity protein PATJ and synaptopodin and modulates directional cell migration (Duning et al., 2008). In Drosophila, KIBRA acts synergistically with Merlin and Expanded as an upstream activator of the Hippo kinase signaling cascade, a pathway involved in organ size control (Baumgartner et al., 2010; Genevet et al., 2010; Yu et al., 2010). The interaction between KIBRA and dynein light chain 1 is critical for linking microtubule motors to other binding partners of KIBRA, which include atypical PKCs, polarity proteins, and vesicular trafficking components (Rayala et al., 2006; Rosse et al., 2009; Traer et al., 2007). The finding that the atypical kinase PKC/Mζ binds to and phosphorylates KIBRA in vitro is of particular interest as PKMζ is implicated in long-term maintenance of synaptic plasticity and memory retention (Buther et al., 2004; Drier et al., 2002; Sacktor et al., 1993). Although a molecular role for KIBRA in distinct contexts and cell types has begun to be defined, its function in neurons is unknown.
Here we report that KIBRA directly binds PICK1 in vitro and in vivo. In addition, KIBRA interacts with GluA1, GluA2, and several other synaptic proteins in an in vivo protein complex. Using pHluorin-GluA2 fusion proteins to monitor live membrane trafficking of AMPARs following N-methyl-D-aspartate receptor (NMDAR) activation, we found that knockdown (KD) of KIBRA significantly accelerates the rate of pH-GluA2 recycling. Furthermore, we show that LTP and LTD in the adult KIBRA knockout (KO) mouse are reduced while plasticity in juveniles is intact. Finally, we demonstrate that KIBRA is essential for trace and contextual fear conditioning in adult mice. Taken together, our data indicate that KIBRA plays an important role in regulating AMPAR trafficking underlying synaptic plasticity and learning.
Results
To further study the role of PICK1 in synaptic plasticity we performed a yeast two-hybrid screen in a rat hippocampus cDNA library using a PICK1 fragment (aa1-358) as bait and isolated two clones that encode a small region of KIBRA (Fig. 1A). The involvement of KIBRA in higher brain function as well as its binding partners and expression pattern made it an attractive target for further study (Almeida et al., 2008; Bates et al., 2009; Corneveaux et al., 2008; Johannsen et al., 2008; Kremerskothen et al., 2003; Papassotiropoulos et al., 2006; Schaper et al., 2008; Schneider et al., 2010).
Fig. 1.
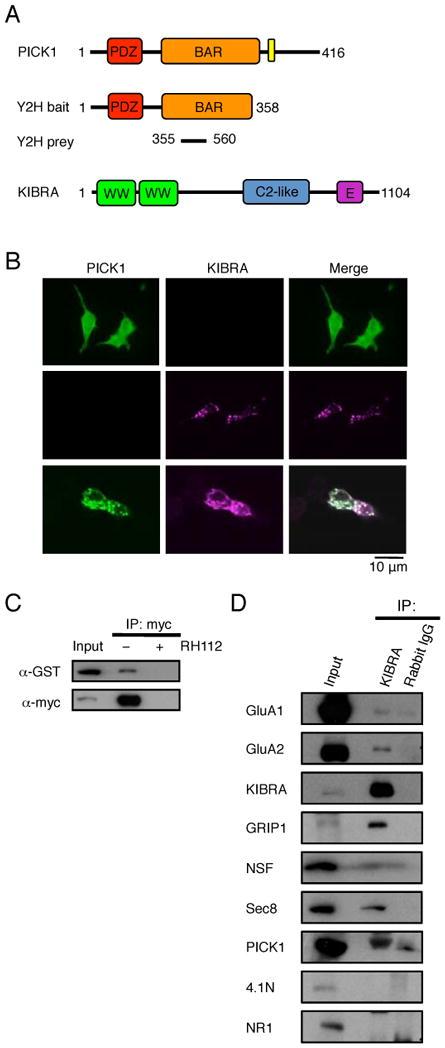
KIBRA directly binds PICK1 and forms a complex with GluA1/2 AMPAR subunits in vivo. (A) Schematic diagrams of yeast two-hybrid bait, prey and full-length proteins. (B) Representative images of HEK293T cells transfected with either HA-PICK1 or GFP-KIBRA alone or in combination. (C) HEK293T cells were transfected with GST-PICK1 and myc-KIBRA, lysed and immunoprecipitated with anti-myc antibodies in the presence or absence of antigenic blocking peptide. Proteins were resolved by Western blot and probed with anti-myc or anti-GST antibodies. (D) Mouse P2 brain fractions were immunoprecipitated with either anti-KIBRA antibodies or normal rabbit IgG. Samples were subjected to Western blot analyses using specific antibodies as indicated.
