Abstract
Transglutaminase is a calcium-dependent enzyme that posttranslationally modifies proteins by cross-linking between glutamine and lysine residues or attachment of a primary amine to specific polypeptide-bound glutamine residues. Eight isozymes play essential roles in various mammalian biological processes. The authors have recently identified 12–amino acid preferred substrate peptide sequences that are highly reactive and act in an isozyme-specific manner. In this study, a rapid, isozyme-specific, and sensitive detection of active keratinocyte type (TGase 1) and tissue type (TGase 2) was successful using fluorescence-labeled peptides. This procedure involved using whole-body sections of a mouse to extensively analyze the tissue distribution of both enzymes that revealed clearly distinct patterns. Strong active TGase 1 was observed in epithelial tissues such as tongue, developing teeth, forestomach, and skin epidermis. Significantly active TGase 2 was observed in various types of tissues as predicted and at particularly higher levels in the intestinal mucosa, muscle membrane, and whole veins in the liver. This manuscript contains online supplemental material at http://www.jhc.org. Please visit this article online to view these materials.
Keywords: transglutaminase, calcium, epithelium, connective tissue
Transglutaminase (TGase, EC 2. 3. 2.13) is a calcium-dependent enzyme that catalyzes the formation of isopeptide cross-links between glutamine and lysine residues in a variety of proteins (Griffin et al. 2002; Lorand and Graham 2003). In addition, TGase catalyzes incorporation of primary amines into protein-bound glutamine residue or conversion into glutamic acid by deamidation. In mammals, eight TGase isozymes have been identified (TGases 1-7, factor XIII), comprising a protein family with unique tissue distribution and physiological roles.
TGase 1, TGase 3, and TGase 5 are involved in skin formation possibly by cooperatively cross-linking structural proteins in keratinocytes (Aeschlimann et al. 1998; Eckert et al. 2005; Hitomi 2005). Among the three isozymes expressing in the epidermis, TGase 1 (keratinocyte type) has been characterized as an essential isozyme because its knockout mice exhibit impaired epidermis (Matsuki et al. 1998). In differentiating keratinocytes, TGase 1 cross-links structural proteins such as involucrin and loricirn to form a highly specialized protein structure, called a cornified envelop, which contributes to the barrier function of the outermost layers (Kalinin et al. 2002; Candi et al. 2005).
TGase 2 (tissue-type TGase) is widely expressed and involved in multiple events such as cell growth, differentiation, and apoptosis via enzymatic modification of extracellular matrix proteins, transcription factors, and signaling molecules (Fesus and Piacentini 2002; Beninati and Piacentini 2004; Jeon and Kim 2006; Mehta et al. 2006; Tatsukawa et al. 2009). The phenotype of TGase 2 knockout mice shows abnormal phagocytosis, apoptosis, and wound healing (Sarang et al. 2009). Despite extensive studies, substrate proteins of physiological relevance acted upon by TGase 2 and tissue distribution of the enzyme are still under investigation.
Factor XIII, which stabilizes fibrin clots, is essential for blood coagulation (Ichinose 2001). TGase 4 is expressed in the prostate and reported to be involved in the formation of plug in rodents (Esposito et al. 1996). Little information is available for TGase 6 and TGase 7 (Grenard et al. 2001).
In the catalytic reaction of TGase, the glutamine-donor substrate and the enzyme form the reaction intermediate and then react to glutamine-acceptor molecules (protein-bound lysine and/or primary amine). Therefore, the recognition of reactive glutamine residue(s) in substrates is crucial for the enzymatic reaction. Recently, using a random peptide library, we have characterized the preferred glutamine-donor substrate sequences for TGases that shows a unique reaction tendency to each isozyme (Sugimura et al. 2006). In these studies, several 12-mer substrate peptide sequences for major TGases have been identified: factor XIII, TGase 1, and TGase 2 (Sugimura et al. 2008; Hitomi, Kitamura, and Sugimura 2009). The most favorable sequence for each isozyme appeared to act as a substrate with high reactivities and isozyme specificities, even in the peptide form. Because the preferred substrate peptides can efficiently react with protein-bound lysine residue and primary amines, these peptides have been used in several applications, including sensitive in vitro assays (Sugimura et al. 2007; Perez Alea et al. 2009; Hitomi, Kitamura, Perez Alea, et al. 2009).
