Abstract
Following initial standard chemotherapy (platinum/taxol), more than 75% of those patients with advanced stage epithelial ovarian cancer (EOC) experience a recurrence. There are currently no accurate prognostic tests that, at the time of the diagnosis/surgery, can identify those patients with advanced stage EOC who will respond to chemotherapy. Using a novel mathematical theory, we have developed three prognostic biomarker models (complex mathematical functions) that—based on a global gene expression analysis of tumor tissue collected during surgery and prior to the commencement of chemotherapy—can identify with a high accuracy those patients with advanced stage EOC who will respond to the standard chemotherapy [long-term survivors (>7 yrs)] and those who will not do so [short-term survivors (<3 yrs)]. Our three prognostic biomarker models were developed with 34 subjects and validated with 20 unknown (new and different) subjects. Both the overall biomarker model sensitivity and specificity ranged from 95.83% to 100.00%. The 12 most significant genes identified, which are also the input variables to the three mathematical functions, constitute three distinct gene networks with the following functions: 1) production of cytoskeletal components, 2) cell proliferation, and 3) cell energy production. The first gene network is directly associated with the mechanism of action of anti-tubulin chemotherapeutic agents, such as taxanes and epothilones. This could have a significant impact in the discovery of new, more effective pharmacological treatments that may significantly extend the survival of patients with advanced stage EOC.
Keywords: ovarian cancer, biomarkers, mathematical models, prognostic biomarker models, treatment response, survival, global gene expression analysis
Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecological malignancy in the U.S.A. Each year, approximately 21,880 women are diagnosed with EOC, and approximately 13,850 women succumb to the disease.1 Over 60% of the patients with EOC are diagnosed at an advanced stage, ie, III or IV [International Federation of Gynecology and Obstetrics (FIGO) staging system]1,2 due to the lack of early symptoms. The five-year survival rate for those patients with stage III or IV EOC is less than 30%.2 Maximal peritoneal cytoreduction surgery followed by platinum and taxol chemotherapy constitutes the current standard care for those patients with an advanced stage of EOC.3,4 Approximately, 70%–80% of the patients with advanced-stage EOC, initially, respond favorably to the platinum/taxol chemotherapy, but more than 75% of them soon experience a recurrence.5,6 Clearly, prognostic tests that could accurately prognose, at the time of the diagnosis/surgery, the response to the standard treatment of care and, therefore, the survival of those patients are in great need. Such prognostic tests would be invaluable in: 1) providing the physicians with the ability to identify responders from non-responders at the outset (immediately after surgery), 2) providing alternative therapies to non-responders of platinum/taxol, and 3) helping pharmaceutical companies to test and develop new analogs of chemotherapeutic agents that may be more effective for the non-responders.
In this study, by analyzing the global gene expression data of the tumor tissue obtained during surgery (and, therefore, prior to the administration of chemotherapy) from 54 patients with advanced-stage EOC (III–IV), we developed three prognostic biomarker models that were able to identify with a high accuracy (overall sensitivity: 95.83%–100.00% and overall specificity: 95.83%–100.00%) both the responders [long-term survivors (LTS)] and the non-responders [short-term survivors (STS)] to the platinum/taxol chemotherapy. We developed all three prognostic biomarker models using 34 patients [14 R/LTS (responders/long-term survivors) and 20 NR/STS (non-responders/short-term survivors)], and we validated all three of them with 20 unknown patients [10 R/LTS and 10 NR/STS] that were new and different from those 34 used in the development of the models.
Each of our three prognostic biomarker models is a complex mathematical function of a number of genes. Five genes are common input variables to all three prognostic biomarker models, and they are deemed highly significant in the process of the response to treatment and, thus, to the survival of patients with stage III or IV EOC. Of those five genes, one is directly associated with the mechanism of action of taxol—one of the two anti-cancer agents of the standard chemotherapy—and one gene is indirectly associated with the mechanism of action of taxol. Moreover, from the remaining most significant genes, one is also indirectly associated with the mechanism of action of taxol.
Materials and Methods
Data acquisition and clinical sample information
We used the raw intensity microarray data (CEL files) by Berchuck et al7 posted at the Duke Institute for Genome Sciences and Policy [Clinical cancer research 11, 3686–3696 (2005)→ (http://data.genome.duke.edu/clinicalcancerresearch.php)].
Briefly, according to Berchuck et al7 tumor tissue was harvested from 54 EOC patients with stages III and IV during surgery (48 with stage III and 6 with stage IV) and prior to the commencement of platinum/taxol chemotherapy. Total RNA was extracted from each tumor tissue sample and was analyzed for global gene expression using the GeneChip array U133A by Affymetrix. Following platinum/taxol chemotherapy, all 54 patients were followed for a period greater than seven years. Thirty patients survived for a period less than 3 years [NR/STS (non-responders/short-term survivors)–(median survival = 17.5 months)], and 24 patients survived for a period greater than 7 years [R/LTS (responders/long-term survivors)–(median survival = 107.5 months)]. None of the 30 NR/STS subjects died of causes other than EOC. At the time of the diagnosis, there was no significant age difference between the NR/STS subjects (median age = 59 yrs) and the R/LTS subjects (median age = 62 yrs). All 6 patients with stage IV EOC turned out to be NR/STS subjects. For more demographic and clinical details, please see the study by Berchuck et al.7
Discovery and validation studies
Of the 54 patients, we randomly selected 34 of them (14 R/LTS and 20 NR/STS) for the development and training of the prognostic biomarker models. The remaining 20 subjects (10 R/LTS and 10 NR/STS) constituted the unknown subjects with which all prognostic biomarker models were tested. This validation method provided us with the means to test our prognostic biomarker models with 20 new and real unknowns that were different from the subjects used for—and, therefore, completely extraneous to—the development and training of the models.
Statistical methods
We processed the original raw intensity data (CEL files) using the MAS5 algorithm (510 K FDA approved). The Affymetrix U133A chip has 22,283 probe sets that can interrogate an equal number of transcripts.
In order to reduce the dimensionality of the data and zero in on those variables (transcripts) that are most significant in the process of treatment-response/survival in the case of EOC patients, we applied our bioinformatic methods that we have developed, presented, and explained in a great detail in our previous studies.8–10 Briefly, we performed ROC curve analysis on the entire data matrix, ie, on all variables (22,283 transcripts × 54 subjects) in order to assess the discriminating capability of all variables with respect to our two groups, namely, R/LTS and NR/STS. In the final round, we selected only those variables with an AUC ≥ 0.80. Eighty four variables (transcripts) fulfilled this criterion, and they constituted the final pool of the most significant variables.
