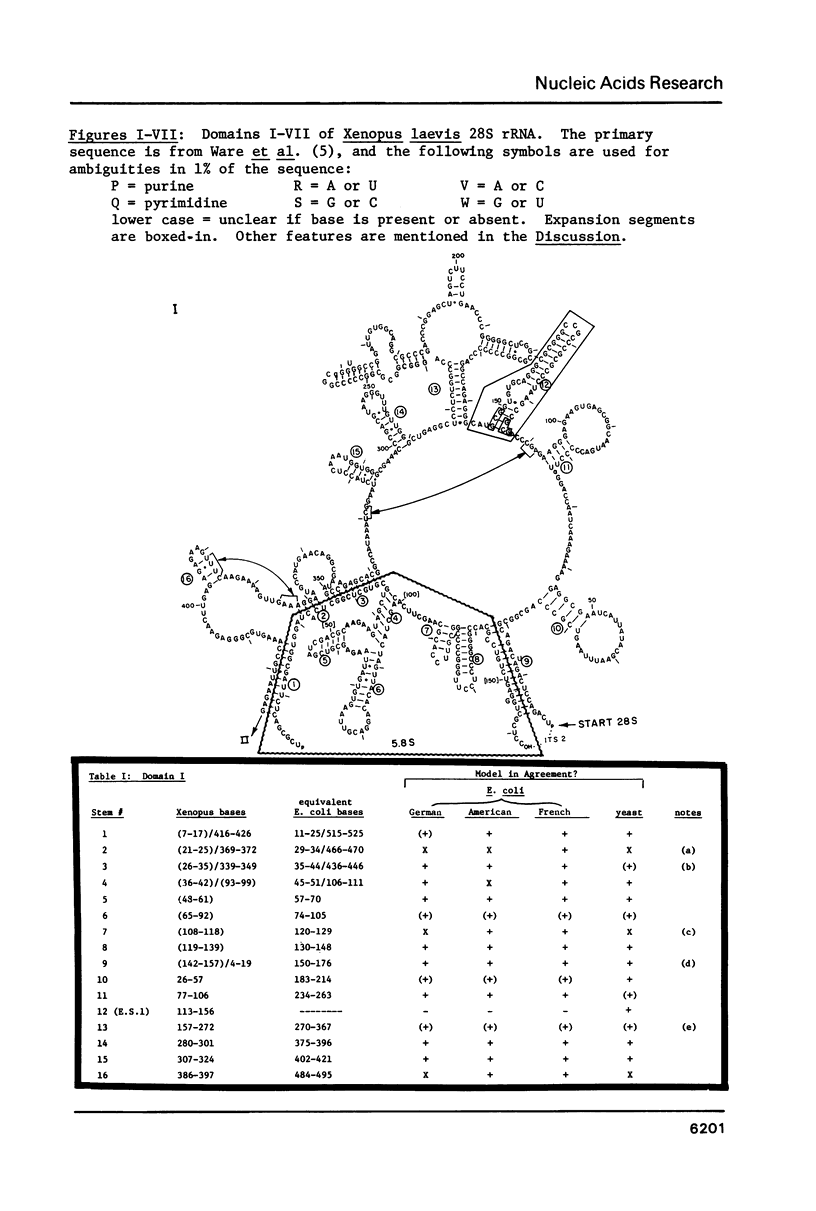
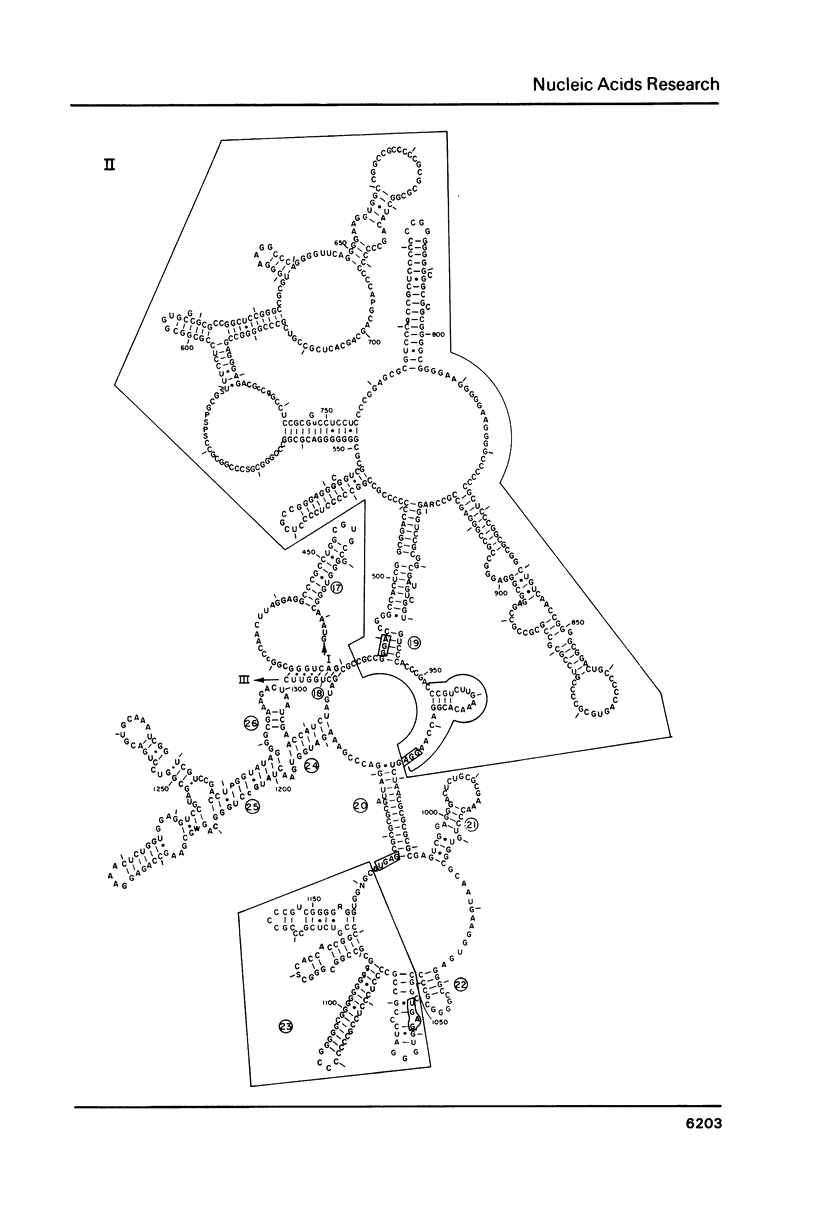
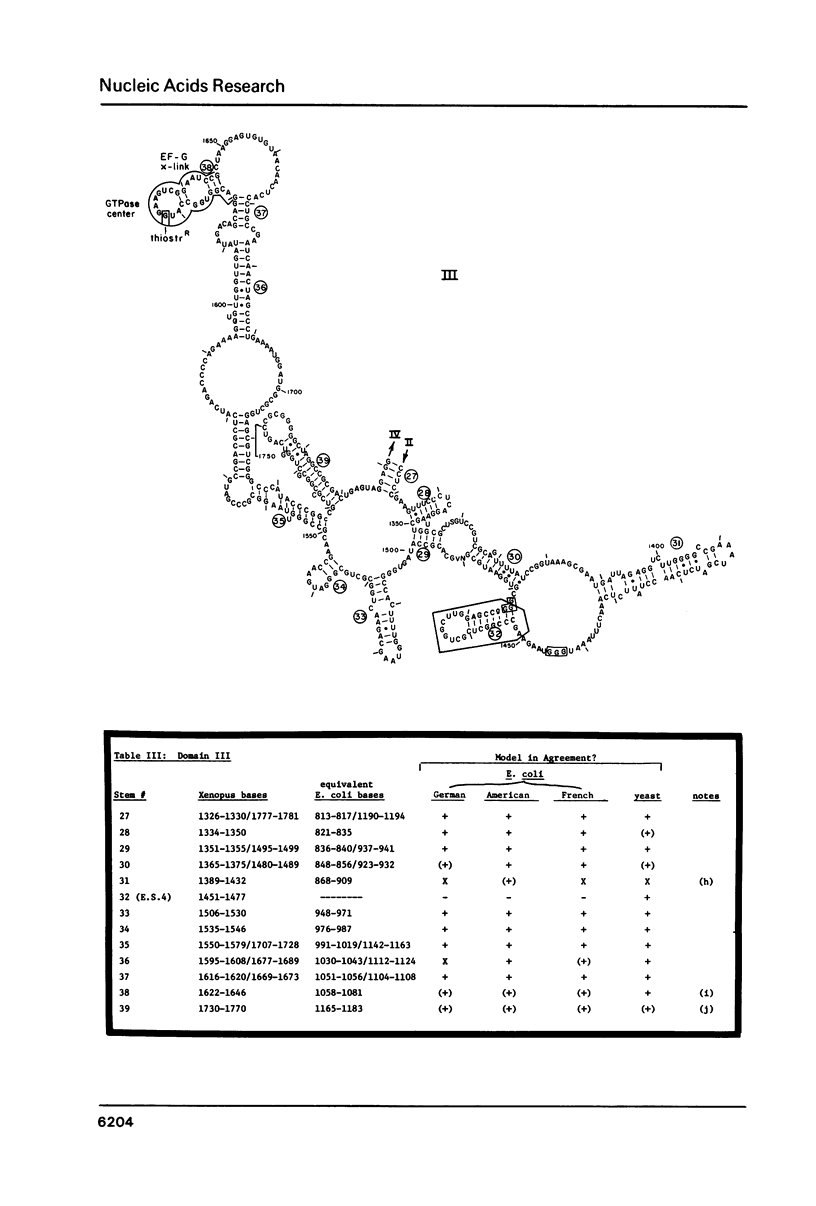
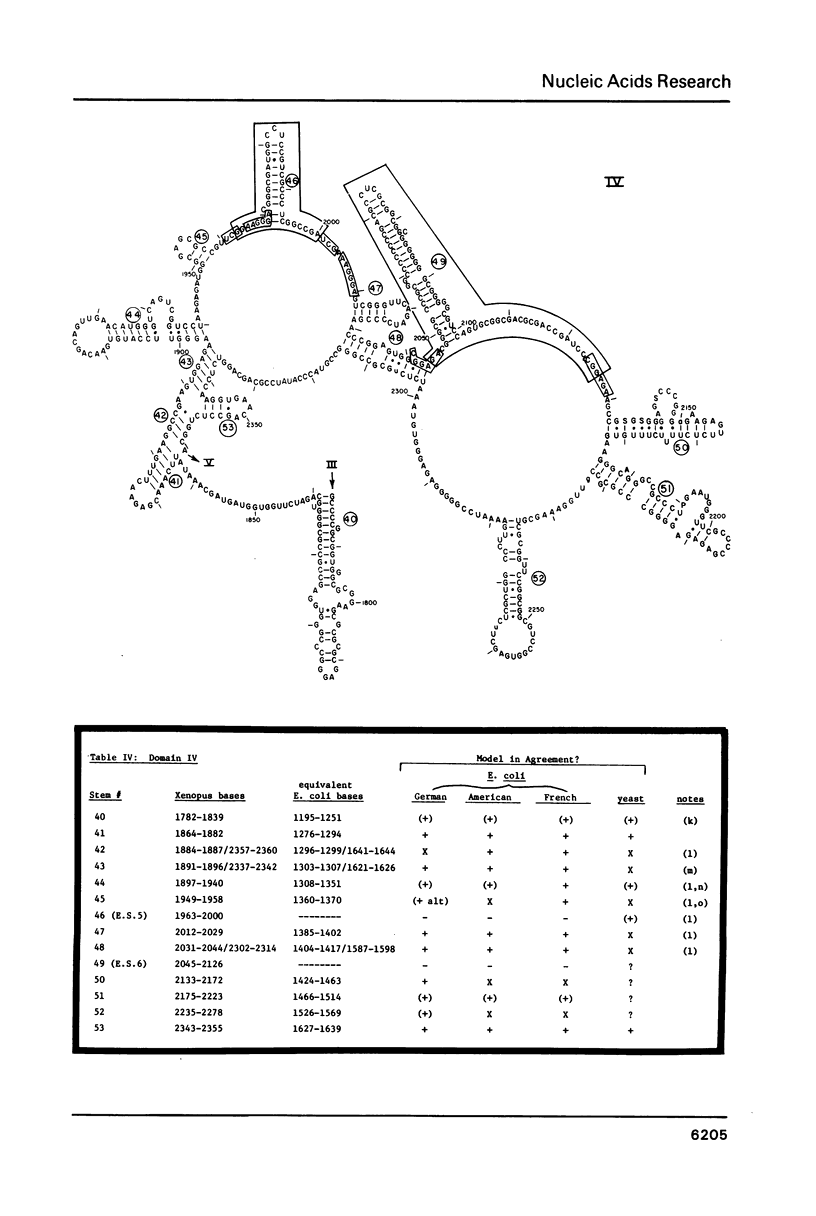
Abstract
Based upon the three experimentally derived models of E. coli 23S rRNA (1-3) and the partial model for yeast 26S rRNA (4), which was deduced by homology to E. coli, we derived a secondary structure model for Xenopus laevis 28S rRNA. This is the first complete model presented for eukaryotic 28S rRNA. Compensatory base changes support the general validity of our model and offer help to resolve which of the three E. coli models is correct in regions where they are different from one another. Eukaryotic rDNA is longer than prokaryotic rDNA by virtue of introns, expansion segments and transcribed spacers, all of which are discussed relative to our secondary structure model. Comments are made on the evolutionary origins of these three categories and the processing fates of their transcripts. Functionally important sites on our 28S rRNA secondary structure model are suggested by analogy for ribosomal protein binding, the GTPase center, the peptidyl transferase center, and for rRNA interaction with tRNA and 5S RNA. We discuss how RNA-RNA interactions may play a vital role in translocation.