Furthermore, using fluorescence-labeled preferred substrate peptide (pepK5), we were also successful in detecting active TGase 1 in the human and mouse skin epidermis (Sugimura et al. 2008; Akiyama et al. 2010). The location of the active TGase 1 was observed by the analysis of the outmost layer of the epidermis. Because this detection method was simple, sensitive, and isozyme specific, we attempted to expand the analysis to the whole-tissue sections and to the detection of active TGase 2.
In this study, frozen sections of the whole mouse were used to perform rapid and sensitive enzymatic reactions for the analysis of tissue distribution in mice, similar to that observed in mouse skin. Using the sections, both active enzymes for TGase 1 and TGase 2 could be detected for the extensive evaluation of tissue distribution. Moreover, areas where both isozymes were enzymatically active have been characterized in several tissues, showing distinct patterns between the two enzymes. This study first describes the procedure for in situ detection of active TGases in an isozyme-specific manner with high sensitivity and provides novel information regarding tissue distribution of TGase 1 and TGase 2.
Materials and Methods
Materials
Fluorescein isothiocyanate (FITC)–labeled oligopeptides were synthesized by Biologica Co. (Nagoya, Japan). Each peptide was dissolved in dimethylsulfoxide at 50 mM as a stock solution. Embedding medium for the tissue sections and whole mouse body was Tissue medium from Chiba Medical (Chiba, Japan) and SCEM (Leica Microsystems; Tokyo, Japan), respectively. Other chemical reagents were obtained from Sigma-Aldrich (St Louis, MO) or WAKO Chemicals (Osaka, Japan).
Preparation of Tissue Sections and Staining by Hematoxylin and Eosin
All animal experiments were carried out according to the guidelines of each institute (Nagoya University, Hyogo College of Medicine, RIKEN Institute, and Tsurumi University). TGase 1 knockout mouse was established previously (Matsuki et al. 1998), and TGase 2 knockout mouse was provided by Dr. Robert Graham (Nanda et al. 2001).
Frozen tissue sections of skin and liver in TGase-deficient mice were prepared without fixation at a 4- to 6-µm thickness according to the standard method. The sections were stored at −80C until use.
In the case of whole mouse body, 9-day-old mouse (male) was used. According to the method described by Kawamoto (Kawamoto and Shimizu 2000; Kawamoto 2003), the whole-body sections were prepared using a multipurpose cryosection preparation kit (Section Lab Co. Ltd.; Hiroshima, Japan). First, the mouse was frozen in cold hexane (−93C) and then freeze-embedded with SCEM. Ten-µm thick sections were cut with a cryomicrotome (Leica CM3050S; Leica Co. Ltd., Wetzlar, Germany) from the frozen specimen block and collected with cryofilm. After drying the section, the sections were used for in situ enzymatic reaction without fixation. For hematoxylin and eosin (H&E) staining, tissue sections were fixed by 4% paraformaldehyde and then mounted with SCEM.
In Situ Detection of Active TGase
Sections were dried and then blocked with 1% bovine serum albumin (Sigma-Aldrich) in PBS (10 mM Na-phosphate, pH 8.0, 150 mM NaCl) at room temperature for 30 min. The sections were incubated for 90 min at 37C in a solution containing 100 mM Tris/HCl (pH 8.0), 1 mM dithiothreitol, and 5 mM CaCl2 in the presence of FITC-labeled peptide at the final concentration of 1 µM. In the reaction mixture, calcium ion was contained at higher than the physiological level to enhance the products. After enzymatic reaction, PBS containing 25 mM EDTA was added to stop the reaction via chelating calcium ion. Then, after washing with PBS three times at room temperature for 5 min, antifading solution was added to the sections for observation.
Microscopic Observation and Analyses
Samples were mainly observed under a Keyence fluorescence microscope (BZ-9000; Osaka, Japan) for analyses of whole-mouse section using a ×4 lens (NA 0.20). Samples for H&E staining were also observed with the same lens. For the skin and liver sections in TGase-deficient mice (Fig. 1), the samples were observed under a LSM5 PASCAL confocal laser-scanning microscope, using a combination of ×20 (NA 0.50) or ×40 water immersion lens (NA 1.2w) and differential interference contrast (Zeiss, Göttingen, Germany).
Figure 1.
