Abstract
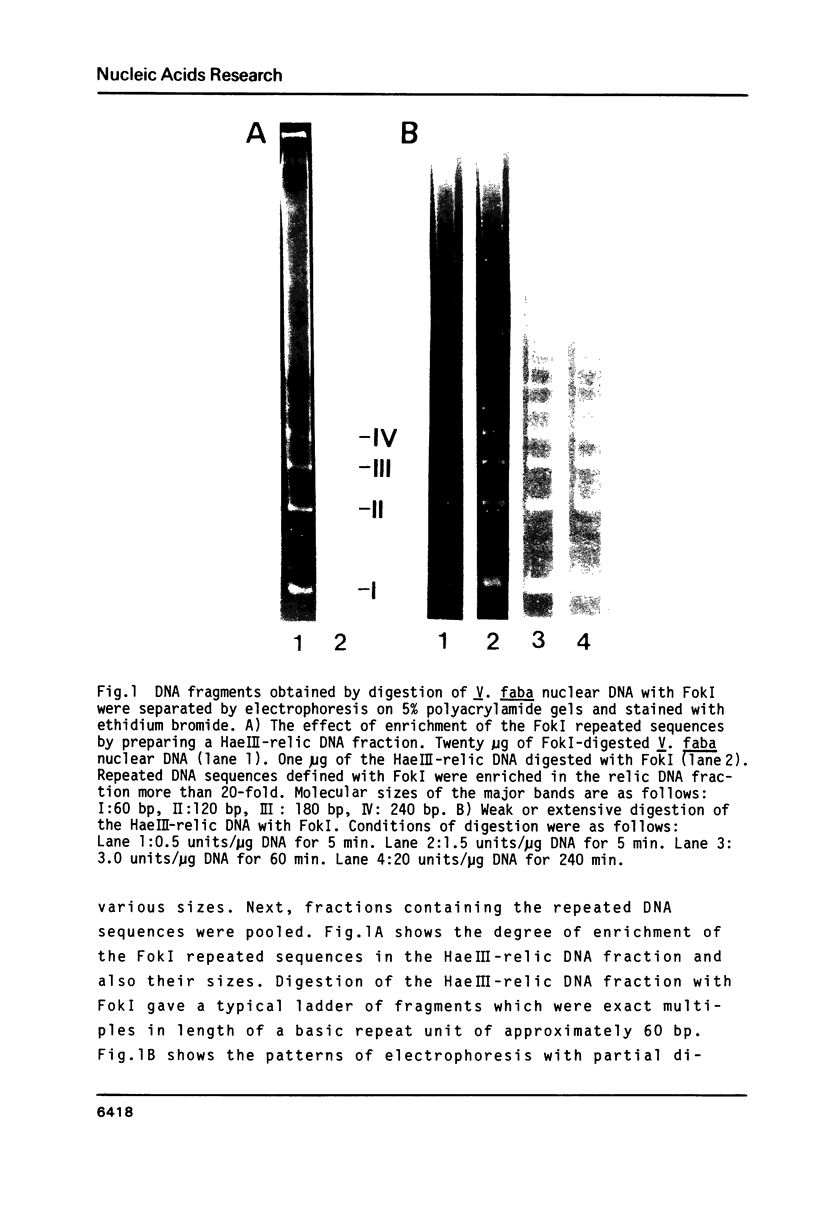
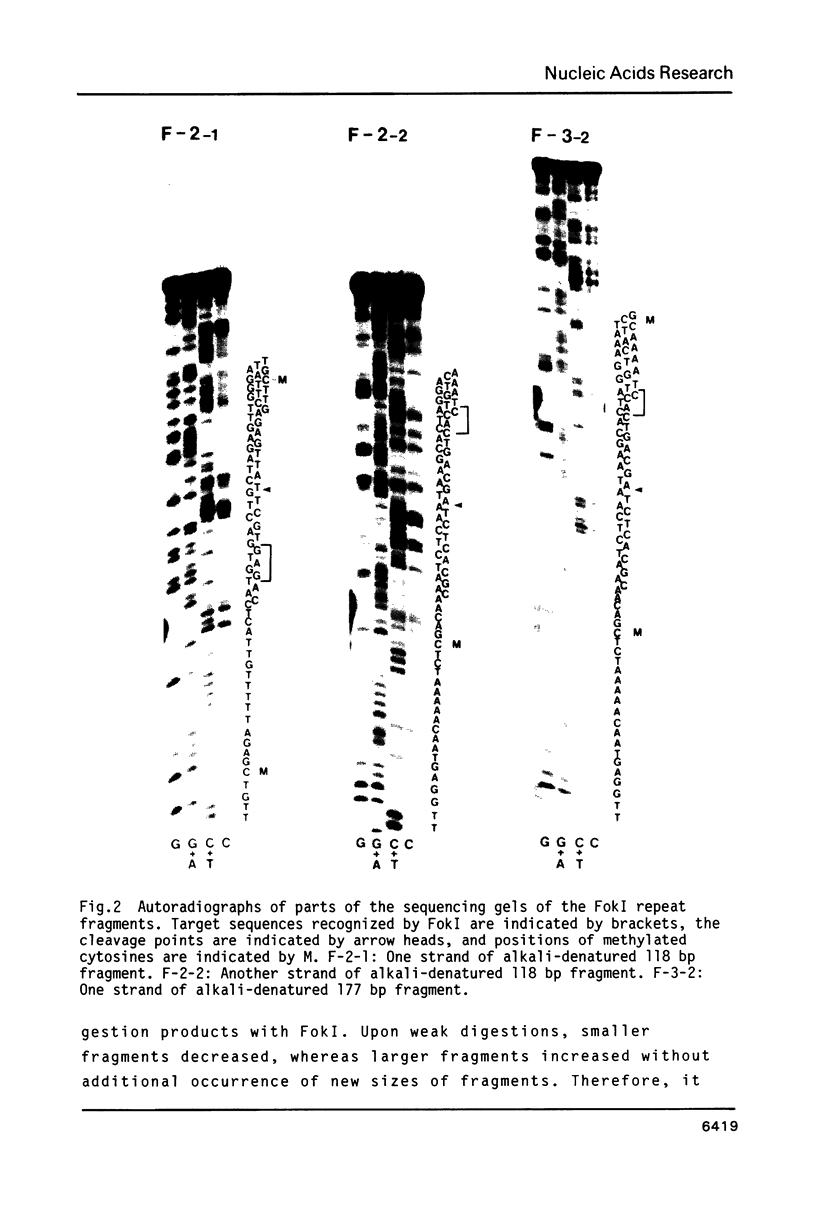
A type of highly repeated DNA sequences present in the genome of Vicia faba was detected by digestion its nuclear DNA with FokI endonuclease and fractionating the digests on polyacrylamide gels. Four fragments of 59, 108, 177 and 246 bp of the FokI repeated sequences were collected from the gels and their primary structures were determined by the method of Maxam and Gilbert. These repeated DNA sequences were shown to be a multiple tandem array of a 59 bp sequence element. And its nucleotide sequence was almost completely conserved among all the sequence members of each the size class and also among these classes. This sequence element consists of a duplet of an about the duplet has an incomplete dyad symmetrical structure.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedbrook J. R., Jones J., O'Dell M., Thompson R. D., Flavell R. B. A molecular description of telometic heterochromatin in secale species. Cell. 1980 Feb;19(2):545–560. doi: 10.1016/0092-8674(80)90529-2. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Britten R. J. Regulation of gene expression: possible role of repetitive sequences. Science. 1979 Jun 8;204(4397):1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Deumling B. Sequence arrangement of a highly methylated satellite DNA of a plant, Scilla: A tandemly repeated inverted repeat. Proc Natl Acad Sci U S A. 1981 Jan;78(1):338–342. doi: 10.1073/pnas.78.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle W. F., Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980 Apr 17;284(5757):601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Endow S. A. Analysis of Drosophila melanogaster satellite IV with restriction endonuclease MboII. J Mol Biol. 1977 Aug 15;114(3):441–449. doi: 10.1016/0022-2836(77)90261-3. [DOI] [PubMed] [Google Scholar]