Full text
PDF















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Rochaix J. D. Structure analysis at the ends of the intervening DNA sequences in the chloroplast 23S ribosomal genes of C. reinhardii. Cell. 1979 Sep;18(1):55–60. doi: 10.1016/0092-8674(79)90353-2. [DOI] [PubMed] [Google Scholar]
- Baer R. J., Dubin D. T. Methylated regions of hamster mitochondrial ribosomal RNA: structural and functional correlates. Nucleic Acids Res. 1981 Jan 24;9(2):323–337. doi: 10.1093/nar/9.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt A., Ebel J. P. The secondary structure of the protein L1 binding region of ribosomal 23S RNA. Homologies with putative secondary structures of the L11 mRNA and of a region of mitochondrial 16S rRNA. Nucleic Acids Res. 1981 Jan 24;9(2):293–307. doi: 10.1093/nar/9.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt M. A., Pouyet J., Ebel J. P., Edwards K., Kössel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981 Sep 11;9(17):4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Sriwdada J., Ebel J. P., Sloof P., Garrett R. A. The binding site of protein L1 ON 23-S ribosomal RNA of Escherichia coli. 2. Identification of the rna region contained in the L1 ribonucleoproteins and determination of the order of the RNA subfragments within this region. Eur J Biochem. 1976 Nov 15;70(2):457–469. doi: 10.1111/j.1432-1033.1976.tb11037.x. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Sriwidada J., Brimacombe R. RNA sequences associated with proteins L1, L9, and L5, L18, L25, in ribonucleoprotein fragments isolated from the 50-S subunit of Escherichia coli ribosomes. Eur J Biochem. 1976 Nov 15;70(2):483–492. doi: 10.1111/j.1432-1033.1976.tb11039.x. [DOI] [PubMed] [Google Scholar]
- Branlant C., Sri Widada J., Krol A., Ebel J. P. RNA sequences in ribonucleoprotein fragments of the complex formed from ribosomal 23-S RNA and ribosomal protein L24 of Escherichia coli. Eur J Biochem. 1977 Mar 15;74(1):155–170. doi: 10.1111/j.1432-1033.1977.tb11377.x. [DOI] [PubMed] [Google Scholar]
- Branlant C., Widada J. S., Krol A., Ebel J. P. Studies on the primary structure of the ribosomal 23S RNA of Escherichia coli: II. A characterisation and an alignment of 24 sections spanning the entire molecule and its application to the localisation of specific fragments. Nucleic Acids Res. 1977 Dec;4(12):4323–4345. doi: 10.1093/nar/4.12.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe R., Maly P., Zwieb C. The structure of ribosomal RNA and its organization relative to ribosomal protein. Prog Nucleic Acid Res Mol Biol. 1983;28:1–48. doi: 10.1016/s0079-6603(08)60081-1. [DOI] [PubMed] [Google Scholar]
- Brow D. A., Noller H. F. Protection of ribosomal RNA from kethoxal in polyribosomes. Implication of specific sites in ribosome function. J Mol Biol. 1983 Jan 5;163(1):27–46. doi: 10.1016/0022-2836(83)90028-1. [DOI] [PubMed] [Google Scholar]
- Burke J. M., RajBhandary U. L. Intron within the large rRNA gene of N. crassa mitochondria: a long open reading frame and a consensus sequence possibly important in splicing. Cell. 1982 Dec;31(3 Pt 2):509–520. doi: 10.1016/0092-8674(82)90307-5. [DOI] [PubMed] [Google Scholar]
- Chan Y. L., Endo Y., Wool I. G. The sequence of the nucleotides at the alpha-sarcin cleavage site in rat 28 S ribosomal ribonucleic acid. J Biol Chem. 1983 Nov 10;258(21):12768–12770. [PubMed] [Google Scholar]