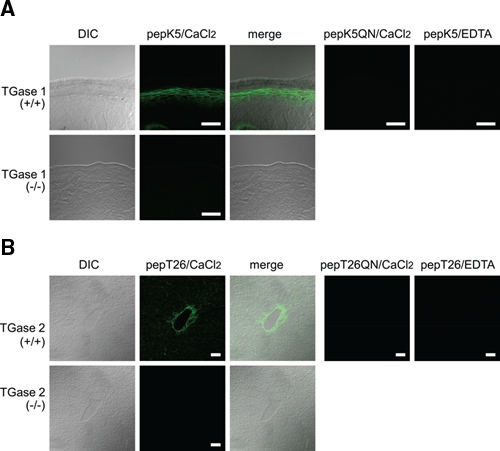
In situ detection of active TGase 1 and TGase 2 in mouse tissue sections. (A) Skin sections from the wild-type (+/+) and TGase 1−null (−/−) mice were reacted with 1 µM FITC-pepK5 in the presence of 5 mM CaCl2. The differential interference images (DIC), the fluorescence images, and their merged images were obtained. As negative controls, reaction samples using 1 µM FITC-pepK5QN or with addition of 5 mM EDTA were carried out. (B) Liver sections from the wild-type (+/+) and the TGase 2−null (−/−) mice were subjected to in situ reaction using 1 µM FITC-pepT26 or 1 µM FITC-pepT26QN as described in (A). The bars indicate 50 µm.
The free software ImageJ (version 1.43u; image processing and analyzing java; http://rsbweb.nih.gov/ij/) was used for linear adjustment of fluorescent images. This software was also used for the image mapping using pseudo-colors from the fluorescent images as a standard procedure.
Results
Reactions of Fluorescence-Labeled Favorable Substrate Peptides for TGase 1 (pepK5) and TGase 2 (pepT26)
We previously identified favorable substrate sequences that were screened from the M13 phage-displayed peptide library (Hitomi, Kitamura, and Sugimura 2009). The respective sequences of pepK5 and pepT26 for TGase 1 and TGase 2 are shown in Table 1. Taking advantage of the isozyme-specific reactivities of these peptides, in situ active TGase 1 and TGase 2 were detected using FITC-labeled peptides. In this procedure, the peptides reacted to the enzyme and covalently cross-linked to glutamine-acceptor substrate proteins in tissues, which provide fluorescence signals. Mutant peptides, in which each reactive glutamine residue had been replaced by asparagine, were also prepared as negative controls (pepK5QN, pepT26QN).
Table 1.
Isozyme-Specific Substrate Peptides for TGase 1 (pepK5) and TGase 2 (pepT26)
| Peptide | Target Enzyme | Sequence |
|---|---|---|
| pepK5 | TGase 1 | YEQHKLPSSWPF |
| pepK5QN | YENHKLPSSWPF | |
| pepT26 | TGase 2 | HQSYVDPWNLDH |
| pepT26QN | HNSYVDPWNLDH |
pepK5QN and pepT26QN are the mutant peptides in which the reactive glutamine residue was substituted with asparagine. These peptides were labeled with FITC at the N-terminus.
To confirm the efficiency of staining, we initially investigated detection using the sections from knockout mice for each TGase (Fig. 1). Because highly active TGase 1 and TGase 2 are respectively expressed in the skin and the liver (Lorand and Graham 2003), these tissue sections in both wild-type and knockout mice were subjected to the reactions.
In the skin section from wild-type mice, a significant fluorescence signal resulting from cross-linked FITC-pepK5 was obtained in cells below the outermost layers, which is consistent with previous results (Fig. 1A; Sugimura et al. 2008). No apparent signal was observed in the section from the TGase 1–deficient mouse or in the reaction using FITC-pepK5QN. In addition, no signal was obtained in the presence of EDTA, suggesting that the reaction is calcium dependent. These results indicated that active TGase 1 was specifically detected by the procedure.
In the liver section, a signal for wild-type TGase 2 was observed around the intimal and medial area at the central vein (Fig. 1B). This signal was not observed in the section from TGase 2 knockout mice. No signal was obtained in the reaction either in the presence of EDTA or after use of the mutant peptide, indicating that the detection was specific to active TGase 2.
Detection of In Situ Active TGases Using Whole-Mouse Section
To obtain the tissue distribution patterns of the active enzymes, the same analyses were carried out using sections of whole-mouse body. Midline and sideline sections of 9-day-old mice were prepared for the above-mentioned analyses and were then subjected to in situ detection of active TGase as well as H&E staining.
In both midline and sideline sections, apparent signals were obtained for FITC-pepK5 and FITC-pepT26 reactions (Fig. 2A, midline; Fig 2B, sideline). No significant fluorescence was observed when using the mutant peptide (FITC-pepK5QN and FITC-pepT26QN), indicating that the signals are specifically derived from the enzymatic reaction. Distinct fluorescence patterns for each cross-linked peptide indicate that the active forms of each enzyme were detected, reflecting the different tissue distribution of the two enzymes. In addition, this reaction is calcium dependent (Suppl. Fig. S1A,B).