Generation of super variables
From the aforementioned 84 most significant variables, 13 became the input variables to the three complex mathematical functions (F1, F2, and F3), which we were able to generate, and which we term—and henceforward refer to as—super variables. Those three super variables (complex mathematical functions) are the final prognostic biomarker models. We should point out here that several other super variables were generated employing the remaining of the aforementioned 84 most significant variables, but, following final assessment, they proved to be not as robust as the F1, F2, or F3, and they are consequently not presented here. The F1 super variable is a function of 7 of the 13 aforementioned significant variables/transcripts, and all of those 7 transcripts correspond to 7 different genes. The F1 super variable, therefore, is a function of the following 7 genes:
| (1.1) |
The F2 super variable is a function of 10 of the 13 aforementioned significant variables/transcripts, and 8 of those 10 transcripts correspond to 8 different genes. The remaining two transcripts correspond to the same gene, namely ACTB. Statistically, those two variables/transcripts were determined to be the same (P > 0.90), and that accords with the fact that two different probe sets are probing the same gene. The F2 super variable, therefore, is a function of 10 different transcripts, or 9 different genes.
| (1.2) |
The F3 super variable is a function of 8 of the 13 aforementioned significant variables/transcripts, and 6 of those 8 transcripts correspond to 6 different genes. The remaining two transcripts, which are the same as the two mentioned in connection with the F2 super variable, correspond to the same gene, namely ACTB, as was the case with the F2 super variable. The F3 super variable, therefore, is a function of 8 different transcripts, or 7 different genes.
| (1.3) |
The three super variables constitute three different and independent complex mathematical functions (Supplementary Data), and, therefore, three different and independent prognostic biomarker models of treatment-response/survival of patients with stage III or IV EOC. As can be seen from Equations (1.1), (1.2), and (1.3), five genes are common to all three super variables and are, thus, very important to the process of interest, ie, treatment-response/survival of patients with stage III or IV EOC. Those five genes are: ACTB, EED, LYPLA2, MED13L, and TUBA3C. Of those, the genes TUBA3C and ACTB are directly and indirectly (respectively) associated with the mechanism of action of taxol. Furthermore, the gene CDC42, a constituent variable of the F3 super variable, is also indirectly associated with the mechanism of action of taxol. As can also be seen from Equations (1.1), (1.2), and (1.3), the three super variables are collectively composed of 12 different genes (or 13 different transcripts), all of which are listed in Table 1, along with their name, relative differential expression, and other properties.
Table 1.
The 12 genes (constituent variables) of the three prognostic biomarker models (F1, F2, and F3), ranked according to their ROC AUC value. Their significant differential expression [over-expression (↑) or under-expression (↓)] as observed in the NR/STS group relative to the R/LTS group is shown, along with their Affymetrix transcript number, symbol, name, function/process, known gene interactions, and known drug/chemical/hormone interactions.
| Rank | ROC AUC | Affymetrix transcript no | Gene symbol | Gene name | Gene function/process | Gene signif. diff. expr. (NR/STS) | Known interactions | Known Drugs/Chemicals/Hormones |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.88889 | 215566_x_at | LYPLA2 | lysophospholipase II | lipid metabolism | ↑ | MED31, DKC1, SCMH1, HNF4A | calcium, calmodulin |
| 2 | 0.85556 | 200801_x_at | ACTB | actin, beta | cell formation, growth, motility, and spreading | ↑ | MYC, MYCN,TP53, RAC1, RAC2, CDC42, TUBA1A, TUBB, TUBB2C | β-estradiol, progesterone, Lh, FSH, hCG, EPO, TRH, insulin |
| 3 | 0.85417 | 212209_at | MED13L | mediator complex subunit 13-like | regulation of transcription from RNA polymerase II promoter | ↓ | MYC, TP53, ESRRB | topotecan, camptothecin |
| 4 | 0.85139 | 210527_x_at | TUBA3C | tubulin, alpha 3c | microtubule generation | ↑ | MYC, FYN, NMI, TNFRSF1 A-B, TRAF1, TRAF6, RELA, RELB, BCL6, TGFB1, TRADD | paclitaxel, milataxel, docetaxel, vinblastine, vinchristine, podophyllotaxin, EC145, NPI-2358, TTI-237, TPI 287, XRP9881, colchicine, eribulin, vinorelbine, vinflunine, epothilon B, ixabepilone, |
| 5 | 0.84306 | 208728_s_at | CDC42 | cell division cycle 42 (GTP binding protein) | cell formation, growth, cycle progression and spreading; actin polymerization | ↑ | ARP2–3, F Actin, TP53, RAC1, BCAR1, HERC2, Ras, TNF, EDN1, Rac, Filamin | PP1, simvastatin, imatinib, benzo(a) pyrene, sulindac, sirolimus, atorvastatin, bestatin, indomethacin, Ins1, IGF1 |
| 6 | 0.84167 | 212585_at | OSBPL8 | oxysterol binding protein-like 8 | lipid binding and transport; steroid metabolic process | ↓ | HOXA9, SQSTM1, TFCP2, HTT | topotecan, valproic acid |
| 7 | 0.83056 | 200891_s_at | SSR1 | signal sequence receptor, alpha | protein processing (folding, sorting, and degradation) in ER | ↓ | VHL, PTN, MAPK3, XBP1 | GnRH analog, lipopolysaccharide, β-estradiol |
| 8 | 0.82500 | 201929_s_at | PKP4 | plakophilin 4 | cell adhesion | ↓ | ERBB2IP, PTPRJ, PTPRC, OGT, PDZD2, PSEN1 | imatinib |
| 9 | 0.82361 | 209572_s_at | EED | embryonic ectoderm development | negative regulation of transcription; repression of gene activity through histone deacetylation | ↓ | MYC, EGFR, RBBP7, RBBP4, PPP1R8, PPP1CA, DDB1, CDKN2A, PDCD6, HOXC12, HOXB4, HOXB13 | infliximab, microcystin, AGN194204 |
| 10 | 0.82361 | 206031_s_at | USP5 | ubiquitin specific peptidase 5 (isopeptidaseT) | positive regulation of ubiquitin-dependent protein degradation | ↑ | ubiquitin, USP13,RNF11, RAD23B, TNFRSF10D, PRKAB1, DVL2, TADA3, PMS1 | WP1130 |
| 11 | 0.80972 | 209399_at | HLCS | holocarboxylase synthetase | biotin metabolism | ↑ | histone, ACACB, PCCA | biotin,8-bromoguanosine 3′,5′-cyclic monophosphate |
| 12 | 0.80556 | 206790_s_at | NDUFB1 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex | electron transport; oxidative phosphorylation | ↓ | VHL, EWSR1, NDUFA5, HNF4A, | GnRH analog |
Computer programs
Computer programs were written using MATLAB R2010b by The MathWorks, Inc., Natick, MA, USA.
Results
Discovery study
As was mentioned earlier, from the total number of 54 subjects (24 R/LTS and 30 NR/STS) used in this study, we randomly selected 34 subjects (14 R/LTS and 20 NR/STS) for the development and training of the three prognostic biomarker models (F1, F2, and F3); and we will henceforward refer to those 34 subjects as the 34 original subjects. After the development of those three prognostic biomarker models, we assessed their prognostic accuracy using the aforementioned 34 original subjects, which were employed for their development. This constitutes an important first step in the assessment of a prognostic (or a diagnostic) test.
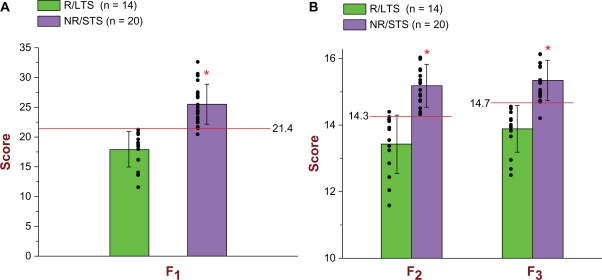
The cut-off score of the F1 prognostic biomarker model, as well as those of the other two models, was determined by taking into account the results of the following two analyses: 1) calculation of the optimal point on the ROC curve based on the 34 scores of the 34 original subjects used in the discovery study [optimal point is defined as the point with the highest sensitivity and the lowest false positive rate (1-specificity)] and 2) calculation of the 99.99% confidence intervals for the mean F1 scores of the two groups (R/LTS and NR/STS) and their respective standard deviations. Based on that, the cut-off score of the F1 model was determined to be 21.388. If a subject has an F1 score less than 21.388, then that subject is classified as an R/LTS; otherwise, that subject is classified as an NR/STS. As can be seen from Figure 1A, the F1 model correctly identified all (14/14) R/LTS subjects and 19/20 NR/STS subjects. In terms of treatment response, since we would like to identify those subjects that will respond to the platinum/taxol chemotherapy, our target group is the R/LTS and our reference group is the NR/STS. Therefore, for the discovery study, insofar as response to treatment is concerned, the F1 model exhibited a sensitivity = 1.000 and a specificity = 19/20 = 0.950. In the case of survival, given that we are interested in identifying the subjects that will be short-term survivors, our target group is the NR/STS and our reference group is the R/LTS. With regard to survival, therefore, for the discovery study, the F1 model exhibited a sensitivity = 0.950 and a specificity = 1.000. Both Figure 1A and Tables 2A and 2B show all pertinent statistical results of the F1 prognostic biomarker model in connection with the discovery study in great detail.