To examine the KIBRA-PICK1 interaction in mammalian cells, we transfected HEK293T cells with full-length constructs encoding HA-PICK1 and GFP-KIBRA individually and in combination. Overexpression of HA-PICK1 alone showed a diffuse cytoplasmic distribution (Xia et al., 1999); however, when cotransfected with GFP-KIBRA, the two proteins colocalized in large cytoplasmic clusters observed upon transfection of GFP-KIBRA alone (Fig. 1B). In addition, GST-PICK1 was coimmunoprecipitated with myc-KIBRA when coexpressed in HEK293T cells and this immunoprecipitation was abolished in the presence of myc epitope blocking peptide, confirming the specificity of the interaction between KIBRA and PICK1 in vitro (Fig.1C). Immunoprecipitation from mouse P2 brain fractions using a specific anti-KIBRA antibody revealed that PICK1, GluA1, and GluA2 are associated with KIBRA in vivo (Fig. 1D). Moreover, other known AMPAR trafficking regulators such as Glutamate Receptor Interacting Protein 1 (GRIP1), N-ethylmaleimide-sensitive factor (NSF) and Sec8 were also present in KIBRA complexes (Fig. 1D) (Dong et al., 1997; Mao et al., 2010; Song et al., 1998), while 4.1N protein and the NR1 subunit of NMDA receptors were not part of this complex. These data suggest that KIBRA may play a role in the regulation of AMPAR trafficking in neurons.
To test this hypothesis, we generated specific KIBRA shRNAs (Fig. S1B) and analyzed the cell surface expression of AMPARs. Knock down of KIBRA had no effect on the steady state level of AMPA receptor subunits analyzed using cell surface biotinylation assays (Fig. S1C-D). We then examined the role of KIBRA in activity-dependent trafficking of AMPARs in cultured hippocampal neurons using an established pH-sensitive GFP-GluA2 (pH-GluA2) live receptor recycling assay (Ashby et al., 2004; Lin and Huganir, 2007). Perfusion of N-methyl-D-aspartate (NMDA) for 5 minutes induced robust internalization of surface pH-GluA2 from the soma and dendrites as we have previously observed (Lin and Huganir, 2007) in both control and shRNA transfected neurons (Fig. 2A-D). However, the rate of pH-GluA2 recycling following NMDA washout was significantly accelerated in KIBRA KD neurons (Fig. 2A, B, C and E), reminiscent of the AMPAR trafficking phenotype in PICK1 KO neurons (Lin and Huganir, 2007). A similar result was obtained with a second independent KIBRA shRNA construct (Fig. S2A-D). Co-transfection of KIBRA shRNA and shRNA-resistant KIBRA constructs fully rescued the recycling phenotype, ruling out the possibility of off-target effects of the shRNA (Fig. 2A-E). These results indicate that KIBRA regulates the activity-dependent recycling but not the initial internalization of AMPARs, demonstrating a role for KIBRA in retaining internalized GluA2. It is possible that KIBRA does this by inhibiting the exocyst complex as overexpression of KIBRA localizes to sec8-containing vesicles (Fig, S2E).
Fig. 2.
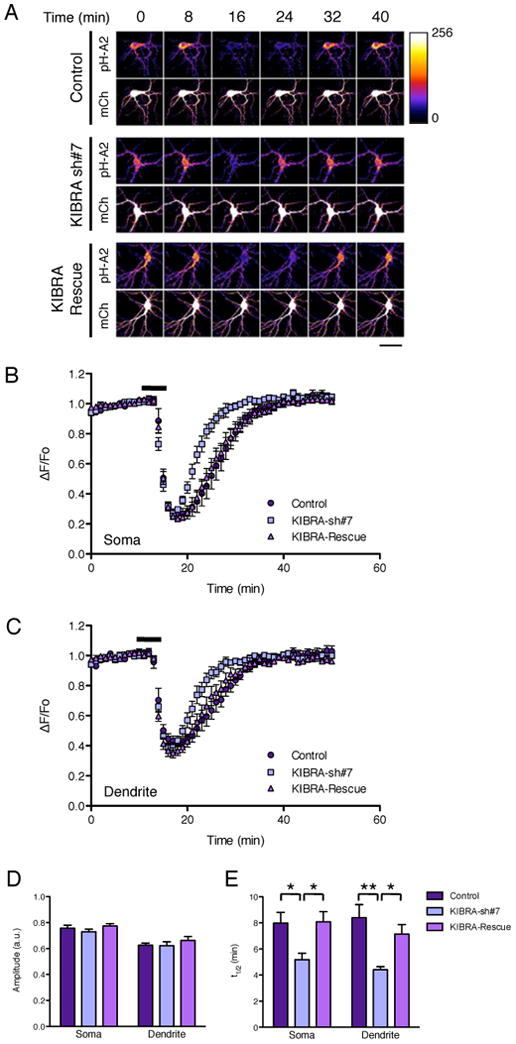
KIBRA KD accelerates the rate of GluA2 recycling following NMDAR-induced internalization. Cultured hippocampal neurons were transfected with empty vectors (control), pSuper-KIBRA-shRNA-7, or pSuper-KIBRA-shRNA-7 and pRK5-shRNA resistant KIBRA rescue constructs together with the pH-GluA2 reporter and mCherry at DIV15. At DIV17, neurons were stimulated with 20 μM NMDA for 5 min and changes in pH-GluA2 fluorescence intensity were monitored by live-cell confocal microscopy. (A) Representative time-series images from control, KIBRA KD and KIBRA rescued neurons (scale bar: 50μm). Average time course of normalized pH-GluA2 fluorescence change (AF/Fo) in the soma (B) and (C) dendrite. Histograms of changes in pH-GluA2 fluorescence amplitude in response to NMDA (D) and recycling rate t1/2 after NMDA washout (E). *P < 0.05, **P < 0.01, ANOVA (n = 8 to 14 cells) in the soma and dendrite.