Figure 2.
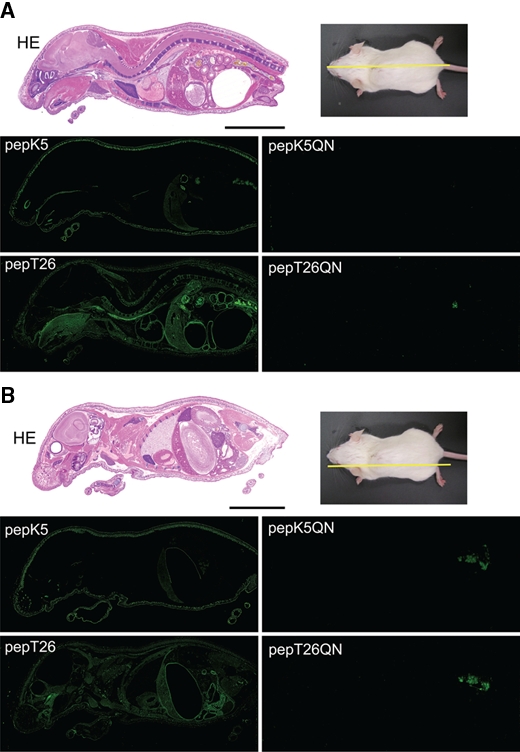
In situ reaction patterns of whole-mouse body sections using FITC-pepK5 and FITC-pepT26. Whole-body sections were prepared and stained with hematoxylin and eosin (HE). The sections were reacted with 1 µM FITC-labeled peptides (pepK5, pepK5QN, pepT26, and pepT26QN) in the presence of CaCl2. (A) Midline and (B) sideline. The bars indicate 1 cm. Nonspecific signals in the pictures for the QN mutants may be due to excrement in the bowel.
To evaluate the localization of active enzymes using FITC-peptide incorporation, fluorescence intensity was expressed as pseudo-colors in the region of interest (Fig. 3). For TGase 1, which is mainly involved in skin formation, much highly active enzyme was observed in the epidermis. Furthermore, other epithelial tissues such as tongue and forestomach showed the existence of relatively high active enzyme. Although they had weaker signals than those of above tissues, hepatocytes showed a significant level. On the other hand, wide tissue distribution patterns were obtained for TGase 2 with various intensities. Digestive tissues such as tongue, esophagus, stomach, and intestine appeared to contain remarkably high active enzyme. Significant active TGase 2 was also observed in veins in the liver in addition to spine and muscles. The brain membrane showed significantly active TGase 2, whereas fewer enzymes were observed in the thymus, spleen, and kidney.
Figure 3.
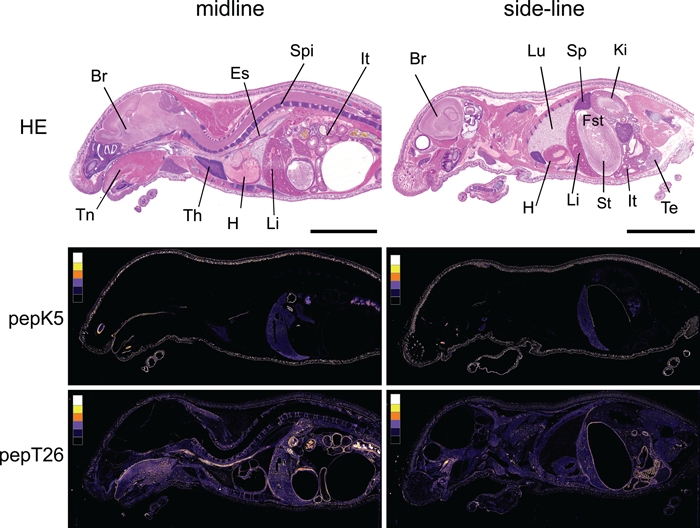
Color images of fluorescence signals detected in the sections reacted with FITC-pepK5 and FITC-pepT26. Based on the fluorescence images of Figure 2, pseudo-color images were produced using ImageJ (version 1.43u). To indicate the fluorescence intensity, we divided level of brightness into six classes (from background to maximum area). The six colors, as shown in the box (left in each picture), indicate the intensity in five grades (white, yellow, orange, purple, dark blue) and black as background. Tissues are indicated as follows: Br, brain; Es, esophagus; Fst, forestomach; H, heart; It, intestine; Ki, kidney; Li, liver; Lu, lung; Sp, spleen; Spi, spine; St, stomach; Te, testis; Th, thymus; Tn, tongue. HE, hematoxylin and eosin. The bars indicate 1 cm.
