Abstract
In the title compound, C18H18ClN3O4, the dihedral angle between the pyrimidine ring and the N-bonded ester grouping is 56.27 (7)° and the dihedral angle between the aromatic rings is 11.23 (7)°.
Related literature
For background to the biological activities of pyrimidine compounds, see: Patil et al. (2003 ▶); Siddiqui et al. (2007 ▶).
Experimental
Crystal data
C18H18ClN3O4
M r = 375.80
Monoclinic,
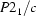
a = 11.530 (2) Å
b = 12.384 (2) Å
c = 14.010 (3) Å
β = 119.820 (4)°
V = 1735.6 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.25 mm−1
T = 173 K
0.23 × 0.20 × 0.16 mm
Data collection
MM007-HF CCD (Saturn 724+) diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku, 2007 ▶) T min = 0.945, T max = 0.961
8909 measured reflections
3925 independent reflections
3518 reflections with I > 2σ(I)
R int = 0.038
Refinement
R[F 2 > 2σ(F 2)] = 0.058
wR(F 2) = 0.142
S = 1.09
3925 reflections
236 parameters
H-atom parameters constrained
Δρmax = 0.65 e Å−3
Δρmin = −0.70 e Å−3
Data collection: CrystalClear (Rigaku, 2007 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811034313/hb6350sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811034313/hb6350Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811034313/hb6350Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The author thanks Shandong Provincial Natural Science Foundation, China (Y2008B29) and Yuandu Scholar of Weifang City for support.
supplementary crystallographic information
Comment
Pyrimidine is a widespread heterocyclic moiety present in numerous natural products. Pyrimidines are important not only because they are an integral part of genetic materials, but also they have important biodynamic properties and biological activities (e.g. Siddiqui et al. (2007); Patil et al. (2003).
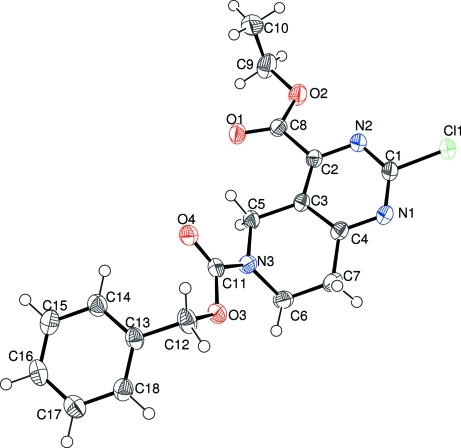
In continuation of our interest in the synthesis of the biologically active heterocyclic compound, here we report the sigle crystal structure of the title compound, (I). In the molecule (Fig. 1), atoms N1,C1,N2,C2,C3 and C4 lie in a plane (p1), with a maximum deviation of 0.0312 (13) Å, atoms N3,C11,O3,O4, and C12 lie in a plane (p2) too, the maximum deviation is 0.0647 (13) Å. The dihedral angle between p1 and p2 is 56.27 (7)°. The dihedral angles made by the phenyl ring with p1 and p2 are 11.23 (7)° and 60.25 (8)°, respectively.
Experimental
5-tert-butyl 3-ethyl 1-isopropyl-6,7-dihydro-1H-pyrazolo[4,3-c] pyridine-3,5(4H)-dicarboxylate was synthesized with 6-benzyl 4-ethyl 2-hydroxy-7,8-dihydropyrido[4,3-d] pyrimidine-4,6(5H)- dicarboxylate (1 eq), andN,N-dimethylaniline (2 eq) in POCl3 (as solvent) in refluxing for 3 hrs. Colourless blocks of (I) were obtained by recrystallization from ethanol at room temperature.
Refinement
All H atoms were fixed geometrically and allowed to ride on their attached atoms, with C—H distances in the range 0.95–0.98 Å, and with Uiso(H) = 1.2Ueq(C) or Uiso(H) = 1.5Ueq(Cmethyl).