- Frommer M., Prosser J., Tkachuk D., Reisner A. H., Vincent P. C. Simple repeated sequences in human satellite DNA. Nucleic Acids Res. 1982 Jan 22;10(2):547–563. doi: 10.1093/nar/10.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Naveh-Many T., Cedar H., Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981 Aug 27;292(5826):860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Hemleben V., Leweke B., Roth A., Stadler J. Organization of highly repetitive satellite DNA of two Cucurbitaceae species (Cucumis melo and Cucumis sativus). Nucleic Acids Res. 1982 Jan 22;10(2):631–644. doi: 10.1093/nar/10.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981 Feb 11;9(3):683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb V. F., Glasser S., King D., Lingrel J. B. A cluster of repetitive elements within a 700 base pair region in the mouse genome. Nucleic Acids Res. 1983 Apr 11;11(7):2177–2184. doi: 10.1093/nar/11.7.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. Dictyostelium 17S, 25S, and 5S rDNAs lie within a 38,000 base pair repeated unit. Cell. 1976 Nov;9(3):431–438. doi: 10.1016/0092-8674(76)90088-x. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Consensus sequence of mouse satellite DNA indicates it is derived from tandem 116 basepair repeats. FEBS Lett. 1981 Jun 29;129(1):25–28. doi: 10.1016/0014-5793(81)80746-6. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Nucleotide sequence definition of a major human repeated DNA, the Hind III 1.9 kb family. Nucleic Acids Res. 1982 May 25;10(10):3211–3219. doi: 10.1093/nar/10.10.