Tissue Distribution of In Situ Active TGases
In Figure 4, we show enlarged patterns for the nine selected tissues. Results from five epithelial tissues are depicted in Figure 4A. In the tongue, both active TGase 1 and TGase 2 are expressed mainly in the filiform papilla, where TGase 1 is not expressed in the outermost cornified layer but highly expressed in underlying cell layers. TGase 2 is also present in the muscle layers. Bright signals in developing teeth were obtained only for active TGase 1 and were mainly located in odontoblast layers and sterate reticulum.
Figure 4.
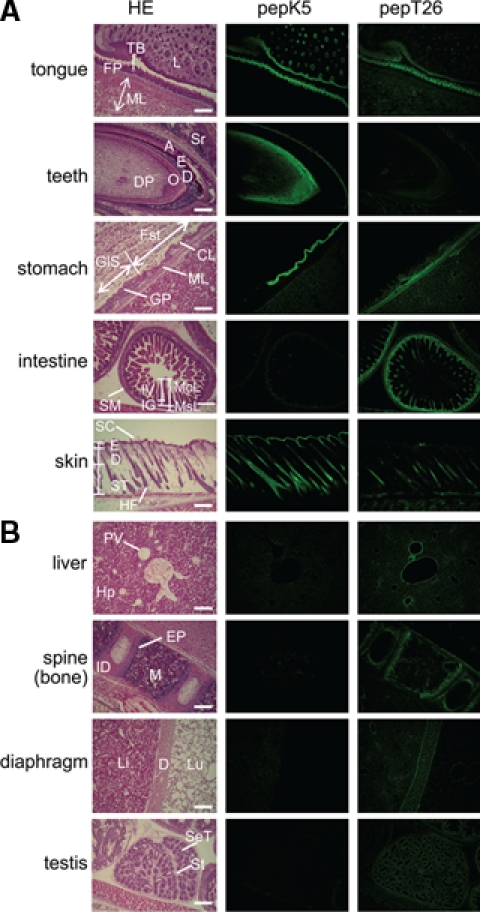
Tissue distribution of in situ active area of TGase 1 and TGase 2. Enlarged pictures of hematoxylin and eosin (HE) staining (left), fluorescence imaging of FITC-pepK5 (center), and FITC-pepT26 (right) are shown for each tissue. (A) Tongue: FP, filiform papilla; ML, muscle layer; L, lip, TB, taste bud. Teeth: DP, dental papilla; O, odontoblast; D, dentin; E, enamel; A, ameloblast; Sr, sterate reticulum. Stomach: Fst, forestomach (stratified); GlS, Glandular stomach (monolayer); CL, cornified layer; GP, gastric pits; ML, muscle layer. Intestine: McL, mucosal layer; MsL, muscle layer; IV, intestinal villi; IG, intestinal glands; SM, seronous membrane. Skin: SC, skin cornified layer; E, epidermis; D, dermis; ST, subcutaneous tissue; HF, hair follicle. (B) Liver: PV, interlobular portal vein; Hp, hepatocyte. Spine (bone): M, bone marrow; ID, intervertebral disk; EP, epiphyseal plate. Diaphragm (D): Li, liver; Lu, lung. Testis: St, stroma; SeT, semiferous cells. The bars indicate 200 µm.
In the stomach of rodents, the glandular stomach is positioned after the forestomach, in which epithelial cells undergo an organized progressive differentiation to form a mature stratified squamous epithelium. Interestingly, active TGase 1 was significantly detected beneath the outer cornified layer limited to the forestomach area, whereas TGase 2 was weakly expressed in both the glandular and forestomach muscle layers. In the intestine, strong signals were obtained only for TGase 2, and they were found in the mucosal and muscle layers. Enlarged observation of mucosal layers showed that the extracellular matrix area beneath the epithelial cells on the surface was stained (data not shown). In the skin, active TGase 1 was expectedly detected in the epidermal layer and hair follicles. In hair follicles, a weak level of active TGase 2 was detected in the central region, which is different from the pattern of TGase 1.