We next generated KIBRA KO mice (Fig. S3A) to examine its role in synaptic transmission, plasticity and behavior in vivo. Correct homologous recombination, germline transmission, and genotype were confirmed by Southern blot using the indicated probe after PCR genotyping (Fig. S3B). Homozygous KO animals are viable and have no gross developmental defects or anatomical abnormalities (Fig. S3C). Western blot analyses using specific anti-KIBRA antibodies on total brain lysates verified the absence of KIBRA protein in KO animals (Fig. S3D). Although we did not observe any significant changes in the total expression of many synaptic proteins in KIBRA KO mice, there was a downregulation of NSF in juvenile animals. Interestingly, there is a marked compensation for the absence of KIBRA by its highly homologous family member, WWC2, in young animals, which diminishes by adulthood (Fig. S3D-F).
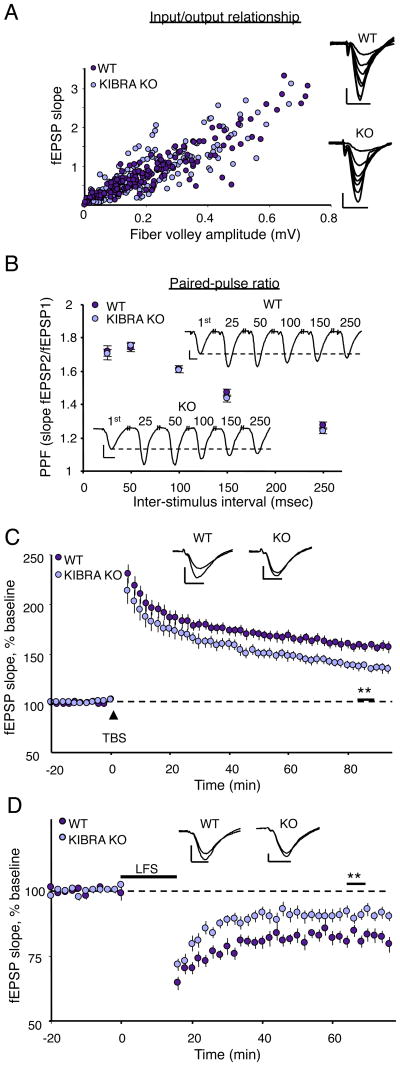
Analysis of baseline synaptic transmission in juvenile (3-4-week-old) or adult (2-3.5-month-old) KIBRA KO mice revealed no significant differences in input/output relationships compared to wild-type littermates (Fig. 3A, S4A). The lack of change in surface GluA1/2 expression in KIBRA KO neurons is consistent with this finding (Fig. S1E-F). Presynaptic function assessed by paired-pulse facilitation was also normal in KIBRA KO mice (Fig. 3B, S4B). Consistent with a role in activity-dependent trafficking of AMPARs, adult KIBRA KO mice displayed significant deficits in both LTP induced with theta-burst stimulation and NMDA-dependent LTD induced with low-frequency stimulation at hippocampal Schaffer collateral-CA1 synapses (Fig. 3C, D). Surprisingly, these forms of plasticity are intact in juvenile KIBRA KO mice (Fig. S4C, D). This selective impairment in synaptic plasticity in adult mice is strikingly similar to the phenotype observed in mice lacking PICK1 (Volk et al., 2010).
Fig. 3.
Impaired synaptic plasticity in adult KIBRA KO mice. (A) The slope of the input output curve is not altered in adult KIBRA KO mice. P > 0.05, two-tailed student's t-test (WT, n=29, average I-O slope = 3.4± 0.2 ms-1; KO, n=28, 3.3 ± 0.2 ms-1). (B) Paired pulse facilitation is not altered in adult KIBRA KO mice. A two-way repeated measures ANOVA revealed no significant genotype × inter-stimulus interval interaction or main effect of genotype. (WT n = 20, KO n = 17; p > 0.05 at all inter-stimulus intervals) (C) LTP induced with theta-burst stimulation is reduced in adult KIBRA KO mice. **P < 0.01, two-tailed student's t-test (wild-type, n = 13, 158±4%; KO, n = 10, 136±4%). (D) LTD induced with 900 stimuli at 1Hz is reduced in adult KIBRA KO mice. **P < 0.01, two-tailed student's t-test (wild-type, n = 11, 82±2%; KO, n = 12, 91±2%). Scale bars for all trace insets: 0.5mv, 5msec.