Figures
Fig. 1.
The molecular structure of (I) with displacement ellipsoids drawn at the 50% probability level.
Crystal data
| C18H18ClN3O4 | F(000) = 784 |
| Mr = 375.80 | Dx = 1.438 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 6088 reflections |
| a = 11.530 (2) Å | θ = 1.6–27.5° |
| b = 12.384 (2) Å | µ = 0.25 mm−1 |
| c = 14.010 (3) Å | T = 173 K |
| β = 119.820 (4)° | Block, colorless |
| V = 1735.6 (6) Å3 | 0.23 × 0.20 × 0.16 mm |
| Z = 4 |
Data collection
| MM007-HF CCD (Saturn 724+) diffractometer | 3925 independent reflections |
| Radiation source: rotating anode | 3518 reflections with I > 2σ(I) |
| Confocal | Rint = 0.038 |
| ω scans at fixed χ = 45° | θmax = 27.5°, θmin = 2.0° |
| Absorption correction: multi-scan (CrystalClear; Rigaku, 2007) | h = −14→12 |
| Tmin = 0.945, Tmax = 0.961 | k = −12→16 |
| 8909 measured reflections | l = −16→18 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.058 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.142 | H-atom parameters constrained |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0565P)2 + 1.1725P] where P = (Fo2 + 2Fc2)/3 |
| 3925 reflections | (Δ/σ)max < 0.001 |
| 236 parameters | Δρmax = 0.65 e Å−3 |
| 0 restraints | Δρmin = −0.70 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.70799 (5) | 0.21924 (5) | 0.19746 (6) | 0.0470 (2) | |
| O1 | 0.11289 (14) | 0.13056 (12) | 0.00647 (13) | 0.0367 (4) | |
| O2 | 0.29267 (15) | 0.02745 (12) | 0.10881 (13) | 0.0405 (4) | |
| O3 | 0.04486 (14) | 0.60281 (12) | −0.14053 (12) | 0.0329 (3) | |
| O4 | −0.02266 (14) | 0.43250 (12) | −0.20318 (12) | 0.0349 (4) | |
| N1 | 0.51941 (17) | 0.36384 (15) | 0.13641 (14) | 0.0320 (4) | |
| N2 | 0.45847 (16) | 0.17871 (14) | 0.12649 (14) | 0.0299 (4) | |
| N3 | 0.12864 (17) | 0.46466 (14) | −0.02346 (14) | 0.0316 (4) | |
| C1 | 0.5421 (2) | 0.25868 (18) | 0.14647 (17) | 0.0315 (4) | |
| C2 | 0.33018 (19) | 0.20875 (16) | 0.08209 (15) | 0.0256 (4) | |
| C3 | 0.28806 (18) | 0.31618 (16) | 0.05975 (14) | 0.0246 (4) | |
| C4 | 0.3906 (2) | 0.39277 (17) | 0.09374 (16) | 0.0280 (4) | |
| C5 | 0.14314 (19) | 0.35043 (16) | 0.00482 (16) | 0.0282 (4) | |
| H5A | 0.0904 | 0.3072 | −0.0627 | 0.034* | |
| H5B | 0.1076 | 0.3363 | 0.0550 | 0.034* | |