Figure 1.
(A) Scatter plot and bar graph of all 34 original subjects [14 R/LTS (green) and 20 NR/STS (purple)] used in the Discovery Study in connection with the F1 prognostic biomarker model. As can be seen, all 14 R/LTS subjects (green color) had F1 scores less than the determined cut-off score of 21.4, and, therefore, they were all identified correctly [for treatment response: sensitivity = 1.000; for survival: specificity = 1.000]. Regarding the NR/STS group (purple color), 19/20 had F1 scores greater than the determined cut-off score of 21.4, and, therefore, 19/20 were identified correctly [for treatment response: specificity = 19/20 = 0.950; for survival: sensitivity = 19/20 = 0.950]. For the Discovery Study, the ROC AUC of the F1 is 0.98929 with a 95% CI = [0.90449, 0.99884]. The mean F1 score of the 14 R/LTS subjects was 17.9358 (top of green bar) and the standard deviation (whisker above or below the top of the green bar) was 2.9622; whereas the mean F1 score of the 20 NR/STS subjects was 25.4697 (top of purple bar) and the standard deviation (whisker above or below the top of the purple bar) was 3.3651. The significance level was set at α = 0.001 (two-tailed), and the probability of significance was P = 1.30 × 10−7 (independent t-Test with T-value = −6.7405). The F1 is parametrically distributed with respect to both groups. (B) Scatter plot and bar graph of all 34 original subjects (14 R/LTS and 20 NR/STS) used in the Discovery Study in connection with the F2 and F3 prognostic biomarker models. As can be seen, 13/14 R/LTS subjects (green color) had F2 scores less than the determined cut-off score of 14.3, and, therefore, 13/14 subjects were identified correctly [for treatment response: sensitivity = 13/14 = 0.929; for survival: specificity = 13/14 = 0.929]. Regarding the NR/STS group (purple color), all 20 subjects had F2 scores greater than the determined cut-off score of 14.3, and, therefore, they were all identified correctly [for treatment response: specificity = 1.000; for survival: sensitivity = 1.000]. As can also be seen, all 14 R/LTS subjects (green color) had F3 scores less than the determined cut-off score of 14.7, and, therefore, they were all identified correctly [for treatment response: sensitivity = 1.000; for survival: specificity = 1.000]. Regarding the NR/STS group (purple color), 19/20 subjects had F3 scores greater than the determined cut-off score of 14.7, and, therefore, 19/20 subjects were identified correctly [for treatment response: specificity = 19/20 = 0.950; for survival: sensitivity = 19/20 = 0.950]. For the Discovery Study, the ROC AUC of the F2 is 0.98929 with a 95% CI = [0.90321, 0.99886], whereas the ROC AUC of the F3 is 0.98214 with a 95% CI = [0.86165, 0.99782]. The mean F2 score of the 14 R/LTS subjects was 13.4223 (top of green bar) and the standard deviation (whisker above or below the top of the green bar) was 0.8905; whereas the mean F2 score of the 20 NR/STS subjects was 15.1843 (top of purple bar) and the standard deviation (whisker above or below the top of the purple bar) was 0.6407. The mean F3 score of the 14 R/LTS subjects was 13.8864 and the standard deviation was 0.7017; whereas the mean F3 score of the 20 NR/STS subjects was 15.3433 and the standard deviation was 0.6082. The significance level was set at α = 0.001 (two-tailed), and the probability of significance for the F2 was P = 1.37 × 10−7 (independent t-Test with T-value = −6.7217), whereas the probability of significance for the F3 was P = 2.93 × 10−7 (independent t-Test with T-value = −6.4541). Both the F2 and the F3 are parametrically distributed with respect to both groups.
Table 2.
Statistical results of the three prognostic biomarker models (F1, F2, and F3)with respect to both treatment response and survival in the Discovery Study (identification of the 34 original subjects) and in the Validation Study (identification of the 20unknown subjects, which were new and different from the 34 original subjects). (A) The ROC AUC value, the 95% confidence interval of the ROC AUC value, the T value and probability of significance of the independent t-Test, the 99.99% confidence interval for the mean score of the R/LTS group and that of the NR/STS group, along with their respective standard deviations, of the F1, F2, and F3 prognostic biomarker models in the Discovery Study are shown. (B) The sensitivity and the specificity of the F1, F2, and F3 prognostic biomarker models in the Discovery Study with respect to both treatment response and survival are shown. (C) The ROC AUC value, the sensitivity, and the specificity of the F1, F2, and F3 prognostic biomarker models in the Validation Study with respect to both treatment response and survival are shown. (D) The T value and probability of significance of the independent t-Test and the mean score of the R/LTS group and that of the NR/STS group, along with their respective standard deviations, of the F1, F2, and F3 prognostic biomarker models in the Validation Study are shown. As can be seen, all six of those group mean scores, as observed in the validation study with the 20 unknown subjects, fall within the 99.99% confidence intervals of the respective group mean scores as predicted in the discovery study (A).
|
A (Discovery study) | ||||||
|---|---|---|---|---|---|---|
| Prognostic test | ROC AUC | 95% CI of AUC | T-Value | P (2-tailed)α = 0.001 | R/LTS Group [99.99% CI of mean] (SD) | NR/STS Group [99.99% CI of mean] (SD) |
| F1 | 0.98929 | [0.90449, 0.99884] | −6.7405 | 1.30 × 10−7 | [15.8665, 20.6631] (2.9622) | [22.9236, 27.6717] (3.3651) |
| F2 | 0.98929 | [0.90321, 0.99886] | −6.7217 | 1.37 × 10−7 | [12.7822, 14.2173] (0.8905) | [14.6982, 15.6030] (0.6407) |
| F3 | 0.98214 | [0.86165, 0.99782] | −6.4541 | 2.93 × 10−7 | [13.3940, 14.5352] (0.7017) | [14.8956, 15.7599] (0.6082) |
|
B (Discovery study) | |||||
|---|---|---|---|---|---|
| Prognostic test | ROC AUC |
Sensitivity |
Specificity |
||
| Treatment response | Survival | Treatment response | Survival | ||
| F1 | 0.98929 | 1.0000 | 0.9500 | 0.9500 | 1.0000 |
| F2 | 0.98929 | 0.9286 | 1.0000 | 1.0000 | 0.9286 |
| F3 | 0.98214 | 1.0000 | 0.9500 | 0.9500 | 1.0000 |
|
C (Validation study) | |||||
|---|---|---|---|---|---|
| Prognostic test | ROC AUC |
Sensitivity |
Specificity |
||
| Treatment response | Survival | Treatment response | Survival | ||
| F1 | 1.00000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| F2 | 1.00000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| F3 | 1.00000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
|
D (Validation study) | ||||
|---|---|---|---|---|
| Prognostic test | T-Value | P (2-tailed) α = 0.001 | R/LTS group Mean ± SD | NR/STS group Mean ± SD |
| F1 | −7.8523 | 3.19 × 10−7 | 16.1301 ± 2.9288 | 24.3990 ± 1.5847 |
| F2 | −6.8447 | 2.09 × 10−6 | 13.0048 ± 0.7932 | 14.9212 ± 0.3933 |
| F3 | −6.2992 | 6.14 × 10−6 | 13.6150 ± 0.6979 | 15.0952 ± 0.2552 |
The cut-off score of the F2 prognostic biomarker model was determined to be 14.259. If a subject has an F2 score less than 14.259, then that subject is classified as an R/LTS; otherwise, that subject is classified as an NR/STS. As can be seen from Figure 1B, the F2 model correctly identified 13/14 R/LTS subjects and all (20/20) NR/STS subjects. In connection with treatment response, therefore, and with regard to the discovery study, the F2 model exhibited a sensitivity = 13/14 = 0.929 and a specificity = 1.000; whereas in connection with survival, its sensitivity and specificity were 1.000 and 0.929, respectively. Figure 1B and Tables 2A and 2B show all pertinent statistical results of the F2 prognostic biomarker model in connection with the discovery study in great detail.