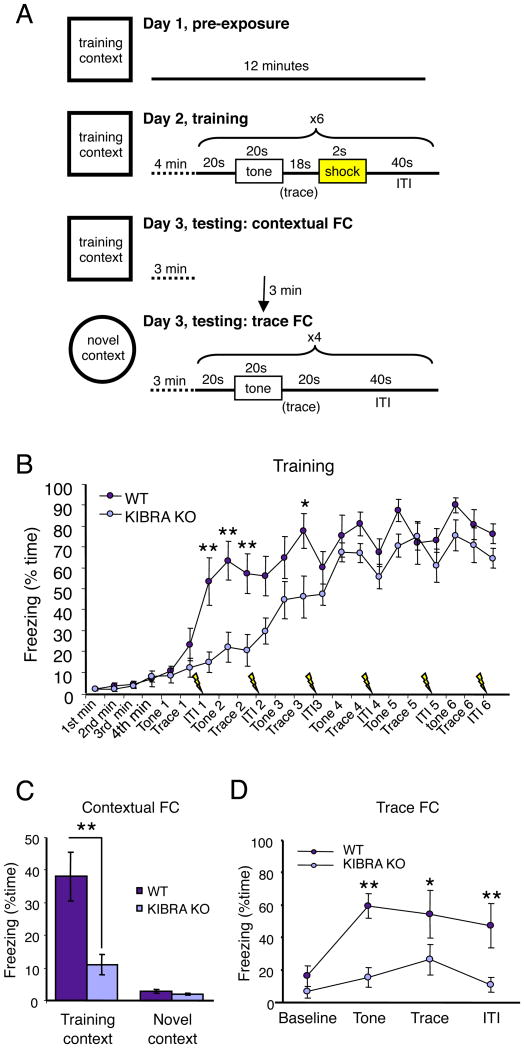
After discovering marked deficits in hippocampal LTP and LTD in adult mice lacking KIBRA, we investigated the requirement for KIBRA in learning and memory. We trained adult (2.5-3.5 month-old) male WT and KIBRA KO mice using a trace fear conditioning protocol (Fig. 4A). Intact hippocampal function is critical for eliciting both trace (freezing in response to tone presentation) and contextual (freezing in response to training context exposure) fear conditioning in response to this training protocol (McEchron et al., 1998; Smith et al., 2007). Compared to WT animals, KIBRA KO mice learn the association between shock and tone/context more slowly indicated by a delayed increase in freezing over the course of the six training trials (Fig. 4B); however, by the end of the training session, KIBRA KO mice exhibit freezing levels comparable to WT. When tested 24 hours after training, KIBRA KO mice showed a dramatic reduction in retention of both contextual and trace fear memory (Fig. 4C, D). These data provide strong evidence that KIBRA is a novel regulator of AMPAR trafficking and synaptic plasticity required for normal hippocampus-dependent learning in adult mice.
Fig. 4.
Impaired learning and memory in adult KIBRA KO mice. (A) Schematic of fear conditioning protocol. (B) KIBRA KO mice exhibit delayed freezing to a 6 trial training protocol, but show levels of freezing comparable to wild-type by the end of the training session. A two-way repeated measures ANOVA revealed a significant genotype × interval interaction, P < 0.01, F21,336 = 3.59 (wild-type, n = 8, KO, n = 10). *P < 0.05, **P < 0.01, Bonferroni post-hoc test, wild-type vs. KO. ITI, inter-train interval. (C) Freezing during the first minute of testing in the training context (**P < 0.01, two-tailed student's t-test) but not in a novel context (**P > 0.1, two-tailed student's t-test) is significantly impaired in KIBRA KO mice (wild-type, n = 8, KO, n = 10). (D) Trace fear conditioning during the first test block is impaired in KIBRA KO mice. A two-way repeated measures ANOVA revealed a significant genotype × interval interaction, P < 0.01, F6,72 = 5.81 (wild-type, n = 5, KO, n = 9). *P < 0.05, **P < 0.01, Bonferroni post-hoc test, wild-type vs. KO.