| C6 | 0.2223 (2) | 0.53565 (17) | 0.06382 (17) | 0.0353 (5) | |
| H6A | 0.2174 | 0.5234 | 0.1315 | 0.042* | |
| H6B | 0.1992 | 0.6120 | 0.0415 | 0.042* | |
| C7 | 0.3628 (2) | 0.51207 (17) | 0.08584 (19) | 0.0365 (5) | |
| H7A | 0.3741 | 0.5431 | 0.0258 | 0.044* | |
| H7B | 0.4284 | 0.5473 | 0.1555 | 0.044* | |
| C8 | 0.2320 (2) | 0.11846 (16) | 0.06036 (16) | 0.0280 (4) | |
| C9 | 0.2049 (2) | −0.0631 (2) | 0.0968 (2) | 0.0466 (6) | |
| H9A | 0.1219 | −0.0353 | 0.0924 | 0.056* | |
| H9B | 0.2500 | −0.1104 | 0.1623 | 0.056* | |
| C10 | 0.1705 (3) | −0.1273 (2) | −0.0041 (2) | 0.0520 (7) | |
| H10A | 0.1185 | −0.0824 | −0.0694 | 0.078* | |
| H10B | 0.1175 | −0.1905 | −0.0072 | 0.078* | |
| H10C | 0.2528 | −0.1511 | −0.0019 | 0.078* | |
| C11 | 0.04534 (18) | 0.49442 (17) | −0.12869 (16) | 0.0279 (4) | |
| C12 | −0.0290 (2) | 0.6427 (2) | −0.25290 (18) | 0.0364 (5) | |
| H12A | −0.0266 | 0.5874 | −0.3030 | 0.044* | |
| H12B | 0.0157 | 0.7083 | −0.2592 | 0.044* | |
| C13 | −0.1726 (2) | 0.66913 (17) | −0.28908 (16) | 0.0293 (4) | |
| C14 | −0.2715 (2) | 0.59019 (18) | −0.32842 (17) | 0.0343 (5) | |
| H14 | −0.2482 | 0.5171 | −0.3310 | 0.041* | |
| C15 | −0.4040 (2) | 0.6170 (2) | −0.36400 (18) | 0.0390 (5) | |
| H15 | −0.4707 | 0.5624 | −0.3894 | 0.047* | |
| C16 | −0.4380 (2) | 0.7237 (2) | −0.36211 (18) | 0.0388 (5) | |
| H16 | −0.5285 | 0.7426 | −0.3874 | 0.047* | |
| C17 | −0.3409 (2) | 0.8033 (2) | −0.32367 (18) | 0.0370 (5) | |
| H17 | −0.3647 | 0.8766 | −0.3228 | 0.044* | |
| C18 | −0.2084 (2) | 0.77566 (18) | −0.28631 (17) | 0.0321 (4) | |
| H18 | −0.1416 | 0.8302 | −0.2586 | 0.039* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0225 (3) | 0.0441 (3) | 0.0666 (4) | −0.0014 (2) | 0.0163 (3) | −0.0168 (3) |
| O1 | 0.0234 (7) | 0.0309 (8) | 0.0474 (9) | −0.0025 (6) | 0.0112 (6) | 0.0032 (7) |
| O2 | 0.0296 (8) | 0.0273 (8) | 0.0499 (9) | −0.0021 (6) | 0.0085 (7) | 0.0081 (7) |
| O3 | 0.0283 (7) | 0.0276 (7) | 0.0328 (7) | 0.0048 (6) | 0.0077 (6) | 0.0067 (6) |
| O4 | 0.0286 (7) | 0.0335 (8) | 0.0307 (7) | 0.0053 (6) | 0.0057 (6) | −0.0014 (6) |
| N1 | 0.0244 (8) | 0.0322 (9) | 0.0354 (9) | −0.0054 (7) | 0.0118 (7) | −0.0068 (7) |
| N2 | 0.0231 (8) | 0.0311 (9) | 0.0313 (8) | −0.0004 (7) | 0.0103 (7) | −0.0049 (7) |