Regarding the F3 prognostic biomarker model, the cut-off score was determined to be 14.694, signifying that a score less than 14.694 belongs to an R/LTS subject, whereas a score greater than 14.694 belongs to an NR/STS subject. As can be seen from Figure 1B, the F3 model correctly identified all (14/14) R/LTS subjects and 19/20 NR/STS subjects. For the discovery study, therefore, with regard to treatment response, the sensitivity and specificity of the F3 model were 1.000 and 0.950, respectively; with regard to survival, its sensitivity and specificity were 0.950 and 1.000, respectively. Figure 1B and Tables 2A and 2B show all pertinent statistical results of the F3 prognostic biomarker model in connection with the discovery study in great detail.
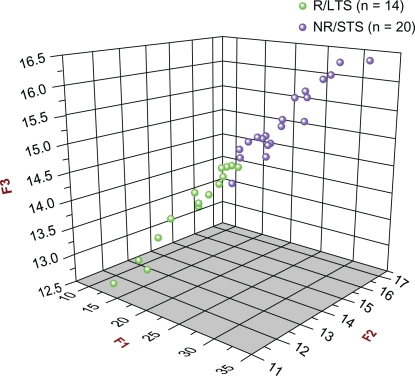
Figure 2 shows the 3D scatter plot of the F1 vs. F2 vs. F3 scores of all 34 original subjects, providing, thus, a visual depiction of the prognostic accuracy of all three models with respect to the discovery study.
Figure 2.
3D Scatter plot of all 34 original subjects [14 R/LTS (green) and 20 NR/STS (purple)] used in the Discovery Study in connection with the F1, F2, and F3 prognostic biomarker models. The F1, F2, and F3 scores of all 34 original subjects are plotted against each other (F1 vs. F2 vs. F3). As can be seen, there are two distinct, separate clusters: the green one (R/LTS group) is at the front and at a lower level, whereas the purple one (NR/STS group) is at the back and at a higher level. It can also be seen that one subject from the NR/STS group was misclassified.
Validation study
As was mentioned earlier, from the total number of 54 subjects (24 R/LTS and 30 NR/STS) used in this study, we had randomly segregated 20 subjects (10 R/LTS and 10 NR/STS) for the sole and express purpose of testing our three prognostic biomarker models. Those 20 unknown subjects were completely extraneous to all three models, that is to say they were new and different from the original 34 subjects, and they had never before been encountered by any of the three models. This constitutes the most important test in the assessment of a prognostic (or diagnostic) test.
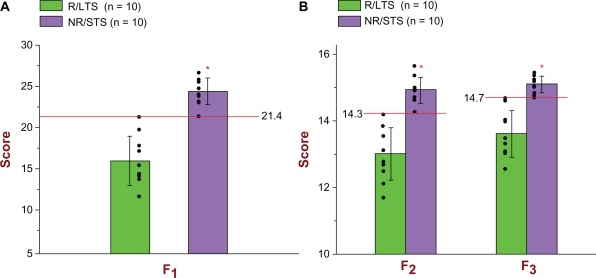
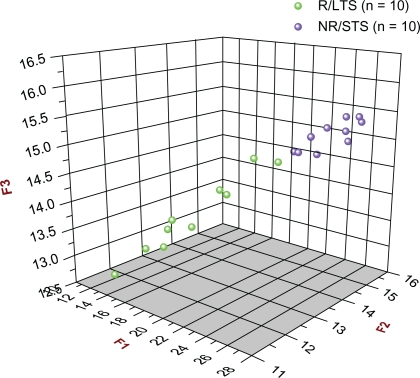
As can be seen from Figures 3A and 3B and Table 2C, all three prognostic biomarker models correctly prognosed all of the 20 unknown subjects. More specifically, all 10 R/LTS subjects had F1, F2, and F3 scores that were less than the respective cut-off scores (21.388, 14.259, 14.694); whereas all 10 NR/STS subjects had F1, F2, and F3 scores that were greater than the respective aforementioned cutoff scores. Therefore, with regard to both response to treatment and survival, and in connection with the validation study, both the sensitivity and the specificity of all three prognostic biomarker models were 1.000. Figure 4 shows the 3D scatter plot of the F1 vs. F2 vs. F3 scores of all 20 unknown subjects, providing, thus, a visual depiction of the prognostic accuracy of all three models with respect to the validation study.
Figure 3.
(A) Scatter plot and bar graph of all 20 unknown (new and different) subjects [10 R/LTS (green) and 10 NR/STS (purple)] used in the Validation Study in connection with the F1 prognostic biomarker model. As can be seen, all 10 R/LTS subjects (green color) had F1 scores less than the determined cut-off score of 21.4, and, therefore, they were all identified correctly [for treatment response: sensitivity = 1.000; for survival: specificity = 1.000]. Regarding the NR/STS group (purple color), all 10 of them had F1 scores greater than the determined cut-off score of 21.4, and, therefore, they were all identified correctly [for treatment response: specificity = 1.000; for survival: sensitivity = 1.000]. For the Validation Study, the ROC AUC of the F1 is 1.000. The mean F1 score of the 10 R/LTS subjects was 16.1301 (top of green bar) and the standard deviation (whisker above or below the top of the green bar) was 2.9288; whereas the mean F1 score of the 10 NR/STS subjects was 24.3990 (top of purple bar) and the standard deviation (whisker above or below the top of the purple bar) was 1.5847. The significance level was set at α = 0.001 (two-tailed), and the probability of significance was P = 3.19 × 10−7 (independent t-Test with T-value = −7.8523). The F1 is parametrically distributed with respect to both groups. (B) Scatter plot and bar graph of all 20 unknown (new and different) subjects (10 R/LTS and 10 NR/STS) used in the Validation Study in connection with the F2 and F3 prognostic biomarker models. As can be seen, all 10 R/LTS subjects (green color) had F2 scores less than the determined cut-off score of 14.3, and, therefore, they were all identified correctly [for treatment response: sensitivity = 1.000; for survival: specificity = 1.000]. Regarding the NR/STS group (purple color), all 10 of them had F2 scores greater than the determined cut-off score of 14.3,and, therefore, they were all identified correctly [for treatment response: specificity = 1.000; for survival: sensitivity = 1.000]. As can also be seen, all 10 R/LTS subjects (green color) had F3 scores less than the determined cut-off score of 14.7, and, therefore, they were all identified correctly [for treatment response: sensitivity = 1.000; for survival: specificity = 1.000]. Regarding the NR/STS group (purple color), all 10 of them had F3 scores greater than the determined cut-off score of 14.7,and, therefore, they were all identified correctly [for treatment response: specificity = 1.000; for survival: sensitivity = 1.000]. For the Validation Study, therefore, the ROC AUC of both F2 and F3 is 1.000. The mean F2 score of the 10 R/LTS subjects was 13.0048 (top of green bar) and the standard deviation (whisker above or below the top of the green bar) was 0.7932; whereas the mean F2 score of the 10 NR/STS subjects was 14.9212 (top of purple bar) and the standard deviation (whisker above or below the top of the purple bar) was 0.3933. The mean F3 score of the 10 R/LTS subjects was 13.6150 and the standard deviation was 0.6979; whereas the mean F3 score of the 10 NR/STS subjects was 15.0952 and the standard deviation was 0.2552. The significance level was set at α = 0.001 (two-tailed), and the probability of significance for the F2 was P = 2.09 × 10−6 (independent t-Test with T-value = −6.8447), whereas the probability of significance for the F3 was P = 6.14 × 10−6 (independent t-Test with T-value = −6.2992). Both the F2 and the F3 are parametrically distributed with respect to both groups.
