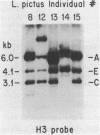
Abstract
We have isolated and characterized a third nonallelic tandemly arrayed histone cluster (LpE) from the sea urchin Lytechinus pictus. Although this tandem array is not intermingled with the other two early histone gene families also found in the L. pictus genome, the order and polarity of the five histone coding sequences in this family are the same as every other well characterized sea urchin early histone gene family. Heteroduplex analysis and restriction endonuclease mapping experiments indicate that the LpE family is more closely related to the B-C than the A-D family of early histone genes. Examination of several individual sperm DNA samples has revealed considerable polymorphism in each of the three tandem repeat families. Within an individual, however, each family is remarkably homogeneous. Thus, our results indicate that rapid fixation of variants acts to homogenize the members of a single tandem array at a considerably faster rate within a family than between families. However, at least some exchange of sequences between families is evident based on the conservation of many restriction endonuclease recognition sites and from analysis of a a cosmid clone in which the A-D and E tandem repeats are found adjacent to one another. These differences in the rate of fixation of variants within and between these families are likely to be responsible for the maintenance of diversity between the different families.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnheim N., Krystal M., Schmickel R., Wilson G., Ryder O., Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Busslinger M., Portmann R., Irminger J. C., Birnstiel M. L. Ubiquitous and gene-specific regulatory 5' sequences in a sea urchin histone DNA clone coding for histone protein variants. Nucleic Acids Res. 1980 Mar 11;8(5):957–977. doi: 10.1093/nar/8.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., Rusconi S., Birnstiel M. L. An unusual evolutionary behaviour of a sea urchin histone gene cluster. EMBO J. 1982;1(1):27–33. doi: 10.1002/j.1460-2075.1982.tb01119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs G., Levy S., Kedes L. H. Rapid purification of biologically active individual histone messenger RNAs by hybridization to cloned DNA linked to cellulose. Biochemistry. 1979 Jan 9;18(1):208–213. doi: 10.1021/bi00568a032. [DOI] [PubMed] [Google Scholar]
- Childs G., Maxson R., Cohn R. H., Kedes L. Orphons: dispersed genetic elements derived from tandem repetitive genes of eucaryotes. Cell. 1981 Mar;23(3):651–663. doi: 10.1016/0092-8674(81)90428-1. [DOI] [PubMed] [Google Scholar]
- Childs G., Nocente-McGrath C., Lieber T., Holt C., Knowles J. A. Sea urchin (lytechinus pictus) late-stage histone H3 and H4 genes: characterization and mapping of a clustered but nontandemly linked multigene family. Cell. 1982 Dec;31(2 Pt 1):383–393. doi: 10.1016/0092-8674(82)90132-5. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Dover G. A. Unequal exchanges and the coevolution of X and Y rDNA arrays in Drosophila melanogaster. Cell. 1983 Jul;33(3):849–855. doi: 10.1016/0092-8674(83)90027-2. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Thoday J. M., Dover G. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature. 1982 Feb 18;295(5850):564–568. doi: 10.1038/295564a0. [DOI] [PubMed] [Google Scholar]
- Coen E., Strachan T., Dover G. Dynamics of concerted evolution of ribosomal DNA and histone gene families in the melanogaster species subgroup of Drosophila. J Mol Biol. 1982 Jun 15;158(1):17–35. doi: 10.1016/0022-2836(82)90448-x. [DOI] [PubMed] [Google Scholar]
- Cohn R. H., Kedes L. H. Nonallelic histone gene clusters of individual sea urchins (Lytechinus pictus): mapping of homologies in coding and spacer DNA. Cell. 1979 Nov;18(3):855–864. doi: 10.1016/0092-8674(79)90137-5. [DOI] [PubMed] [Google Scholar]