Discussion
Despite several studies implicating KIBRA in human memory performance, the role of KIBRA in brain function was unknown. We identified KIBRA in a search for PICK1 interacting proteins and found that it binds to PICK1 in vitro and in vivo and is part of an AMPAR protein complex associated with the regulation of AMPAR membrane trafficking. This data is consistent with previous results reporting that KIBRA is involved in membrane trafficking in non-neuronal cells and is associated with other neuronally expressed proteins, including dynein light chain 1 and synaptopodin, that are important in membrane trafficking and synaptic spine structure (Duning et al., 2008; Rayala et al., 2006; Rosse et al., 2009; Traer et al., 2007).
We report that KIBRA and PICK1 are associated and that they bind AMPARs along with other members of the AMPAR-associated complex including GRIP1, NSF, and Sec8. KIBRA regulates the membrane trafficking of AMPARs and plays an important role in modulating the recycling of AMPARs after activity dependent internalization, similarly to previously studied members of this complex (Shepherd and Huganir, 2007; Lin and Huganir, 2007, Mao et al., 2010). GRIP1/2 accelerates AMPAR recycling while PICK1 inhibits AMPAR recycling (Lin and Huganir, 2007, Mao et al., 2010; Citri et al., 2010). This protein complex is also important for synaptic plasticity in several brain regions (Shepherd and Huganir, 2007). Deletion of the GRIP1 and 2 genes eliminates cerebellar LTD (Takamiya et al., 2008) while deletion of the PICK1 gene eliminates cerebellar LTD (Steinberg et al., 2006) and also produces deficits in hippocampal LTP and LTD (Terashima et al., 2008; Volk et al., 2010). The specific role of KIBRA in this complex is unknown but it is likely to play an active role in the regulation of this scaffolding complex. KIBRA has two WW domains, protein-protein interaction motifs that bind to proline repeat sequences, and a C2 domain, a calcium-binding domain (Kremerskothen et al., 2003; Rizo and Sudhof, 1998)), that could be involved in the regulation of KIBRA's function. Intriguingly, KIBRA is an interacting partner and substrate of the atypical PKC isoform PKC-ζ that has been implicated in the maintenance of LTP and memory retention (Buther et al., 2004). It is possible that phosphorylation of KIBRA by PKC-ζ may be important for the regulation of AMPAR trafficking during the maintenance of LTP.
We also show that KIBRA is critical for synaptic plasticity and learning and memory. KIBRA KO mice have significant deficits in hippocampal LTP and LTD and have profound learning and memory defects. Interestingly, the functional effects of KIBRA KD and KO are very reminiscent of loss of function phenotypes previously reported for PICK1 KOs (Lin and Huganir, 2007; Volk et al., 2010). KIBRA and PICK1 interact robustly and the PICK1 and KIBRA KOs share similar cellular and behavioral phenotypes, suggesting that the two proteins act in the same pathway to regulate trafficking of GluA2-containing AMPARs. Adding to this complexity is the existence of a highly homologous relative of KIBRA, WWC2. Elucidation of the role of WWC2 in the brain may reveal a more broad function of WWC family members in AMPAR trafficking. The fact that basal synaptic transmission and surface expression of AMPARs is unaffected in mice lacking KIBRA may indicate that WWC2 is largely able to compensate for chronic loss of KIBRA, resulting in normal steady-state levels of synaptic AMPAR expression.
Loss of either PICK1 (Volk et al., 2010) or KIBRA results in synaptic plasticity and learning deficits in adult, but not in young animals, supporting a developmentally regulated requirement for KIBRA and PICK1 in normal brain function. If KIBRA and WWC2 are functionally similar, the high expression levels of these proteins early in development may render one homolog expendable in young animals. However, when levels of both KIBRA and WWC2 are low (as in adult animals), wild-type levels of both homologs may be required for normal brain function.
Finally, although our studies have been conducted in mice, the link between KIBRA and human memory suggests that KIBRA impacts human memory by regulating AMPAR membrane trafficking and synaptic plasticity. Considering the association between KIBRA and Alzheimer's disease (Corneveaux et al., 2008; Schneider et al., 2010), these results also provide support for the concept that altered AMPAR trafficking is a critical component of the cognitive deficits observed in Alzheimer's disease (Hsieh et al., 2006). The elucidation of KIBRA's role in the regulation of AMPAR function and synaptic plasticity provides insight into the molecular basis of natural variation in human memory performance. It will be interesting to analyze the potential genetic association of other members of the AMPAR protein complex with human memory performance to help dissect the molecular basis of cognitive function.
Experimental Procedures
Subjects
Wild-type and KIBRA KO mice were of the 129/C57BL6 hybrid background. Sprague Dawley rats were used for E18 hippocampal cultures. All animals were treated in accordance with the Johns Hopkins University Animal Care and Use Committee guidelines.