Figure 4.
3D Scatter plot of all 20 unknown (new and different) subjects [10 R/LTS (green) and 10 NR/STS (purple)] used in the Validation Study in connection with the F1, F2, and F3 prognostic biomarker models. The F1, F2, and F3 scores of all 20 unknown subjects are plotted against each other (F1 vs. F2 vs. F3). As can be seen, there are two distinct, separate clusters: the green one (R/LTS group) is at the front and at a lower level, whereas the purple one (NR/STS group) is at the back and at a higher level. It can also be seen that there were no misclassifications.
Table 2D, in addition to other pertinent statistical results of our three prognostic biomarker models, shows the observed mean F1, F2, and F3 scores of the two groups (R/LTS and NR/STS) of the 20 unknown subjects. As can be seen, all six of those group mean scores, as observed in the validation study with the 20 unknown subjects, fall within the 99.99% confidence intervals of the respective group mean scores as predicted in the discovery study (Table 2A).
Overall prognostic biomarker model performance
If we combined the discovery study results with those of the validation study, then the overall performance of our three prognostic biomarker models would be as follows. With regard to treatment response, both the F1 and F3 exhibited a sensitivity = 1.000 (24/24 R/LTS subjects) and a specificity = 0.967 (29/30 NR/STS subjects); whereas the F2 exhibited a sensitivity = 0.958 (23/24 R/LTS subjects) and a specificity = 1.000 (30/30 NR/STS subjects). With regard to survival, both the F1 and F3 exhibited a sensitivity = 0.967 (29/30 NR/STS subjects) and a specificity = 1.000 (24/24 R/LTS subjects); whereas the F2 exhibited a sensitivity = 1.000 (30/30 NR/STS subjects) and a specificity = 0.958 (23/24 R/LTS subjects).
Gene networks
In connection with the 12 constituent genes of all three super variables, we conducted an Ingenuity Pathways Analysis (IPA) search. We sought to ascertain all that was known about those 12 genes pertaining to their function/process, their known interactions with other genes, and their known interactions with drugs, chemicals, and/or hormones as derived from the findings of scientific, peer-reviewed studies. The IPA results are listed in Table 1, along with the direction of the statistically significant differential expression (over-expression or under-expression) of those 12 genes in the NR/STS group relative to that of the R/LTS group.
The aforementioned 12 genes can be categorized into three general groups: 1) genes that regulate the expression of cytostructural proteins, 2) genes that regulate cell proliferation, and 3) genes that regulate metabolism.
The genes ACTB, TUBA3C, and CDC42 have functions that pertain to the cytoskeleton, and as such, they compose the first group.
Cancer proliferation and metastasis relies on cytostructural materials, ie, cytoskeletal proteins, such as microfilaments (actin) and microtubules.11,12 The first two genes promote the expression of actin and microtubules, respectively. CDC42 promotes the polymerization of actin into microfilaments13,14; reorganization of the actin cytoskeleton15; and cell formation, growth, andspreading.16,17 There is also evidence that CDC42 can regulate the polarization of both the actin and the microtubule cytoskeleton.13,18 In our study all three of the aforementioned cytostructural genes were significantly over-expressed in the NR/STS group relative to the R/LTS group.
The following genes, whose function pertains to cell proliferation in general, compose the second group: MED13L, SSR1, PKP4, EED, and USP5.
The MED13L protein (also known as, among other names, THRAP2, TRAP240L, and KIAA1025) is a member of the Mediator complex, a group of about 30 transcriptional co-activators that play various regulatory roles in the induction of RNA polymerase II transcription.19 Compositional differences may account for different functions among the Mediator proteins; for instance some promote transcription, whereas others act as transcriptional repressors.19,20 A number of those Mediator proteins are novel, and, consequently, their exact function is not known, including that of MED13L.21 Regarding specifically the MED13L gene, it has been observed that over-expression of the TP53 gene (p53) in human colon carcinoma cell lines relative to controls suppresses the expression of MED13L (KIAA1025).22 That could very well explain our finding that the MED13L gene was significantly under-expressed in the NR/STS group relative to the R/LTS group by affirming the existence of a more aggressive EOC cancer in the case of the former group in comparison with the latter one.
SSR1 is an ER (endoplasmic reticulum) receptor part of the translocon-associated protein (TRAP) complex. The function of the TRAP complex remains unclear,23,24 and that is even more so in the case of disease, such as cancer. In general, it is thought that the TRAP complex proteins are involved in protein translocation from the ER and in protein processing (folding, sorting, and degradation) in the ER. The SSR1, more specifically, has been hypothesized to be involved in the unfolded protein response (UPR) in the ER, ie, protein degradation, when there is a large accumulation of abnormal proteins.25 In our study, the SSR1 gene was significantly under-expressed in the NR/STS group relative to the R/LTS group.
The PKP4 protein (aka p0071) belongs to the family of arm-repeat proteins,26 which are involved in cell adhesion.27 There is little known about both the structural and the functional role of the PKP4 protein.27 According to the results of our analysis, the PKP4 gene was significantly under-expressed in the NR/STS group relative to the R/LTS group, and that accords with the observation that metastatic cancer cells rely on greater cell mobility and, thus, lower cell adhesion.
The EED protein is part of the Polycomb-group (PcG) proteins involved in repressive transcriptional control28 mediated via histone deacetylation.29 We found that the EED gene was significantly under-expressed in the NR/STS group relative to the R/LTS group, indicating that in the case of more aggressive EOC, inhibitory control of gene activity was more diminished.
USP5 belongs to the largest class of deubiquitinating enzymes (USPs) that regulate protein ubiquitination, a post-translational modification of cellular proteins. Compounds that inhibit the regulation of protein ubiquitination, such as bortezomib, have been approved by the FDA and are used for the treatment of certain types of cancer.30,31 Moreover, other compounds that specifically suppress the activity of USP5, such as WP1130, lead to apoptosis of tumor cells.32 Those findings are in agreement with our results: we found that the USP5 gene was significantly over-expressed in the NR/STS group relative to the R/LTS group. There is also evidence that malignant tumors, via the release of certain factors, may promote the expression of USPs and other deubiquitinating enzymes in order to induce major alterations in the metabolism, such as increased proteolysis and lipolysis, for energy purposes.33,34
The third group comprises genes whose function is involved in metabolism in general and lipid metabolism in particular. Those genes are: LYPLA2, OSBPL8, HLCS, and NDUFB1.