- Cohn R. H., Kedes L. H. Nonallelic histone gene clusters of individual sea urchins (Lytechinus pictus): polarity and gene organization. Cell. 1979 Nov;18(3):843–853. doi: 10.1016/0092-8674(79)90136-3. [DOI] [PubMed] [Google Scholar]
- Ford P. J., Brown R. D. Sequences of 5S ribosomal RNA from Xenopus mulleri and the evolution of 5S gene-coding sequences. Cell. 1976 Aug;8(4):485–493. doi: 10.1016/0092-8674(76)90216-6. [DOI] [PubMed] [Google Scholar]
- Grosveld F. G., Lund T., Murray E. J., Mellor A. L., Dahl H. H., Flavell R. A. The construction of cosmid libraries which can be used to transform eukaryotic cells. Nucleic Acids Res. 1982 Nov 11;10(21):6715–6732. doi: 10.1093/nar/10.21.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Schedl P., Kedes L. Sequence analysis and evolution of sea urchin (Lytechinus pictus and Strongylocentrotus purpuratus) histone H4 messenger RNAs. J Mol Biol. 1976 Jun 25;104(2):351–369. doi: 10.1016/0022-2836(76)90276-x. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Hohn B., Murray K. Packaging recombinant DNA molecules into bacteriophage particles in vitro. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3259–3263. doi: 10.1073/pnas.74.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Cohn R. H., Kedes L. H., Davidson N. Positions of sea urchin (Strongylocentrotus purpuratus) histone genes relative to restriction endonuclease sites on the chimeric plasmids pSp2 and pSp17. Biochemistry. 1977 Apr 5;16(7):1504–1512. doi: 10.1021/bi00626a040. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes L. H., Cohn R. H., Lowry J. C., Chang A. C., Cohen S. N. The organization of sea urchin histone genes. Cell. 1975 Nov;6(3):359–369. doi: 10.1016/0092-8674(75)90185-3. [DOI] [PubMed] [Google Scholar]
- Kedes L. H. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- Lund T., Grosveld F. G., Flavell R. A. Isolation of transforming DNA by cosmid rescue. Proc Natl Acad Sci U S A. 1982 Jan;79(2):520–524. doi: 10.1073/pnas.79.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxson R., Mohun T., Gormezano G., Childs G., Kedes L. Distinct organizations and patterns of expression of early and late histone gene sets in the sea urchin. Nature. 1983 Jan 13;301(5896):120–125. doi: 10.1038/301120a0. [DOI] [PubMed] [Google Scholar]
- Overton G. C., Weinberg E. S. Length and sequence heterogeneity of the histone gene repeat unit of the sea urchin, S. purpuratus. Cell. 1978 Jun;14(2):247–257. doi: 10.1016/0092-8674(78)90111-3. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts J. M., Axel R. Gene amplification and gene correction in somatic cells. Cell. 1982 May;29(1):109–119. doi: 10.1016/0092-8674(82)90095-2. [DOI] [PubMed] [Google Scholar]
- Roberts S. B., Weisser K. E., Childs G. Sequence comparisons of non-allelic late histone genes and their early stage counterparts. Evidence for gene conversion within the sea urchin late stage gene family. J Mol Biol. 1984 Apr 25;174(4):647–662. doi: 10.1016/0022-2836(84)90088-3. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Smith M., Lambie E. J. Intrachromosomal movement of genetically marked Saccharomyces cerevisiae transposons by gene conversion. Mol Cell Biol. 1984 Apr;4(4):703–711. doi: 10.1128/mcb.4.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Vitelli L., Weinberg E. S. An inverted sea urchin histone gene sequence with breakpoints between TATA boxes and mRNA cap sites. Nucleic Acids Res. 1983 Apr 11;11(7):2135–2153. doi: 10.1093/nar/11.7.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]
- Yagura T., Yagura M., Muramatsu M. Drosophila melanogaster has different ribosomal RNA sequences on S and Y chromosomes. J Mol Biol. 1979 Oct 9;133(4):533–547. doi: 10.1016/0022-2836(79)90406-6. [DOI] [PubMed] [Google Scholar]