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mullins J. I., Blumenfeld M. Satellite Ic: a possible link between the satellite DNAs of D. virilis and D. melanogaster. Cell. 1979 Jul;17(3):615–621. doi: 10.1016/0092-8674(79)90269-1. [DOI] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. Selfish DNA: the ultimate parasite. Nature. 1980 Apr 17;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Peacock W. J., Dennis E. S., Rhoades M. M., Pryor A. J. Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4490–4494. doi: 10.1073/pnas.78.7.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Płucienniczak A., Skowroński J., Jaworski J. Nucleotide sequence of bovine 1.715 satellite DNA and its relation to other bovine satellite sequences. J Mol Biol. 1982 Jun 25;158(2):293–304. doi: 10.1016/0022-2836(82)90434-x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Deininger P. L., Houck C. M., Schmid C. W. A dimer satellite sequence in bonnet monkey DNA consists of distinct monomer subunits. J Mol Biol. 1980 Jan 15;136(2):151–167. doi: 10.1016/0022-2836(80)90310-1. [DOI] [PubMed] [Google Scholar]
- Sakamaki T., Fukuei K., Takahashi N., Tanifuji S. Rapidly labeled intermediates in DNA replication in higher plants. Biochim Biophys Acta. 1975 Jul 7;395(3):314–321. doi: 10.1016/0005-2787(75)90202-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Streeck R. E. Inserted sequences in bovine satellite DNA's. Science. 1981 Jul 24;213(4506):443–445. doi: 10.1126/science.6264600. [DOI] [PubMed] [Google Scholar]
- Taparowsky E. J., Gerbi S. A. Sequence analysis of bovine satellite I DNA (1.715 gm/cm3). Nucleic Acids Res. 1982 Feb 25;10(4):1271–1281. doi: 10.1093/nar/10.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis J. N., Deumling B., Ingle J. Localisation of satellite DNA sequences in nuclei and chromosomes of two plants. Nature. 1975 Sep 11;257(5522):152–155. doi: 10.1038/257152a0. [DOI] [PubMed] [Google Scholar]