- Chan Y. L., Olvera J., Wool I. G. The structure of rat 28S ribosomal ribonucleic acid inferred from the sequence of nucleotides in a gene. Nucleic Acids Res. 1983 Nov 25;11(22):7819–7831. doi: 10.1093/nar/11.22.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C. G., Gerbi S. A. Ribosomal RNA evolution by fragmentation of the 23S progenitor: maturation pathway parallels evolutionary emergence. J Mol Evol. 1982;18(5):329–336. doi: 10.1007/BF01733899. [DOI] [PubMed] [Google Scholar]
- Cox R. A., Godwin E., Hastings J. R. Spectroscopic evidence for the uneven distribution of adenine and uracil residues in ribosomal ribonucleic acid of Drosophila melanogaster and of Plasmodium knowlesi and its possible evolutionary significance. Biochem J. 1976 Jun 1;155(3):465–475. doi: 10.1042/bj1550465a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg J. E., Kintner C., Lund E. Specific binding of tRNAMet to 23S rRNA of Escherichia coli. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1071–1075. doi: 10.1073/pnas.75.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B., Rebbert M. L. Nucleotide sequences at the boundaries between gene and insertion regions in the rDNA of Drosophilia melanogaster. Nucleic Acids Res. 1981 Oct 10;9(19):5011–5020. doi: 10.1093/nar/9.19.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K. A reinvestigation of 5' leads to 3' polarity in 40S ribosomal RNA precursor of Xenopus laevis. Cell. 1976 Jul;8(3):443–448. doi: 10.1016/0092-8674(76)90157-4. [DOI] [PubMed] [Google Scholar]
- Delihas N. Liver ribosomal ribonucleic acid structural studies. Characterization of fragments from partial nuclease digestion. Biochemistry. 1967 Nov;6(11):3356–3362. doi: 10.1021/bi00863a004. [DOI] [PubMed] [Google Scholar]
- Dijk J., Garrett R. A., Müller R. Studies on the binding of the ribosomal protein complex L7/12-L10 and protein L11 to the 5'-one third of 23S RNA: a functional centre of the 50S subunit. Nucleic Acids Res. 1979 Jun 25;6(8):2717–2729. doi: 10.1093/nar/6.8.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohme F., Nierhaus K. H. Role of 5S RNA in assembly and function of the 50S subunit from Escherichia coli. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2221–2225. doi: 10.1073/pnas.73.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell. 1980 May;20(1):185–197. doi: 10.1016/0092-8674(80)90246-9. [DOI] [PubMed] [Google Scholar]
- Eckerman D. J., Symons R. H. Sequence at the site of attachment of an affinity-label derivative of puromycin on 23-S ribosomal RNA of Escherichia coli ribosomes. Eur J Biochem. 1978 Jan 2;82(1):225–234. doi: 10.1111/j.1432-1033.1978.tb12015.x. [DOI] [PubMed] [Google Scholar]
- Edwards K., Kössel H. The rRNA operon from Zea mays chloroplasts: nucleotide sequence of 23S rDNA and its homology with E.coli 23S rDNA. Nucleic Acids Res. 1981 Jun 25;9(12):2853–2869. doi: 10.1093/nar/9.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Wool I. G. The site of action of alpha-sarcin on eukaryotic ribosomes. The sequence at the alpha-sarcin cleavage site in 28 S ribosomal ribonucleic acid. J Biol Chem. 1982 Aug 10;257(15):9054–9060. [PubMed] [Google Scholar]
- Fernandez-Puentes C., Vazquez D. Effects of some proteins that inactivate the eukaryotic ribosome. FEBS Lett. 1977;78(1):143–146. doi: 10.1016/0014-5793(77)80292-5. [DOI] [PubMed] [Google Scholar]