Electrophysiology
Hippocampal slices were prepared from KIBRA WT and KO mice ranging from 3-4 weeks (juvenile) or 2-3.5 months (adult) in age. Prior to recording, a cut was made between CA3 and CA1 to minimize recurrent activity. Field excitatory postsynaptic potentials (fEPSPs) were evoked at 0.033Hz with a 125μm platinum/ iridium concentric bipolar electrode (FHC, Bowdoinham, ME) placed in the middle of stratum radiatum of CA1. A 1-2MΩ glass recording electrode filled with ACSF was positioned ∼200μm away (orthodromic) from the stimulating electrode. Input-output curves were obtained for each slice and responses were set to ∼40% max for LTP experiments and ∼55% max for LTD experiments. Theta burst LTP: 4 trains of 10 bursts at 5Hz, with each burst consisting of 4 stimuli given at 100Hz, 10sec inter-train interval. LFS LTD: 900 single pulses at 1Hz. For LTD in the presence of AP5, AP5 was included throughout the experiment. All plasticity experiments are presented as responses normalized to the average of the 20min baseline.
Behavior
Training/testing
Mice were handled for 3 min on each of 5 consecutive days prior to beginning experiments. On Day 1 mice were habituated to the training chamber for 12 minutes. Training occurred on Day 2 as follows: Mice were allowed to acclimate to the chamber for 4 min prior to the onset of six consecuative training blocks, each consisting of a 20sec baseline, followed by a 20sec, 2KHz, 80dB tone (conditioned stimulus, CS), followed by an 18sec trace interval of silence, followed by a 2 second scrambled 1mA foot shock (unconditioned stimulus, US), followed by a 40sec inter-trial interval (ITI). On Day 3 mice were tested. Mice were first placed in the training chamber for 3min to assess contextual fear conditioning, after which they were returned to their home cage for 3min. Testing for trace fear conditioning took place in a novel chamber, which was distinct from the training chamber. Mice were allowed to acclimate to the novel chamber for 3 min prior to tone presentation to assess % freezing in the novel chamber. Next, mice were presented with four testing blocks consisting of a 20sec baseline followed by a 20sec 2KHz, 80dB tone followed by a 60sec ITI.
Data analysis and statistics
Percentage of time freezing was quantified using automated motion detection software (CleverSys).
Neuronal cell culture, plasmid transfection, and imaging
Hippocampal neurons from E18 rat pups were plated onto poly-L-lysine coated dishes or coverslips in Neurobasal growth medium supplemented with 2% B27, 2 mM Glutamax, 50 U/mL penicillin, 50 μg/mL streptomycin and 5% FBS. Neurons were switched to serum-free Neurobasal medium 24 h post seeding and fed twice a week. Neurons were transfected at DIV 14-15 using lipofectamine 2000 (Invitrogen) and pH-GluA2 recycling live-imaging assays were performed 48 h post transfection as described previously (Lin and Huganir, 2007). Briefly, coverslips containing neurons were assembled onto a closed perfusion chamber and continuously perfused with recording buffer (25 mM HEPES, 120 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM D-glucose, 1 μM TTX, pH 7.4). After 10 min of baseline recording (F0), neurons were perfused with NMDA solution (25 mM HEPES, 120 mM NaCl, 5 mM KCl, 2 mM CaCl2, 0.3 mM MgCl2, 30 mM D-glucose, 1 μM TTX, 20 μM NMDA, 10 μM glycine, pH 7.4) for 5 min before the perfusion was switched back to recording buffer for the remainder of the session. All imaging experiments were performed at room temperature using a Zeiss LSM 510 Meta/NLO system (Carl Zeiss, Thornwood, NY). The pHluorin fluorescence was imaged at 488 nm excitation and collected through a 505-550 nm filter, while the mCherry signal was imaged at 561 nm excitation and 575-615 nm emission. Neurons were imaged through a 63X oil objective (N.A. = 1.40) at a 3μm single optical section and collected at a rate of 1 image per min.
Images were analyzed using ImageJ software (NIH) by calculating the normalized change in average pHluorin over mCherry fluorescence intensities from a somatic and dendritic shaft area (“soma”) and from a purely dendritic area (“dendrite”) defined manually to compensate for x-y movements of the recorded neurons. The fluorescence intensity change is expressed as ΔF/Fo and the amplitude of fluorescence change (ΔFmax/Fo) represents the extent of GluA2 endocytosis. The rate of GluA2 recycling can be calculated as the time taken from fluorescence minima to 50% of the fluorescence maxima (t1/2).