Throughout all its stages, carcinogenesis entails extensive remodeling of lipid metabolism. In many different types of cancer, altered lipid metabolism has been observed.35,36 Owing to increased demand for membrane biosynthesis and energy for the generation and sustenance of new cells, carcinogenesis effects alterations not only in lipid metabolism but also in glycolysis,37,38 with the most drastic alterations observed in the most aggressive tumor cells.35 LYPLA2 is the enzyme that catalyzes the hydrolysis of 2-lysophosphatidylcholine (which, along with arachidonic acid, is derived from the hydrolysis of phosphatidylcholine—a phospholipid that is a major component of cell membranes) to glycerophosphocholine. As was mentioned above, there is evidence that malignant tumors produce certain factors, such as PIF, which promote cell survival under hypoxia and oxidative stress by altering metabolism so that energy can be derived from both proteolysis and lipolysis.33,34 More specifically, PIF induces proteolysis by increasing the expression of deubiquitinating enzymes, such as USPs—a process mediated by the conversion of arachidonic acid to 15-HETE (15-hydroxyeicosatetraenoic acid).39 Moreover, both of the aforementioned hydrolytic reactions result in the release of fatty acids, which may be utilized for energy production via β-oxidation. Cachexia, a condition characterized by progressive body mass loss due to extensive proteolysis and lipolysis, has been observed in connection with cancer, and it has been associated with a poor survival rate, a diminished chemotherapy response, and an increased toxicity intolerance.33 According to our analysis, the LYPLA2 gene was significantly over-expressed in the NR/STS group relative to the R/LTS group; that accords with the aforementioned observations and, along with the significant over-expression of the USP5 gene, suggests greater protein and lipid catabolism for energy purposes (via β-oxidation) on the part of the more aggressive cancer cells (NR/STS group).
The protein OSBPL8 is an intracellular lipid receptor that belongs to the family of oxysterols (oxygenated cholesterol derivatives). Oxysterols activate the liver X receptors (LXR) that regulate the expression of a number of genes whose function pertains to cholesterol metabolism.40 Some of those genes, such as SREBP1, have been shown to promote fatty acid synthesis when activated by the oxysterol/LXR group.41,42 We found that the OSBPL8 gene was significantly under-expressed in the NR/STS group relative to the R/LTS group, suggesting that lipogenesis is more suppressed in the more aggressive cancer cells. That is in agreement with our findings about the over-expression of the LYPLA2 gene in the same group (NR/STS), which points to a greater lipolysis in the case of the more aggressive cancer cells.
HLCS is an enzyme that catalyzes the covalent biotinylation of the five crucial mammalian carboxylase enzymes: pyruvate carboxylase (PC), acetyl-CoA carboxylase 1 and 2 (ACC1 and ACC2), 3-methylcrotonyl-CoA carboxylase (MCC), and propionyl-CoA carboxylase (PCC). From an energy production perspective, the most likely targets of HLCS in connection with advanced-stage EOC are PCC and MCC. The former catalyzes the carboxylation of propionyl CoA, which is the end product of the metabolism of the fatty acids with an odd number of carbons, and which, at the end of β-oxidation, is converted into succinyl-CoA, which then can enter the TCA cycle. MCC is a key enzyme in the breakdown of leucine, which is eventually broken down to acetoacetate and acetyl CoA, the latter of which can enter the TCA cycle. High demand for energy, however, would serve no purpose if it were the only requirement on the part of cancer cells; it is matched by an equally high demand for materials necessary for the generation of new cells—materials such as cytostructural proteins (tubulin, actin, etc.), which require biosynthesis of amino acids. The TCA cycle can be used for the biosynthesis of non-essential amino acids provided it is anaplerotically sustained, something which can be accomplished via the end products of the aforementioned PCC and MCC, or via the aforementioned pyruvate carboxylase (PC) [conversion of pyruvate to oxaloacetate], or via glutaminolysis [breaking down glutamine into α-ketoglutarate]. All those metabolic processes and pathways are regulated by the HLCS gene, something that signifies its immense importance not only in cancer but also in health. We found that the HLCS gene was significantly over-expressed in the NR/STS group relative to the R/LTS group, which suggests higher lipolysis and proteolysis in the case of the more aggressive cancer cells, and which is in accordance with our findings on the previous genes.
Along with 45 other subunits, the NDUFB1dehydrogenase (ubiquinone) 1 beta subcomplex constitutes the mitochondrial Complex I—a very large multiprotein enzyme which is located in the inner mitochondrial membrane, and which catalyzes the first step of the electron transport chain, the redox machinery of the oxidative phosphorylation. It has been observed by multiple studies43 that, owing to their surrounding hypoxic environment, tumor cells rely to a much larger extent on anaerobic glycolysis to produce energy rather than on oxidative phosphorylation. This is in complete agreement with our finding that the NDUFB1 gene was significantly under-expressed in the NR/STS group relative to the R/LTS group, for it points to a lower utilization of oxidative phosphorylation on the part of the more aggressive cancer cells.
Genes related to the mechanism of action of taxol
Taxol is an anti-tubulin chemotherapeutic agent that acts as a mitotic inhibitor. More specifically, it increases polymerization of microtubules from α-β tubulin heterodimers, and it stabilizes microtubules by preventing their depolymerization. This action prevents the formation of the mitotic spindle, a necessary step in the process of mitosis, and that results in the arrest of the mitosis cycle either in the G2 or the M phase.44 How some cancer cells can evade the action of taxol is not clear.45 Over-expressing tubulin, so as to offset the action of taxol, may be one way according to our evidence. As was mentioned earlier, the gene TUBA3C, which encodes the production of α-tubulin, and which is common to all three super variables and one of the top four most significant predictors, was significantly over-expressed in those patients (NR/STS) who did not respond to taxol (platinum/taxol chemotherapy). Furthermore, the fact that taxol binds to the β-tubulin subunit46,47 would render the over-expression of TUBA3C on the part of the more aggressive cancer cells a successful strategy for evading the action of taxol and, thus, for survival.
Actin, a globular protein, is the monomeric component of microfilaments (part of the cytoskeleton) and thin filaments (part of the myofibril). β-Actin, the monomeric component of microfilaments only, belongs to one of the three groups of isoforms of actin (α, β, and γ). The β and γ isoforms compose the actin cytoskeleton (microfilament networks). The actin cytoskeleton is involved, among other important cellular processes, in regulation of gene transcription, chromatin remodeling, cell cytokinesis and mitosis, and cell motility.48 That, therefore, cytoskeletal actin, β-actin in particular, has been linked to numerous types of cancer should be not surprising. Moreover, cytoskeletal actin has been shown to play a role in the control of the polarity of the mitotic spindle (microtubule polarization).49 Taxol, on the other hand, besides inhibiting the depolymerization of microtubules, has been shown to induce multipolar formation of the mitotic spindle,47 preventing thus the organized movement of the microtubules, necessary for the separation of the chromosomes. Over-expression of β-actin could overcome this action of taxol, and that is something to which our evidence points: we found that the ACTB gene, which encodes for the production of β-actin, was significantly over-expressed in the NR/STS group relative to the R/LTS group.
The CDC42 gene not only promotes the polymerization of actin into microfilaments, the reorganization of the actin cytoskeleton, and cell formation, growth, and spreading; but also it can regulate the polarization of both the actin and the microtubule cytoskeleton. Theoretically, therefore, over-expression of the CDC42 gene can overcome the action of taxol, as well; and that is the finding of our analysis: the CDC42 gene was significantly over-expressed in the NR/STS group relative to the R/LTS group.