| N3 | 0.0283 (9) | 0.0249 (9) | 0.0290 (8) | 0.0018 (7) | 0.0047 (7) | 0.0018 (7) |
| C1 | 0.0221 (9) | 0.0352 (11) | 0.0329 (10) | −0.0015 (8) | 0.0105 (8) | −0.0072 (9) |
| C2 | 0.0228 (9) | 0.0274 (10) | 0.0226 (8) | −0.0002 (7) | 0.0083 (7) | −0.0005 (7) |
| C3 | 0.0222 (9) | 0.0273 (9) | 0.0212 (8) | −0.0019 (7) | 0.0083 (7) | −0.0002 (7) |
| C4 | 0.0266 (9) | 0.0289 (10) | 0.0249 (9) | −0.0033 (8) | 0.0100 (8) | −0.0025 (8) |
| C5 | 0.0223 (9) | 0.0274 (10) | 0.0291 (10) | 0.0014 (8) | 0.0083 (8) | 0.0033 (8) |
| C6 | 0.0347 (11) | 0.0264 (10) | 0.0312 (10) | 0.0027 (8) | 0.0061 (9) | −0.0025 (8) |
| C7 | 0.0314 (11) | 0.0259 (10) | 0.0397 (12) | −0.0054 (9) | 0.0084 (9) | −0.0023 (9) |
| C8 | 0.0268 (10) | 0.0259 (10) | 0.0278 (9) | −0.0012 (8) | 0.0110 (8) | 0.0004 (7) |
| C9 | 0.0371 (12) | 0.0343 (12) | 0.0531 (14) | −0.0074 (10) | 0.0108 (11) | 0.0146 (11) |
| C10 | 0.0436 (14) | 0.0340 (13) | 0.0746 (18) | −0.0014 (11) | 0.0264 (13) | −0.0056 (12) |
| C11 | 0.0208 (9) | 0.0292 (10) | 0.0312 (10) | 0.0027 (8) | 0.0110 (8) | 0.0012 (8) |
| C12 | 0.0275 (11) | 0.0420 (13) | 0.0359 (11) | 0.0062 (9) | 0.0130 (9) | 0.0148 (9) |
| C13 | 0.0252 (9) | 0.0332 (11) | 0.0262 (9) | 0.0037 (8) | 0.0103 (8) | 0.0092 (8) |
| C14 | 0.0298 (10) | 0.0340 (11) | 0.0331 (10) | 0.0035 (9) | 0.0110 (9) | 0.0066 (9) |
| C15 | 0.0290 (11) | 0.0460 (13) | 0.0351 (11) | −0.0053 (10) | 0.0107 (9) | 0.0017 (10) |
| C16 | 0.0256 (10) | 0.0527 (14) | 0.0339 (11) | 0.0078 (10) | 0.0116 (9) | 0.0047 (10) |
| C17 | 0.0362 (11) | 0.0393 (12) | 0.0343 (11) | 0.0076 (10) | 0.0167 (9) | 0.0036 (9) |
| C18 | 0.0292 (10) | 0.0344 (11) | 0.0294 (10) | −0.0010 (9) | 0.0120 (8) | 0.0053 (8) |
Geometric parameters (Å, °)
| Cl1—C1 | 1.746 (2) | C6—H6B | 0.9900 |
| O1—C8 | 1.203 (2) | C7—H7A | 0.9900 |
| O2—C8 | 1.322 (2) | C7—H7B | 0.9900 |
| O2—C9 | 1.463 (3) | C9—C10 | 1.493 (4) |
| O3—C11 | 1.352 (2) | C9—H9A | 0.9900 |
| O3—C12 | 1.454 (2) | C9—H9B | 0.9900 |
| O4—C11 | 1.216 (2) | C10—H10A | 0.9800 |
| N1—C1 | 1.322 (3) | C10—H10B | 0.9800 |
| N1—C4 | 1.344 (3) | C10—H10C | 0.9800 |
| N2—C1 | 1.312 (3) | C12—C13 | 1.508 (3) |
| N2—C2 | 1.341 (2) | C12—H12A | 0.9900 |
| N3—C11 | 1.350 (2) | C12—H12B | 0.9900 |