Generation of KIBRA KO Mice
The KIBRA KO mouse was generated by targeting exons 4 and 5 for excision by Cre recombinase to result in an out-of-frame mutation in the KIBRA genomic DNA. A 13.9kb KIBRA genomic DNA fragment was cloned into the pBlueScript vector with its KpnI site destroyed. A 4.0kb internal Kpn1 fragment was cut and cloned into pNeo-FRT-loxP such that a Neo resistant cassette and KIBRA exons 4/5 were flanked by loxP sequences. The loxP-flanked fragment was subsequently cloned back into the pBlueScript cloning vector. After germline transmission, Neo was deleted with the Cre/loxP system by breeding to CMV-Cre transgenic mice. Initial Southern blots to confirm homologous recombination of the targeting vector were performed using an outer probe (data not shown).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Health (MH64856 and NS36715) and the Howard Hughes Medical Institute (to R.L.H.). V.A. is supported by fellowships from the International Human Frontier Science Program (LT00399/2008-L) and the Australian National Health and Medical Research Council (ID. 477108). L.V. is supported by a training grant from the National Institute of Health (T32MH15330). We thank Min Dai and Monica Coulter for technical support.
Footnotes
Competing Financial Interests Statement: Under a licensing agreement between Millipore Corporation and The Johns Hopkins University, R.L.H. is entitled to a share of royalties received by the University on sales of products described in this article. R.L.H. is a paid consultant to Millipore Corporation. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict–of–interest policies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almeida OP, Schwab SG, Lautenschlager NT, Morar B, Greenop KR, Flicker L, Wildenauer D. KIBRA genetic polymorphism influences episodic memory in later life, but does not increase the risk of mild cognitive impairment. J Cell Mol Med. 2008;12:1672–1676. doi: 10.1111/j.1582-4934.2008.00229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby MC, Ibaraki K, Henley JM. It's green outside: tracking cell surface proteins with pH-sensitive GFP. Trends Neurosci. 2004;27:257–261. doi: 10.1016/j.tins.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Bates TC, Price JF, Harris SE, Marioni RE, Fowkes FG, Stewart MC, Murray GD, Whalley LJ, Starr JM, Deary IJ. Association of KIBRA and memory. Neurosci Lett. 2009 doi: 10.1016/j.neulet.2009.04.050. [DOI] [PubMed] [Google Scholar]
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Buther K, Plaas C, Barnekow A, Kremerskothen J. KIBRA is a novel substrate for protein kinase Czeta. Biochem Biophys Res Commun. 2004;317:703–707. doi: 10.1016/j.bbrc.2004.03.107. [DOI] [PubMed] [Google Scholar]
- Citri A, Bhattacharyya S, Ma C, Morishita W, Fang S, Rizo J, Malenka RC. Calcium binding to PICK1 is essential for the intracellular retention of AMPA receptors underlying long-term depression. J Neurosci. 2010;30:16437–16452. doi: 10.1523/JNEUROSCI.4478-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RL, Anggono V, Huganir RL. PICK1 regulates incorporation of calcium-permeable AMPA receptors during cortical synaptic strengthening. J Neurosci. 2010;30:6360–6366. doi: 10.1523/JNEUROSCI.6276-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Craig DW, et al. Evidence for an association between KIBRA and late-onset Alzheimer's disease. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- Drier EA, Tello MK, Cowan M, Wu P, Blace N, Sacktor TC, Yin JC. Memory enhancement and formation by atypical PKM activity in Drosophila melanogaster. Nat Neurosci. 2002;5:316–324. doi: 10.1038/nn820. [DOI] [PubMed] [Google Scholar]
- Duning K, Schurek EM, Schluter M, Bayer M, Reinhardt HC, Schwab A, Schaefer L, Benzing T, Schermer B, Saleem MA, et al. KIBRA modulates directional migration of podocytes. J Am Soc Nephrol. 2008;19:1891–1903. doi: 10.1681/ASN.2007080916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esclassan F, Coutureau E, Di Scala G, Marchand AR. A cholinergic-dependent role for the entorhinal cortex in trace fear conditioning. J Neurosci. 2009;29:8087–8093. doi: 10.1523/JNEUROSCI.0543-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SM, Takamiya K, Xia J, Suh JG, Johnson R, Yu S, Huganir RL. Calcium-permeable AMPA receptor plasticity is mediated by subunit-specific interactions with PICK1 and NSF. Neuron. 2005;45:903–915. doi: 10.1016/j.neuron.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh H, Boehm J, Sato C, Iwatsubo T, Tomita T, Sisodia S, Malinow R. AMPAR removal underlies Abeta-induced synaptic depression and dendritic spine loss. Neuron. 2006;52:831–843. doi: 10.1016/j.neuron.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J, Heon S, Kim MJ, Son GH, Park Y, Henley JM, Weiss JL, Sheng M, Collingridge GL, Cho K. Metabotropic glutamate receptor-mediated LTD involves two interacting Ca(2+) sensors, NCS-1 and PICK1. Neuron. 2008;60:1095–1111. doi: 10.1016/j.neuron.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen S, Duning K, Pavenstadt H, Kremerskothen J, Boeckers TM. Temporal-spatial expression and novel biochemical properties of the memory-related protein KIBRA. Neuroscience. 2008;155:1165–1173. doi: 10.1016/j.neuroscience.2008.06.054. [DOI] [PubMed] [Google Scholar]
