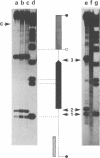
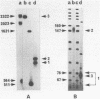
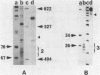
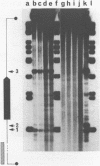
Abstract
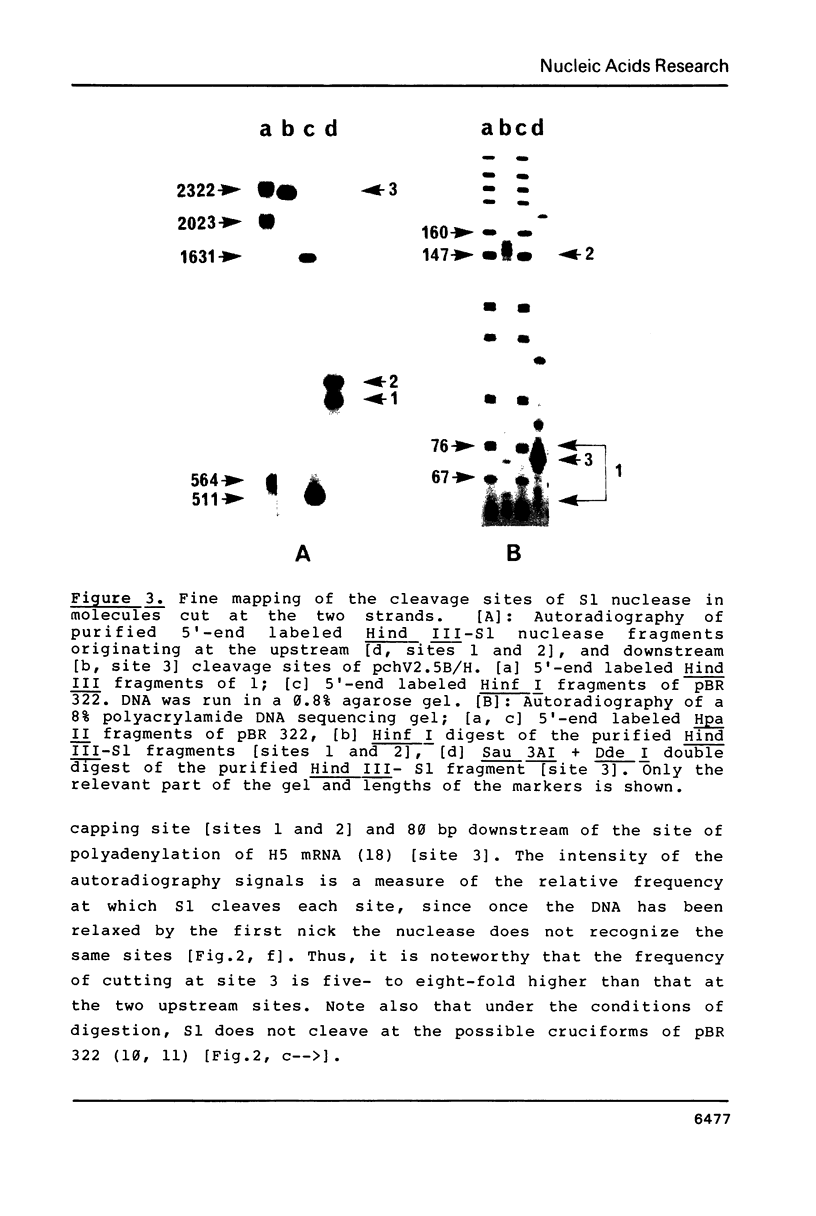
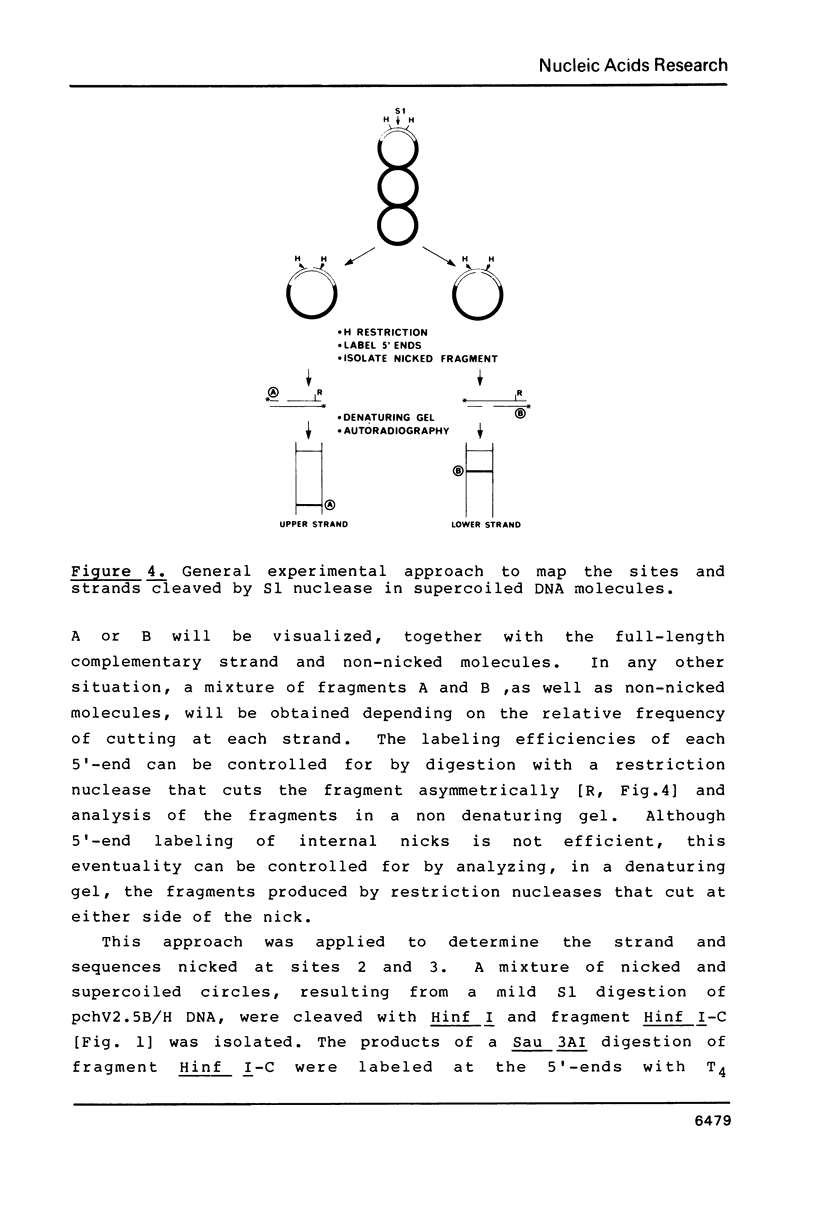
The potential of the cloned histone H5 gene to form altered DNA structures has been examined by S1 nuclease digestion of supercoiled recombinant plasmids containing up to 8.8 kbp of chicken DNa. The three main nicking sites map at the upstream and downstream sequences flanking the structural gene. The cleavage sites share sequence homology, strand specificity, and do not seem to be single-stranded. The sequence of the S1-sensitive sites does not suggest that the fragments can adopt any of the known DNA secondary structures.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cantor C. R. DNA choreography. Cell. 1981 Aug;25(2):293–295. doi: 10.1016/0092-8674(81)90045-3. [DOI] [PubMed] [Google Scholar]
- Dybvig K., Clark C. D., Aliperti G., Schlesinger M. J. A chicken repetitive DNA sequence that is highly sensitive to single-strand specific endonucleases. Nucleic Acids Res. 1983 Dec 10;11(23):8495–8508. doi: 10.1093/nar/11.23.8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. Chromatin structure, DNA structure. Nature. 1982 Dec 2;300(5891):402–403. doi: 10.1038/300402a0. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Glikin G. C., Gargiulo G., Rena-Descalzi L., Worcel A. Escherichia coli single-strand binding protein stabilizes specific denatured sites in superhelical DNA. Nature. 1983 Jun 30;303(5920):770–774. doi: 10.1038/303770a0. [DOI] [PubMed] [Google Scholar]