- Georgiev O. I., Nikolaev N., Hadjiolov A. A., Skryabin K. G., Zakharyev V. M., Bayev A. A. The structure of the yeast ribosomal RNA genes. 4. Complete sequence of the 25 S rRNA gene from Saccharomyces cerevisae. Nucleic Acids Res. 1981 Dec 21;9(24):6953–6958. doi: 10.1093/nar/9.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Gerbi S. A. Fine structure of ribosomal RNA. IV. Extraordinary evolutionary conservation in sequences that flank introns in rDNA. Nucleic Acids Res. 1980 Aug 25;8(16):3623–3637. doi: 10.1093/nar/8.16.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Thurlow D. L., Gerbi S. A., Zimmermann R. A. Specific binding of a prokaryotic ribosomal protein to a eukaryotic ribosomal RNA: implications for evolution and autoregulation. Proc Natl Acad Sci U S A. 1981 May;78(5):2722–2726. doi: 10.1073/pnas.78.5.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell P., Harris R. J., Symons R. H. Affinity labelling of 23-S ribosomal RNA in the active centre of Escherichia coli peptidyl transferase. Eur J Biochem. 1974 Dec 2;49(3):539–544. doi: 10.1111/j.1432-1033.1974.tb03858.x. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A., Georgiev O. I., Nosikov V. V., Yavachev L. P. Primary and secondary structure of rat 28 S ribosomal RNA. Nucleic Acids Res. 1984 Apr 25;12(8):3677–3693. doi: 10.1093/nar/12.8.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobden A. N., Cundliffe E. The mode of action of alpha sarcin and a novel assay of the puromycin reaction. Biochem J. 1978 Jan 15;170(1):57–61. doi: 10.1042/bj1700057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq B. Sequence homologies between eukaryotic 5.8S rRNA and the 5' end of prokaryotic 23S rRNa: evidences for a common evolutionary origin. Nucleic Acids Res. 1981 Jun 25;9(12):2913–2932. doi: 10.1093/nar/9.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan N. C., Gall J. G. The intervening sequence of the ribosomal RNA gene is highly conserved between two Tetrahymena species. Nucleic Acids Res. 1982 May 11;10(9):2809–2822. doi: 10.1093/nar/10.9.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey S. E., Craig I. W. Altered ribosomal RNA genes in mitochondria from mammalian cells with chloramphenicol resistance. Nature. 1981 Apr 16;290(5807):607–608. doi: 10.1038/290607a0. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Salim M., Maden B. E. Extensive homologies between the methylated nucleotide sequences in several vertebrate ribosomal ribonucleic acids. Biochem J. 1978 Mar 1;169(3):531–542. doi: 10.1042/bj1690531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A., Machatt M. A., Branlant C., Ebel J. P. RNA-RNA interactions in the binding site of protein L24 on 23S ribosomal RNA of E. coli. II. Sequence analysis of the interacting fragments. Nucleic Acids Res. 1978 Dec;5(12):4933–4947. doi: 10.1093/nar/5.12.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Machatt M. A., Ebel J. P., Branlant C. The 3'-terminal region of bacterial 23S ribosomal RNA: structure and homology with the 3'-terminal region of eukaryotic 28S rRNA and with chloroplast 4.5s rRNA. Nucleic Acids Res. 1981 Apr 10;9(7):1533–1549. doi: 10.1093/nar/9.7.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E. Methylation map of Xenopus laevis ribosomal RNA. Nature. 1980 Nov 20;288(5788):293–296. doi: 10.1038/288293a0. [DOI] [PubMed] [Google Scholar]
- Maly P., Brimacombe R. Refined secondary structure models for the 16S and 23S ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1983 Nov 11;11(21):7263–7286. doi: 10.1093/nar/11.21.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly P., Rinke J., Ulmer E., Zwieb C., Brimacombe R. Precise localization of the site of cross-linking between protein L4 and 23S ribonucleic acid induced by mild ultraviolet irradiation of Escherichia coli 50S ribosomal subunits. Biochemistry. 1980 Sep 2;19(18):4179–4188. doi: 10.1021/bi00559a007. [DOI] [PubMed] [Google Scholar]