- Kremerskothen J, Plaas C, Buther K, Finger I, Veltel S, Matanis T, Liedtke T, Barnekow A. Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun. 2003;300:862–867. doi: 10.1016/s0006-291x(02)02945-5. [DOI] [PubMed] [Google Scholar]
- Lin DT, Huganir RL. PICK1 and phosphorylation of the glutamate receptor 2 (GluR2) AMPA receptor subunit regulates GluR2 recycling after NMDA receptor-induced internalization. J Neurosci. 2007;27:13903–13908. doi: 10.1523/JNEUROSCI.1750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Cull-Candy SG. Subunit interaction with PICK and GRIP controls Ca2+ permeability of AMPARs at cerebellar synapses. Nat Neurosci. 2005;8:768–775. doi: 10.1038/nn1468. [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- Mao L, Takamiya K, Thomas G, Lin DT, Huganir RL. GRIP1 and 2 regulate activity-dependent AMPA receptor recycling via exocyst complex interactions. Proc Natl Acad Sci U S A. 2010;107:19038–19043. doi: 10.1073/pnas.1013494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEchron MD, Bouwmeester H, Tseng W, Weiss C, Disterhoft JF. Hippocampectomy disrupts auditory trace fear conditioning and contextual fear conditioning in the rat. Hippocampus. 1998;8:638–646. doi: 10.1002/(SICI)1098-1063(1998)8:6<638::AID-HIPO6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- Rayala SK, den Hollander P, Manavathi B, Talukder AH, Song C, Peng S, Barnekow A, Kremerskothen J, Kumar R. Essential role of KIBRA in co-activator function of dynein light chain 1 in mammalian cells. J Biol Chem. 2006;281:19092–19099. doi: 10.1074/jbc.M600021200. [DOI] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- Rosse C, Formstecher E, Boeckeler K, Zhao Y, Kremerskothen J, White MD, Camonis JH, Parker PJ. An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation. PLoS Biol. 2009;7:e1000235. doi: 10.1371/journal.pbio.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacktor TC, Osten P, Valsamis H, Jiang X, Naik MU, Sublette E. Persistent activation of the zeta isoform of protein kinase C in the maintenance of long-term potentiation. Proc Natl Acad Sci USA. 1993;90:8342–8346. doi: 10.1073/pnas.90.18.8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper K, Kolsch H, Popp J, Wagner M, Jessen F. KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol Aging. 2008;29:1123–1125. doi: 10.1016/j.neurobiolaging.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Schneider A, Huentelman MJ, Kremerskothen J, Duning K, Spoelgen R, Nikolich K. KIBRA: A New Gateway to Learning and Memory? Front Aging Neurosci. 2010;2:4. doi: 10.3389/neuro.24.004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL. The Cell Biology of Synaptic Plasticity: AMPA Receptor Trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- Smith DR, Gallagher M, Stanton ME. Genetic background differences and nonassociative effects in mouse trace fear conditioning. Learn Mem. 2007;14:597–605. doi: 10.1101/lm.614807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song I, Kamboj S, Xia J, Dong H, Liao D, Huganir RL. Interaction of the N-ethylmaleimide-sensitive factor with AMPA receptors. Neuron. 1998;21:393–400. doi: 10.1016/s0896-6273(00)80548-6. [DOI] [PubMed] [Google Scholar]
- Steinberg JP, Takamiya K, Shen Y, Xia J, Rubio ME, Yu S, Jin W, Thomas GM, Linden DJ, Huganir RL. Targeted in vivo mutations of the AMPA receptor subunit GluR2 and its interacting protein PICK1 eliminate cerebellar long-term depression. Neuron. 2006;49:845–860. doi: 10.1016/j.neuron.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Takamiya K, Mao L, Huganir RL, Linden DJ. The glutamate receptor-interacting protein family of GluR2-binding proteins is required for long-term synaptic depression expression in cerebellar Purkinje cells. J Neurosci. 2008;28:5752–5755. doi: 10.1523/JNEUROSCI.0654-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima A, Pelkey KA, Rah JC, Suh YH, Roche KW, Collingridge GL, McBain CJ, Isaac JT. An essential role for PICK1 in NMDA receptor-dependent bidirectional synaptic plasticity. Neuron. 2008;57:872–882. doi: 10.1016/j.neuron.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traer CJ, Rutherford AC, Palmer KJ, Wassmer T, Oakley J, Attar N, Carlton JG, Kremerskothen J, Stephens DJ, Cullen PJ. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat Cell Biol. 2007;9:1370–1380. doi: 10.1038/ncb1656. [DOI] [PubMed] [Google Scholar]
- Volk L, Kim CH, Takamiya K, Yu Y, Huganir RL. Developmental regulation of protein interacting with C kinase 1 (PICK1) function in hippocampal synaptic plasticity and learning. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1016103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.