In summary, when over-expressed, the genes TUBA3C, ACTB, and CDC42 can collectively (or even perhaps in a given combination thereof) overcome the exerted actions of taxol and diminish its efficacy. It stands to reason, therefore, to expect that a new pharmacological approach whereby, via a combination of chemotherapeutic agents, all three of those aforementioned genes are targeted will be more successful than the current standard treatment in extending the life span of those women with EOC who, because of the specific pattern of the aforementioned genetic networks, will not respond to the platinum/taxol chemotherapy and will turn out to be short-term survivors.
Discussion
Studies in ovarian cancer or cancer in general, for that matter, typically culminate with the presentation of a list of significantly differentially expressed genes. To the best of our knowledge, we are not aware of any studies in this field that developed actual prognostic tests that could be utilized in the clinic, much less of any studies that developed such tests and validated them with real, unknown (new and different) subjects and demonstrated such high sensitivity and specificity for those tests.
Comparing our results pertaining to the most significant genes responsible for determining long-term vs. short-term survival in patients with advanced stage EOC with those of the original study by Berchuck et al7 we noticed that three of our 12 most significant genes (Table 1), namely SSR1, PKP4, and EED, were also determined to be significant and under-expressed in the short-term survivors relative to the long-term survivors. We should also point out here that Berchuck et al7 used completely different methods from ours for their analysis. They employed statistical methods; more specifically, they used a combination of a Bayesian classification tree with multivariate discriminant analysis. We, on the other hand, performed mathematical modeling to generate functions that can describe the process of interest as accurately as possible.
Having utilized our novel mathematical theory and bioinformatic approach, we were able to 1) identify a group of most significant genes and 2) generate three complex mathematical functions that can prognose with a high degree of accuracy the outcome of treatment response and survival in patients with late stages of EOC at the earliest possible time (diagnosis/surgery). Following validation with 20 unknown (new and different) subjects, and with respect to both treatment response and survival, our three prognostic biomarker models exhibited an overall sensitivity from 95.83% to 100% and an overall specificity also from 95.83% to 100%. Provided there is further and more extensive validation, the clinical significance of our three prognostic biomarker models is evident: physicians will be able to identify from the outset those EOC patients who will and will not respond to the platinum/taxol chemotherapy, and they will consequently administer a different treatment to the latter. This could result in a considerable increase of the survival of those patients who do not respond to the conventional treatment.
Pertaining to the genetic networks that cause a discrimination between the NR/STS and the R/LTS patients with EOC, our findings on the 12 genes employed by the three prognostic biomarker models can be summarized as follows: compared with the R/LTS group, the NR/STS group exhibited a significant increase in 1) generation of cytostructural proteins, 2) cell proliferation, and 3) cell energy production (via a significant increase in anaerobic glycolysis, lipolysis, and proteolysis). Given that our three prognostic biomarker models (complex mathematical functions), by utilizing the aforementioned 12 genes as input variables, can identify the NR/STS and the R/LTS patients with EOC with a high accuracy, it follows that those 12 genes, which compose three distinct genetic networks, are responsible for determining both the treatment-response and the survival outcome in patients with late stage EOC. The fact that the patient tumor tissue was collected during surgery, and that all patients received chemotherapy only after surgery, further reinforces the previous statement, ie, that the genetic patterns of the aforementioned 12 genes as generated by our three prognostic biomarker models can determine a priori whether a patient with advanced stage EOC will be a responder to the platinum/taxol chemotherapy (and therefore a long-term survivor) or otherwise. This in conjunction with the fact that one of the genetic networks, namely, TUBA3C, ACTB, and CDC42, could be influenced by anti-tubulin agents, such as taxanes and epothilones, could have significant pharmacological implications for the treatment of EOC in the future. For example, those EOC patients who, after surgery, are prognosed to be non-responders (short-term survivors) could be treated with a combination of the aforementioned anti-tubulin agents or with new anti-tubulin agents or with other chemotherapeutic agents and be monitored for a successful outcome, ie, for a survival period greater than 3 years. Moreover, the same investigative pharmacological approach could be applied by using a different family of chemotherapeutic agents and targeting any or both of the other two gene networks (cell proliferation and/or cell energy production). As was mentioned earlier, a pharmacological approach that could target all three gene networks at the same time may turn out to be the most successful.
In the end, utilizing our prognostic biomarker tests for the development of new, more effective pharmacological regimens may lead to considerably more successful treatments, ie, to a significant increase in the survival of all patients with advanced stage EOC.
Acknowledgments
We would like to extend our gratitude to Dr. Andrew Berchuck for providing us with clinical information and Dr. Gunda I. Georg for her comments and advice on pharmacological issues.
Footnotes
Author Contributions
JBN conceived and developed the mathematical theory of super variables and generated the three super variables in this study. JBN co-conceived, co-designed, performed the analysis, and executed this project; and wrote and co-edited the manuscript. KLMB co-conceived and co-designed this project; helped with the acquisition of data; and co-edited the manuscript. APNS and WCL co-conceived, co-designed, and provided the necessary support and resources for this project; and co-edited the manuscript.
Disclosure of Potential Conflicts of Interest
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
Grant Support
This study was funded by the National Institutes of Health (grant number: T32 DA007097) and by the National Cancer Institute (grant number: R01 CA106878).