- Mandal R. K., Dawid I. B. The nucleotide sequence at the transcription termination site of ribosomal RNA in Drosophila melanogaster. Nucleic Acids Res. 1981 Apr 24;9(8):1801–1811. doi: 10.1093/nar/9.8.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt O., Roth H. E., Wystup G., Nierhaus K. H. Binding of Escherichia coli ribosomal proteins to 23S RNA under reconstitution conditions for the 50S subunit. Nucleic Acids Res. 1979 Aug 10;6(11):3641–3650. doi: 10.1093/nar/6.11.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Sequence and secondary structure of mouse 28S rRNA 5'terminal domain. Organisation of the 5.8S-28S rRNA complex. Nucleic Acids Res. 1982 Sep 11;10(17):5273–5283. doi: 10.1093/nar/10.17.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N. A 5.8 S rRNA-like sequence in prokaryotic 23 S rRNA. FEBS Lett. 1980 Oct 6;119(2):212–214. doi: 10.1016/0014-5793(80)80254-7. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O. Role of the 5'-terminal sequence in the RNA binding site of yeast 5.8 S rRNA. FEBS Lett. 1980 Jun 16;115(1):71–76. doi: 10.1016/0014-5793(80)80729-0. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Kop J., Wheaton V., Brosius J., Gutell R. R., Kopylov A. M., Dohme F., Herr W., Stahl D. A., Gupta R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981 Nov 25;9(22):6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomiyama H., Sakaki Y., Takagi Y. Nucleotide sequence of a ribosomal RNA gene intron from slime mold Physarum polycephalum. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1376–1380. doi: 10.1073/pnas.78.3.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny V., Nierhaus K. H. Initiator proteins for the assembly of the 50S subunit from Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7238–7242. doi: 10.1073/pnas.79.23.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka T., Nomiyama H., Yoshida H., Kukita T., Kuhara S., Sakaki Y. Complete nucleotide sequence of the 26S rRNA gene of Physarum polycephalum: its significance in gene evolution. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3163–3167. doi: 10.1073/pnas.80.11.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Peattie D. A., Douthwaite S., Garrett R. A., Noller H. F. A "bulged" double helix in a RNA-protein contact site. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7331–7335. doi: 10.1073/pnas.78.12.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C. L., Korn L. J. Computer analysis of nucleic acids and proteins. Methods Enzymol. 1980;65(1):595–609. doi: 10.1016/s0076-6879(80)65062-9. [DOI] [PubMed] [Google Scholar]
- Rae P. M. Coding region deletions associated with the major form of rDNA interruption in Drosophila. Nucleic Acids Res. 1981 Oct 10;9(19):4997–5010. doi: 10.1093/nar/9.19.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M., Kohorn B. D., Wade R. P. The 10 kb Drosophila virilis 28S rDNA intervening sequence is flanked by a direct repeat of 14 base pairs of coding sequence. Nucleic Acids Res. 1980 Aug 25;8(16):3491–3504. doi: 10.1093/nar/8.16.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H., Higashinakagawa T., Miller O., Jr The 5' leads to 3' polarity of the Xenopus Ribosomal RNA precursor molecule. Cell. 1976 Jul;8(3):449–454. doi: 10.1016/0092-8674(76)90158-6. [DOI] [PubMed] [Google Scholar]