- Goding C. R., Russell W. C. S1 sensitive sites in adenovirus DNA. Nucleic Acids Res. 1983 Jan 11;11(1):21–36. doi: 10.1093/nar/11.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983 Apr 14;302(5909):632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., Colman A., Wells J. R. Chicken histone H5 mRNA: the polyadenylated RNA lacks the conserved histone 3' terminator sequence. Nucleic Acids Res. 1982 Nov 11;10(21):6777–6785. doi: 10.1093/nar/10.21.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., D'Andrea R., Wells J. R. The chicken H5 gene is unlinked to core and H1 histone genes. Nucleic Acids Res. 1983 Feb 11;11(3):619–627. doi: 10.1093/nar/11.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace H. A., Pelham H. R., Travers A. A. Association of an S1 nuclease-sensitive structure with short direct repeats 5' of Drosophila heat shock genes. Nature. 1983 Aug 11;304(5926):555–557. doi: 10.1038/304555a0. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Molgaard H. V., Perucho M., Ruiz-Carrillo A. Histone H5 messenger RNA is polyadenylated. Nature. 1980 Jan 31;283(5746):502–504. doi: 10.1038/283502a0. [DOI] [PubMed] [Google Scholar]
- Nickol J. M., Felsenfeld G. DNA conformation at the 5' end of the chicken adult beta-globin gene. Cell. 1983 Dec;35(2 Pt 1):467–477. doi: 10.1016/0092-8674(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Affolter M., Renaud J. Genomic organization of the genes coding for the six main histones of the chicken: complete sequence of the H5 gene. J Mol Biol. 1983 Nov 15;170(4):843–859. doi: 10.1016/s0022-2836(83)80191-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Kilpatrick M. W., Wells R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J Biol Chem. 1984 Feb 10;259(3):1963–1967. [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Goodman T. C., Hillen W., Horn G. T., Klein R. D., Larson J. E., Müller U. R., Neuendorf S. K., Panayotatos N., Stirdivant S. M. DNA structure and gene regulation. Prog Nucleic Acid Res Mol Biol. 1980;24:167–267. doi: 10.1016/s0079-6603(08)60674-1. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]