Results for the other four tissues, where significant signals and mostly distinct patterns for TGase 1 and TGase 2 were observed, are shown in Figure 4B. In the liver, active TGase 2 was expressed around the endothelium in the region of the central and interlobular portal veins. In the spine, diaphragm, and testis, active TGase 2 but not TGase 1 was observed. In the spine, TGase 2 was active at the epiphyseal plate and intervertebral disk. These results suggested that active TGase 1 and TGase 2 were located largely in the epithelial and connective tissues, respectively.
Discussion
Because posttranslational modifications by TGases are essential for various biological processes in mammals, it is important to evaluate their activities with high sensitivity and in an isozyme-specific manner. Although many in vitro assay systems have been established to measure the TGase activity, there is no method for monitoring isozyme-specific activity in situ. We recently obtained highly reactive substrate peptides that act as efficient tools for detecting enzymatic activity of several TGases (Sugimura et al. 2006; Sugimura et al. 2008). More recently, using a fluorescence-labeled peptide (pepK5) specific for TGase 1, we have successfully completed in situ detection of active TGases both in human and mouse skin (Sugimura et al. 2008; Akiyama et al. 2010). In the present study, we demonstrated that this system could be adapted to investigate another major isozyme, TGase 2, and to analyze the whole-mouse body.
To date, tissue distribution of the expression pattern of TGase analysis has been reported by immunochemical analyses (Hiiragi et al. 1999; Griffin et al. 2002). However, immunoblotting of lysates prepared from tissues does not allow precise localization of active TGase. Furthermore, TGases detected by antibodies are not necessarily active because the enzymes may be cross-linked themselves or with other proteins, resulting in inactivation. The procedure performed in this study, however, could detect active TGase located in tissues.
In situ detection of active TGase has been possible by observing reaction products that incorporate FITC-labeled cadaverine or green fluorescent protein (GFP) fused with glutamine-containing proteins (Furutani et al. 2001; Oji et al. 2006). However, isozyme specificity cannot be evaluated by these methods. Two substrate peptides (pepK5 and pepT26), which react specifically with each isozyme in vitro, enable detection of the distribution of active TGase in an isozyme-specific manner as shown in this study.
TGase 1 and TGase 2 are mainly localized in the membrane and the cytoplasm, respectively (Lorand and Graham 2003). During reaction, there may be some leakage of the enzymes from the tissue sections, which might decrease the detection of intracellular active TGase. However, the results of the detection can be evaluated because the signals in the liver and skin as well as other tissues are consistent with the previous results obtained by immunohistochemical analysis.
Although the results of in situ detection of both active TGases were mostly consistent with previous reports, novel findings were observed in several tissues. TGase 1, originally identified as a skin keratinocyte-type TGase, was detected at significant levels in other epithelial tissues such as the tongue and forestomach. Thus, TGase 1 is found in an active form primarily in such stratified epithelial tissues. In these TGase 1–expressing tissues, the enzyme may contribute to sustaining these tissues against the external environment by cross-linking structural proteins. In addition, novel findings include having significant expression in the odontoblast and epiphyseal plate in developing teeth and being weakly active in hepatocytes. Analysis using cultured cells for these cells may provide information on the physiological significance of TGase 1.
In contrast, active TGase 2 was observed primarily around single epithelial cells and in connective tissues. For example, significant signals were obtained beneath epithelial cells in the intestinal mucosa and around veins in the liver. Furthermore, other connective tissues such as muscle membrane and chondrocytes (data not shown) showed apparent signals. TGase 2, which plays a role in stabilizing the extracellular matrix by cross-linking reactions, may maintain the integrity of these connective tissues. Identification of enzyme substrates and observation of signals in these tissues will provide valuable information on the physiological significance of TGases.
In conclusion, in situ sensitive and specific detection of the active form of two major isozymes, TGase 1 and TGase 2, was successful. A number of diseases such as autoimmune disease, neoplastic disease, neuronal degeneration, and aberrant skin formation result from the aberrant expression of these TGases (Huber et al. 1995; Sollid 2002; Ruan and Johnson 2007; Mehta 2009). Our procedure may be applicable for clinical diagnosis of diseases in which there is abnormal TGase activity. Furthermore, it will be applied for several investigations such as intracellular activity by efficiently introducing peptides into cells (Van Nooden 2010).
Acknowledgments
We greatly appreciate Dr. Masatoshi Maki and Dr. Hideki Shibata in our laboratory for providing valuable suggestions. The knockout mouse for TGase 2 was kindly provided by Dr. Robert Graham (Victor Chang Cardiac Institute, Australia).