References
- 1.American Cancer Society . Cancer Facts and Figures 2010. Atlanta: American Cancer Society; 2010. [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–9. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 4.Vergote I, Trope CG, Amant F, Kristensen GB, Ehlen T, Johnson N, et al. Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. N Engl J Med. 2010;363:943–53. doi: 10.1056/NEJMoa0908806. [DOI] [PubMed] [Google Scholar]
- 5.Du Bois A, Luck HJ, Meier W, Adams HP, Mobus V, Costa S, et al. A randomized clinical trial of cisplatin/paclitaxel versus carboplatin/paclitaxel as first-line treatment of ovarian cancer. J Natl Cancer Inst. 2003;95:1320–9. doi: 10.1093/jnci/djg036. [DOI] [PubMed] [Google Scholar]
- 6.Ozols RF, Bundy BN, Greer BE, Fowler JM, Clarke-Pearson D, Burger RA, et al. Phase III trial of carboplatin and paclitaxel compared with cisplatin and paclitaxel in patients with optimally resected stage III ovarian cancer: a Gynecologic Oncology Group study. J ClinOncol. 2003;21:3194–200. doi: 10.1200/JCO.2003.02.153. [DOI] [PubMed] [Google Scholar]
- 7.Berchuck A, Iversen ES, Lancaster JM, Pittman J, Luo J, Lee P, et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 2005;11(10):3686–696. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- 8.Nikas JB, Keene CD, Low WC. Comparison of analytical mathematical approaches for identifying key nuclear magnetic resonance spectroscopy biomarkers in the diagnosis and assessment of clinical change of diseases. Journal of Comparative Neurology. 2010;518:4091–112. doi: 10.1002/cne.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikas JB, Low WC. ROC-supervised principal component analysis in connection with the diagnosis of diseases. American Journal of Translational Research. 2011;3(2):180–96. [PMC free article] [PubMed] [Google Scholar]
- 10.Nikas JB, Low WC. Application of clustering analyses to the diagnosis of Huntington disease in mice and other diseases with well-defined group boundaries. Computer Methods and Programs in Biomedicine. 2011 doi: 10.1016/j.cmpb.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim R, Lappas M, Ahmed N, Permezel M, Quinn MA, Rice GE. 2D-PAGE of ovarian cancer: Analysis of soluble and insoluble fractions using medium-range immobilized pH gradients. Biochemical and Biophysical Research Communications. 2011;406:408–13. doi: 10.1016/j.bbrc.2011.02.056. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko K, Satoh K, Masamune A, Satoh A, Shimosegawa T. Myosin light chain kinase inhibitors can block invasion and adhesion of human pancreatic cancer cell lines. Pancreas. 2002;24:34–41. doi: 10.1097/00006676-200201000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: Integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 14.Albiges-Rizo C, Destaing O, Fourcade B, Planus E, Block MR. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. Journal of Cell Science. 2009;122:3037–49. doi: 10.1242/jcs.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt A, Hall MN. Signaling to the actin cytoskeleton. Annu Rev Cell Dev Biol. 1998;14:305–38. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Adams HC, Whitehead IP. The Rho-specific guanine nucleotide exchange factor Dbs regulates breast cancer cell migration. The Journal of Biological Chemistry. 2009;284(23):15771–80. doi: 10.1074/jbc.M901853200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keely PJ, Westwick JK, Whitehead IP, Der CJ, Parise LV. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature. 1997;390:632–6. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]
- 18.Stowers L, Yelon D, Berg LJ, Chant J. Regulation of the polarization of T cells toward antigen-presenting cells by Ras-related GTPase CDC42. Proc Natl Acad Sci. 1995;92:5027–31. doi: 10.1073/pnas.92.11.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Molecular Cell. 2004;14:685–91. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 20.Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, et al. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Molecular Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 21.Musante L, Bartsch O, Ropers HH, Kalscheuer VM. cDNA cloning and characterization of the human THRAP2 gene which maps to chromosome 12q24, and its mouse ortholog Thrap2. Gene. 2004;332:119–27. doi: 10.1016/j.gene.2004.02.044. [DOI] [PubMed] [Google Scholar]
- 22.Daoud SS, Munson PJ, Reinhold W, Young L, Prabhu VV, Yu Q, et al. Impact of p53 knockout and topotecan treatment on gene expression profiles in human colon carcinoma cells: A pharmacogenomic study. Cancer Research. 2003;63:2782–93. [PubMed] [Google Scholar]
- 23.Hartman E, Prehn S. The N-terminal region of the a-subunit of the TRAP complex has a conserved cluster of negative charges. FEBS Letters. 1994;349:324–326. doi: 10.1016/0014-5793(94)00693-8. [DOI] [PubMed] [Google Scholar]
- 24.Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the trans-locon-associated protein complex during translocation across the ER membrane. The Journal of Cell Biology. 2003;160(4):529–39. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirama T, Miller CW, Koeffler HP. Translocon-associated protein K transcripts are induced by granulocyte-macrophage colony-stimulating factor and exhibit complex alternative polyadenylation. FEBS Letters. 1999;455:223–7. doi: 10.1016/s0014-5793(99)00885-6. [DOI] [PubMed] [Google Scholar]
- 26.Cowin P, Kapprell H-P, Franke WW, Tamkun J, Hynes RO. Plakoglobin: a protein common to different kinds of intercellular adhering junctions. Cell. 1986;46:1063–73. doi: 10.1016/0092-8674(86)90706-3. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann I, Kuhn C, Franke WW. Protein p0071, a major plaque protein of non- desmosomaladhering junctions, is a selective cell-type marker. Cell Tissue Res. 2008;334:381–99. doi: 10.1007/s00441-008-0725-2. [DOI] [PubMed] [Google Scholar]
- 28.Denisenko ON, Bomsztyk K. The product of the murine homolog of the drosophila extra sex combs gene displays transcriptional repressor activity. Molecular and Cellular Biology. 1997;17(8):4707–17. doi: 10.1128/mcb.17.8.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nature Genetics. 1999;23:474–8. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 30.Hideshima T, Richardson P, Chauhan D, et al. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–6. [PubMed] [Google Scholar]
- 31.Goy A, Younes A, McLaughlin P, et al. Phase II study of proteasome inhibitor bortezomib in relapsed or refractory B-cell non-Hodgkin’s lymphoma. J ClinOncol. 2005;323:667–75. doi: 10.1200/JCO.2005.03.108. [DOI] [PubMed] [Google Scholar]
- 32.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome-formation and tumor cell apoptosis. Cancer Res. 2010;70:9265–76. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 33.Tisdale MJ. Tumor–host interactions. Journal of Cellular Biochemistry. 2004;93:871–7. doi: 10.1002/jcb.20246. [DOI] [PubMed] [Google Scholar]
- 34.Bossola M, Muscaritoli M, Costelli P, Grieco G, Bonelli G, Pacelli F, et al. Increased muscle proteasome activity correlates with disease severity in gastric cancer patients. Ann Surg. 2003;237:384–389. doi: 10.1097/01.SLA.0000055225.96357.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilvo M, Denkert C, Lehtinen L, Muller B, Brockmoller S, Seppanen-Laakso T, et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71:3236–45. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- 36.Hirsch HA, Iliopoulos D, Joshi A, Zhang Y, Jaeger SA, Bulyk M, et al. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348–61. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young CD, Anderson SM. Sugar and fat—that’s where it’s at: metabolic changes in tumors. Breast Cancer Research. 2008;10:202. doi: 10.1186/bcr1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metabolism. 2006;3:187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 39.Smith HJ, Lorite MJ, Tisdale MJ. Effect of a cancer cachectic factor on protein synthesis/degradation in murine C2C12 myoblasts: Modulation by eicosapentaenoicacid. Cancer Res. 1999;59:5507–13. [PubMed] [Google Scholar]
- 40.Lehto M, Laitinen S, Chinetti G, Johansson M, Ehnholm C, Staels B, et al. The OSBP-related protein family in humans. J Lipid Res. 2001;42:1203–13. [PubMed] [Google Scholar]
- 41.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JMA, Shimomura I, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 2000;14:2819–30. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, et al. Role of LXRs in control of lipogenesis. Genes and Development. 2000;14:2831–38. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frezza C, Gottlieb E. Mitochondria in cancer: Not just innocent bystanders. Seminars in. Cancer Biology. 2009;19:4–11. doi: 10.1016/j.semcancer.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Rahman A, Korzekwa KR, Grogan J, Gonzalez FJ, Harris JW. Selective biotransformation of taxol to 6a-hydroxytaxol by human cytochrome P450 2C8. Cancer Research. 1994;54:5543–46. [PubMed] [Google Scholar]
- 45.Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7:637–51. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by Taxol. Nature. 1979;277:665–67. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 47.Chen JG, Horwitz SB. Differential mitotic responses to microtubule-stabilizing and -destabilizing drugs. Cancer Research. 2002;62:1935–8. [PubMed] [Google Scholar]
- 48.Cicchillitti L, Della Corte A, Di Michele M, Donati MB, Rotilio D, Scambia G. Characterisation of a multimeric protein complex associated with ERp57 within the nucleus in paclitaxel-sensitive and –resistant epithelial ovarian cancer cells: The involvement of specific conformational states of ß-actin. International Journal of Oncology. 2010;37:445–54. doi: 10.3892/ijo_00000693. [DOI] [PubMed] [Google Scholar]
- 49.Xu FL, Saunders WS. Actin and microtubules: working together to control spindle polarity. Cancer Cell. 2008;14:197–99. doi: 10.1016/j.ccr.2008.08.007. [DOI] [PubMed] [Google Scholar]