| N3—C5 | 1.456 (3) | C13—C18 | 1.389 (3) |
| N3—C6 | 1.456 (3) | C13—C14 | 1.391 (3) |
| C2—C3 | 1.397 (3) | C14—C15 | 1.392 (3) |
| C2—C8 | 1.510 (3) | C14—H14 | 0.9500 |
| C3—C4 | 1.402 (3) | C15—C16 | 1.383 (3) |
| C3—C5 | 1.512 (3) | C15—H15 | 0.9500 |
| C4—C7 | 1.504 (3) | C16—C17 | 1.383 (3) |
| C5—H5A | 0.9900 | C16—H16 | 0.9500 |
| C5—H5B | 0.9900 | C17—C18 | 1.389 (3) |
| C6—C7 | 1.519 (3) | C17—H17 | 0.9500 |
| C6—H6A | 0.9900 | C18—H18 | 0.9500 |
| C8—O2—C9 | 115.82 (17) | O2—C9—C10 | 111.1 (2) |
| C11—O3—C12 | 115.75 (17) | O2—C9—H9A | 109.4 |
| C1—N1—C4 | 115.24 (17) | C10—C9—H9A | 109.4 |
| C1—N2—C2 | 114.46 (18) | O2—C9—H9B | 109.4 |
| C11—N3—C5 | 119.03 (17) | C10—C9—H9B | 109.4 |
| C11—N3—C6 | 125.44 (18) | H9A—C9—H9B | 108.0 |
| C5—N3—C6 | 114.90 (16) | C9—C10—H10A | 109.5 |
| N2—C1—N1 | 129.44 (19) | C9—C10—H10B | 109.5 |
| N2—C1—Cl1 | 114.57 (16) | H10A—C10—H10B | 109.5 |
| N1—C1—Cl1 | 115.97 (16) | C9—C10—H10C | 109.5 |
| N2—C2—C3 | 123.25 (18) | H10A—C10—H10C | 109.5 |
| N2—C2—C8 | 115.55 (17) | H10B—C10—H10C | 109.5 |
| C3—C2—C8 | 121.17 (17) | O4—C11—N3 | 124.76 (19) |
| C2—C3—C4 | 115.31 (18) | O4—C11—O3 | 124.00 (18) |
| C2—C3—C5 | 123.65 (17) | N3—C11—O3 | 111.23 (17) |
| C4—C3—C5 | 121.03 (18) | O3—C12—C13 | 113.06 (17) |
| N1—C4—C3 | 121.96 (19) | O3—C12—H12A | 109.0 |
| N1—C4—C7 | 116.26 (18) | C13—C12—H12A | 109.0 |
| C3—C4—C7 | 121.77 (18) | O3—C12—H12B | 109.0 |
| N3—C5—C3 | 111.06 (16) | C13—C12—H12B | 109.0 |
| N3—C5—H5A | 109.4 | H12A—C12—H12B | 107.8 |
| C3—C5—H5A | 109.4 | C18—C13—C14 | 118.86 (19) |
| N3—C5—H5B | 109.4 | C18—C13—C12 | 119.29 (19) |
| C3—C5—H5B | 109.4 | C14—C13—C12 | 121.8 (2) |
| H5A—C5—H5B | 108.0 | C13—C14—C15 | 120.8 (2) |
| N3—C6—C7 | 108.99 (18) | C13—C14—H14 | 119.6 |
| N3—C6—H6A | 109.9 | C15—C14—H14 | 119.6 |
| C7—C6—H6A | 109.9 | C16—C15—C14 | 119.5 (2) |
| N3—C6—H6B | 109.9 | C16—C15—H15 | 120.3 |
| C7—C6—H6B | 109.9 | C14—C15—H15 | 120.3 |
| H6A—C6—H6B | 108.3 | C15—C16—C17 | 120.4 (2) |
| C4—C7—C6 | 111.75 (18) | C15—C16—H16 | 119.8 |
| C4—C7—H7A | 109.3 | C17—C16—H16 | 119.8 |
| C6—C7—H7A | 109.3 | C16—C17—C18 | 119.8 (2) |
| C4—C7—H7B | 109.3 | C16—C17—H17 | 120.1 |
| C6—C7—H7B | 109.3 | C18—C17—H17 | 120.1 |