Footnotes
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
The author(s) disclosed receipt of the following financial support for the research and/or authorship of this article: This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas (No. 20200072) (to KH) from the Ministry of Education, Sports, Science and Technology (MEXT, Japan). This work was also supported in part by a grant from the Chemical Genomics Research Program from RIKEN (to SK) and by a Grant-in-Aid for Scientific Research (C) from JSPS (No. 20591359) (to KY).
References
- Aeschlimann D, Koeller MK, Allen-Hoffmann BL, Mosher DF. 1998. Isolation of a cDNA encoding a novel member of the transglutaminase gene family from human keratinocytes: detection and identification of transglutaminase gene products based on reverse transcription-polymerase chain reaction with degenerate primers. J Biol Chem. 273:3452-3460 [DOI] [PubMed] [Google Scholar]
- Akiyama M, Sakai K, Yanagi T, Fukushima S, Ihn H, Hitomi K, Shimizu H. 2010. Transglutaminase 1 preferred substrate peptide K5 is an efficient tool in diagnosis of lamellar ichthyosis. Am J Pathol. 176:1592-1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beninati S, Piacentini M. 2004. The transglutaminase family: an overview. Amino Acids. 26:367-372 [DOI] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. 2005. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 6:328-340 [DOI] [PubMed] [Google Scholar]
- Eckert RL, Sturniolo MT, Broome AM, Ruse M, Rorke EA. 2005. Transglutaminase function in epidermis. J Invest Dermatol. 124:481-492 [DOI] [PubMed] [Google Scholar]
- Esposito C, Pucci P, Amoresano A, Marino G, Cozzolino A, Porta R. 1996. Transglutaminase from rat coagulating gland secretion: post-translational modification and activation by phosphatidic acids. J Biol Chem. 271:27416-27423 [DOI] [PubMed] [Google Scholar]
- Fesus L, Piacentini M. 2002. Transglutaminase 2: an enigmatic enzyme with diverse functions. Trends Biochem Sci. 27:534-539 [DOI] [PubMed] [Google Scholar]
- Furutani Y, Kato A, Notoya M, Ghoneim MA, Hirose S. 2001. A simple assay and histochemical localization of transglutaminase activity using a derivative of green fluorescent protein as substrate. J Histochem Cytochem. 49:247-258 [DOI] [PubMed] [Google Scholar]
- Grenard P, Bates MK, Aeschlimann D. 2001. Evolution of transglutaminase genes: identification of a transglutaminase gene cluster on human chromosome 15q15. J Biol Chem. 276:33066-33078 [DOI] [PubMed] [Google Scholar]
- Griffin M, Casadio R, Bergamini CM. 2002. Transglutaminases: nature’s biological glues. Biochem J. 368:377-396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiiragi T, Sasaki H, Nagafuchi A, Sabe H, Shen SC, Matsuki M, Yamanishi K, Tsukita S. 1999. Transglutaminase type 1 and its cross-linking activity are concentrated at adherens junctions in simple epithelial cells. J Biol Chem. 274:34148-34154 [DOI] [PubMed] [Google Scholar]
- Hitomi K. 2005. Transglutaminase in skin epidermis. Eur J Dermatol. 15:313-319 [PubMed] [Google Scholar]
- Hitomi K, Kitamura M, Perez Alea M, Ceylon I, Thomas V, El Alaoui S. 2009. A specific colorimetric assay for measuring transglutaminase 1 and factor XIII activities. Anal Biochem. 394:281-283 [DOI] [PubMed] [Google Scholar]
- Hitomi K, Kitamura M, Sugimura Y. 2009. Preferred substrate sequences for transglutaminase 2: screening using a phage-displayed peptide library. Amino Acids. 36:619-624 [DOI] [PubMed] [Google Scholar]
- Huber M, Rettler I, Bernasconi K, Frenk E, Lavrijsen SPM, Ponec M, Bon A, Lautenschlager S, Schorderet DF, Hohl D. 1995. Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science. 267:525-528 [DOI] [PubMed] [Google Scholar]
- Ichinose A. 2001. Physiopathology and regulation of factor XIII. Thromb Haemost. 86:57-65 [PubMed] [Google Scholar]
- Jeon JH, Kim IG. 2006. Role of protein modification mediated by transglutaminase 2 in human viral diseases. Front Biosci. 11:221-231 [DOI] [PubMed] [Google Scholar]