- Roiha H., Glover D. M. Duplicated rDNA sequences of variable lengths flanking the short type I insertions in the rDNA of Drosophila melanogaster. Nucleic Acids Res. 1981 Nov 11;9(21):5521–5532. doi: 10.1093/nar/9.21.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiha H., Miller J. R., Woods L. C., Glover D. M. Arrangements and rearrangements of sequences flanking the two types of rDNA insertion in D. melanogaster. Nature. 1981 Apr 30;290(5809):749–753. doi: 10.1038/290749a0. [DOI] [PubMed] [Google Scholar]
- Röhl R., Nierhaus K. H. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1982 Feb;79(3):729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Wyler T., Hagenbüchle O. Changes in size and secondary structure of the ribosomal transcription unit during vertebrate evolution. J Mol Biol. 1975 May 25;94(3):503–517. doi: 10.1016/0022-2836(75)90217-x. [DOI] [PubMed] [Google Scholar]
- Schindler D. G., Davies J. E. Specific cleavage of ribosomal RNA caused by alpha sarcin. Nucleic Acids Res. 1977 Apr;4(4):1097–1110. doi: 10.1093/nar/4.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F. J., Thompson J., Lee K., Dijk J., Cundliffe E. The binding site for ribosomal protein L11 within 23 S ribosomal RNA of Escherichia coli. J Biol Chem. 1981 Dec 10;256(23):12301–12305. [PubMed] [Google Scholar]
- Sege R., Söll D., Ruddle F. H., Queen C. A conversational system for the computer analysis of nucleic acid sequences. Nucleic Acids Res. 1981 Jan 24;9(2):437–444. doi: 10.1093/nar/9.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloof P., Hunter J. B., Garrett R. A., Branlant C. RNA-RNA interactions in the binding site of protein L24 on 23S ribosomal RNA of Escherichia coli: 1. Evidence for their occurrence between widely separated sequence regions. Nucleic Acids Res. 1978 Oct;5(10):3503–3513. doi: 10.1093/nar/5.10.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Complete DNA sequence coding for the large ribosomal RNA of yeast mitochondria. Nucleic Acids Res. 1983 Jan 25;11(2):339–348. doi: 10.1093/nar/11.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Identification of two erythromycin resistance mutations in the mitochondrial gene coding for the large ribosomal RNA in yeast. Nucleic Acids Res. 1982 Nov 11;10(21):6571–6577. doi: 10.1093/nar/10.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Schmidt F., Cundliffe E. Site of action of a ribosomal RNA methylase conferring resistance to thiostrepton. J Biol Chem. 1982 Jul 25;257(14):7915–7917. [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Jonge P., Leer R. J., Planta R. J. The transcription termination site of the ribosomal RNA operon in yeast. Nucleic Acids Res. 1980 Nov 25;8(22):5179–5192. doi: 10.1093/nar/8.22.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
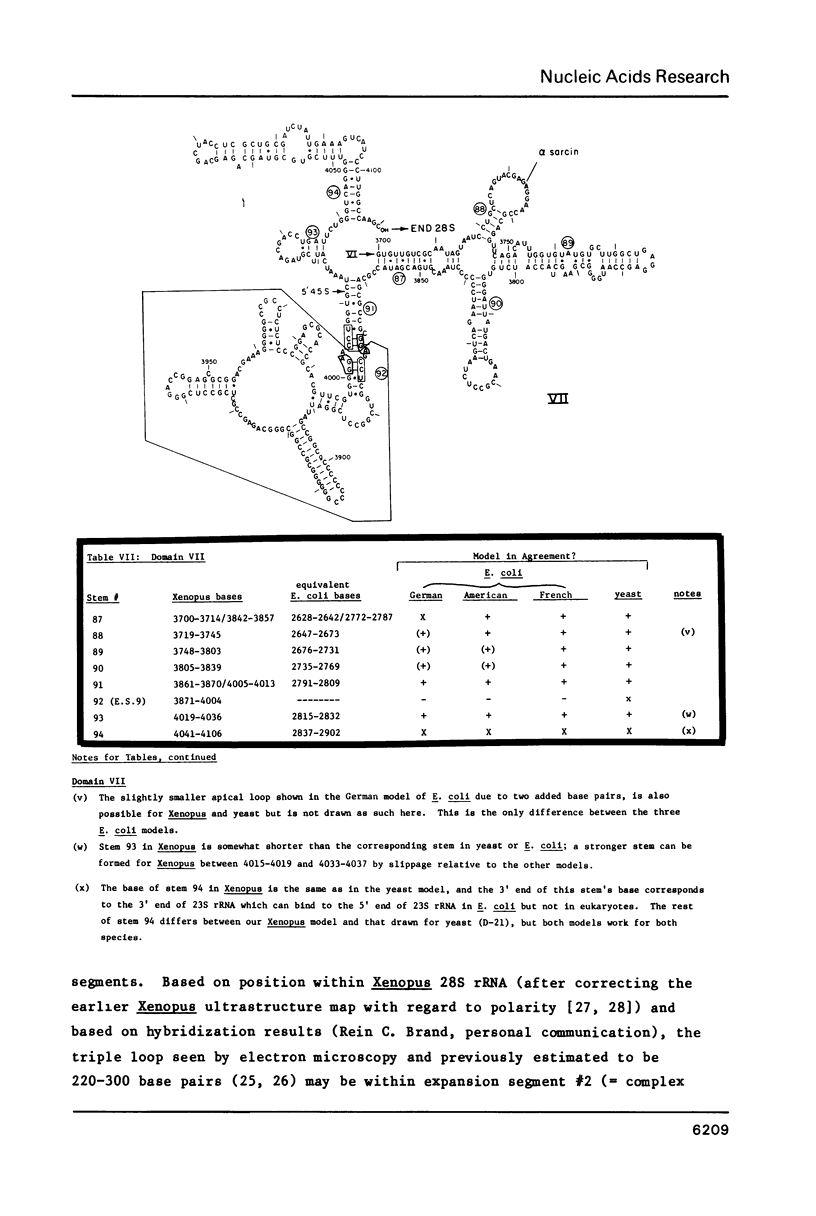
- Ware V. C., Tague B. W., Clark C. G., Gourse R. L., Brand R. C., Gerbi S. A. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1983 Nov 25;11(22):7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Kelley D. E., Perry R. P. Secondary structure maps of ribosomal RNA. II. Processing of mouse L-cell ribosomal RNA and variations in the processing pathway. J Mol Biol. 1974 Oct 25;89(2):397–407. doi: 10.1016/0022-2836(74)90527-0. [DOI] [PubMed] [Google Scholar]
- Wild M. A., Sommer R. Sequence of a ribosomal RNA gene intron from Tetrahymena. Nature. 1980 Feb 14;283(5748):693–694. doi: 10.1038/283693a0. [DOI] [PubMed] [Google Scholar]