| H7A—C7—H7B | 107.9 | C13—C18—C17 | 120.7 (2) |
| O1—C8—O2 | 125.15 (19) | C13—C18—H18 | 119.7 |
| O1—C8—C2 | 122.78 (18) | C17—C18—H18 | 119.7 |
| O2—C8—C2 | 112.07 (16) | ||
| C2—N2—C1—N1 | 5.3 (3) | C9—O2—C8—O1 | −1.8 (3) |
| C2—N2—C1—Cl1 | −176.77 (14) | C9—O2—C8—C2 | 177.02 (18) |
| C4—N1—C1—N2 | −4.1 (3) | N2—C2—C8—O1 | −168.70 (19) |
| C4—N1—C1—Cl1 | 177.96 (14) | C3—C2—C8—O1 | 13.1 (3) |
| C1—N2—C2—C3 | −0.7 (3) | N2—C2—C8—O2 | 12.5 (2) |
| C1—N2—C2—C8 | −178.82 (17) | C3—C2—C8—O2 | −165.70 (18) |
| N2—C2—C3—C4 | −4.1 (3) | C8—O2—C9—C10 | 89.1 (3) |
| C8—C2—C3—C4 | 173.94 (17) | C5—N3—C11—O4 | 1.8 (3) |
| N2—C2—C3—C5 | 177.00 (18) | C6—N3—C11—O4 | 172.3 (2) |
| C8—C2—C3—C5 | −5.0 (3) | C5—N3—C11—O3 | −179.43 (17) |
| C1—N1—C4—C3 | −1.7 (3) | C6—N3—C11—O3 | −9.0 (3) |
| C1—N1—C4—C7 | 177.53 (19) | C12—O3—C11—O4 | −8.4 (3) |
| C2—C3—C4—N1 | 5.3 (3) | C12—O3—C11—N3 | 172.86 (17) |
| C5—C3—C4—N1 | −175.74 (18) | C11—O3—C12—C13 | 91.6 (2) |
| C2—C3—C4—C7 | −173.87 (18) | O3—C12—C13—C18 | 98.8 (2) |
| C5—C3—C4—C7 | 5.1 (3) | O3—C12—C13—C14 | −83.2 (3) |
| C11—N3—C5—C3 | 124.66 (19) | C18—C13—C14—C15 | −0.3 (3) |
| C6—N3—C5—C3 | −46.7 (2) | C12—C13—C14—C15 | −178.33 (19) |
| C2—C3—C5—N3 | −170.82 (17) | C13—C14—C15—C16 | 1.3 (3) |
| C4—C3—C5—N3 | 10.3 (3) | C14—C15—C16—C17 | −1.0 (3) |
| C11—N3—C6—C7 | −104.5 (2) | C15—C16—C17—C18 | −0.3 (3) |
| C5—N3—C6—C7 | 66.2 (2) | C14—C13—C18—C17 | −1.0 (3) |
| N1—C4—C7—C6 | −165.93 (18) | C12—C13—C18—C17 | 177.13 (19) |
| C3—C4—C7—C6 | 13.3 (3) | C16—C17—C18—C13 | 1.2 (3) |
| N3—C6—C7—C4 | −46.0 (2) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6350).
References
- Patil, L. R., Mandhare, P. N., Bondge, S. P., Munde, S. B. & Mane, R. (2003). Indian J. Heterocycl. Chem. 12, 245–248.
- Rigaku (2007). CrystalClear Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Siddiqui, A. A., Rajesh, R. Mojahid-Ul-Islam, Alagarsamy, V., Meyyanathan, S. N., Kumar, B. P. & Suresh, B. (2007). Acta Pol. Pharm. 64, 17–26. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811034313/hb6350sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811034313/hb6350Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811034313/hb6350Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report