- Kalinin AE, Kajava AV, Steinert PM. 2002. Epithelial barrier function: assembly and structural features of the cornified cell envelop. BioEssays. 24:789-800 [DOI] [PubMed] [Google Scholar]
- Kawamoto T. 2003. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch Histrol Cytol. 66:123-143 [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Shimizu M. 2000. A method for preparing 2- to 50-µm-thick fresh frozen section of large samples and undecalcified hard tissues. Histochem Cell Biol. 113:331-339 [DOI] [PubMed] [Google Scholar]
- Lorand L, Graham RM. 2003. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol. 4:140-156 [DOI] [PubMed] [Google Scholar]
- Matsuki M, Yamashita F, Ishida-Yamamoto A, Yamada K, Kinoshita C, Fushiki S, Ueda E, Morishima Y, Tabata K, Yasuno H, Hashida M, Iizuka H, Ikawa M, Okabe M, Kondoh G, Kinoshita T, Takeda J, Yamanishi K. 1998. Defective stratum corneum and early neonatal death in mice lacking the gene for transglutaminase 1. Proc Natl Acad Sci U S A. 95:1044-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta K. 2009. Biological and therapeutic significance of tissue transglutaminase in pancreatic cancer. Amino Acids. 36:709-716 [DOI] [PubMed] [Google Scholar]
- Mehta K, Fok JY, Mangala LS. 2006. Tissue transglutaminase: from biological glue to cell survival cues. Front Biosci. 11:173-185 [DOI] [PubMed] [Google Scholar]
- Nanda N, Iismaa SE, Owens WA, Husain A, Mackay F, Graham RM. 2001. Targeted inactivation of Gh/tissue transglutaminase II. J Biol Chem. 276:20673-20678 [DOI] [PubMed] [Google Scholar]
- Oji V, Oji ME, Adamini N, Walker T, Aufenvenne K, Raghunath M, Traupe H. 2006. Plasminogen activator inhibitor-2 is expressed in different types of congenital ichthyosis: in vivo evidence for its cross-linking into the cornified cell envelope by transglutaminase-1. Br J Dermatol. 154:860-867 [DOI] [PubMed] [Google Scholar]
- Perez Alea M, Kitamura M, Martin G, Thomas V, Hitomi K, El Alaoui 2009. Development of an isozyme-specific colorimetric assay for tissue transglutaminase 2 cross-linking activity. Anal Biochem. 389:150-156 [DOI] [PubMed] [Google Scholar]
- Ruan Q, Johnson GV. 2007. Transglutaminase 2 in neurogenerative disease. Front Biosci. 12:891-904 [DOI] [PubMed] [Google Scholar]
- Sarang Z, Toth B, Balajthy Z, Koroskenyi, Garabuczi E, Fesus L, Szondy Z. 2009. Some lessons from the tissue transglutaminase knockout mouse. Amino Acids. 36:625-631 [DOI] [PubMed] [Google Scholar]
- Sollid LM. 2002. Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol. 2:647-655 [DOI] [PubMed] [Google Scholar]
- Sugimura Y, Hosono M, Kitamura M, Tsuda T, Yamanishi K, Maki M, Hitomi K. 2008. Identification of preferred substrate sequences for transglutaminase 1: development of a novel peptide that can efficiently detect cross-linking enzyme activity in the skin. FEBS J. 275:5667-5677 [DOI] [PubMed] [Google Scholar]
- Sugimura Y, Hosono M, Wada F, Yoshimura T, Maki M, Hitomi K. 2006. Screening for the preferred substrate sequence of transglutaminase using a phage-displayed peptide library: identification of peptide substrates for TGase 2 and factor XIIIa. J Biol Chem. 281:17699-17706 [DOI] [PubMed] [Google Scholar]
- Sugimura Y, Ueda H, Maki M, Hitomi K. 2007. Novel site-specific immobilization of functional protein using a preferred substrate sequence for transglutaminase 2. J Biotechnol. 131:121-127 [DOI] [PubMed] [Google Scholar]
- Tatsukawa H, Fukaya Y, Frampton G, Martinez-Fuentes A, Suzuki K, Kuo TF, Nagatsuma K, Shimokado K, Okuno M, Wu J, Iismaa S, Matsuura T, Tsukamoto H, Zern MA, Graham RM, Kojima S. 2009. Role of transglutaminase 2 in liver injury via cross-linking and silencing of transcription factor Sp1. Gastroenterology. 136:1783-1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nooden CJF. 2010. Imaging enzymes at work: metabolic mapping by enzyme histochemistry. J Histochem Cytochem. 58:481-497 [DOI] [PMC free article] [PubMed] [Google Scholar]


