Abstract
The putative tumor stem cell marker CD133 is the marker of choice for identifying brain tumor stem cells in gliomas, but the use of different CD133 antibody clones possibly recognizing different CD133 splice variants with epitopes of different glycosylation status confuses the field. The aim was to investigate if current inconsistent CD133 observations could be a result of using different CD133 antibodies for immunohistochemical identification of CD133. Ten glioblastomas were immunohistochemically stained with four different CD133 antibody clones (AC133, W6B3C1, C24B9, and ab19898) and analyzed by quantitative stereology. Moreover, the CD133 staining pattern of each antibody clone was investigated in kidney, pancreas, and placenta tissue as well as in glioblastoma and retinoblastoma cultures and cell lines. All antibody clones revealed CD133+ niches and single cells in glioblastomas, but when using different clones, their distribution rarely corresponded. Morphology of identified single cells varied, and staining of various tissues, cultures, and cells lines was also inconsistent among the clones. In conclusion, the authors report inconsistent CD133 detection when using different primary CD133 antibody clones. Thus, direct comparison of studies using different antibody clones and conclusions based on CD133 immunohistochemistry should be performed with caution.
Keywords: comparative, CD133, stem cell, niche, immunohistochemistry, antibodies, glioblastoma, quantitative, stereology
Introduction
CD133 Is the Marker of Choice for Identifying Brain Tumor Stem Cells
Identification of cells with tumor stem cell properties has been reported in several solid tumors, including tumors of the brain (Singh et al. 2004), breast (Al-Hajj et al. 2003), pancreas (Hermann et al. 2007; Li et al. 2007), colon (Ricci-Vitiani et al. 2007; O’Brien et al. 2007), ovary (Zhang S et al. 2008), and prostate (Maitland and Collins 2008). The putative stem cell marker CD133 is the marker of choice for identifying brain tumor stem cells (BTSCs; Singh et al. 2004; Singh et al. 2003), but the use of different CD133 antibody clones possibly recognizing different CD133 splice variants with epitopes of different glycosylation status confuses the field (Mizrak et al. 2008; Bidlingmaier et al. 2008; Fargeas et al. 2004; Singh et al. 2004; Singh et al. 2003). Studies investigating the distribution and prognostic value of CD133 have reported inconsistent findings (Zeppernick et al. 2008; Immervoll et al. 2008; Thon et al. 2010; Beier et al. 2008; Pallini et al. 2008; Christensen et al. 2008; Sulman et al. 2008; Bidlingmaier et al. 2008; Florek et al. 2005; Singh et al. 2004; Singh et al. 2003) and shown that CD133 epitopes can be lacking immunohistochemically despite the presence of CD133 protein (Kemper et al. 2010; Bidlingmaier et al. 2008). Some studies have shown that CD133+ and not CD133– cells possess stem cell–like and tumor-initiating properties (Singh et al. 2004; Singh et al. 2003), whereas others have reported the direct opposite (Clément et al. 2010; Clément et al. 2009; Wang et al. 2008; Beier et al. 2007). The identification or isolation of CD133+ cells is typically carried out using CD133 antibodies, and we hypothesize that some CD133 controversies could be a consequence of using different poorly characterized primary CD133 antibodies to identify or isolate CD133+ cells.
Selection of CD133 Antibody Clones
Initially, five antibody clones were selected for comparative analysis (AC133, W6B3C1, 293C3, C24B9, and ab19898). According to the manufacturers, AC133 and W6B3C1 target a glycosylated extracellular epitope 1 of CD133, 293C3 targets a glycosylated extracellular epitope 2 of CD133, C24B9 targets a non-glycosylated extracellular epitope of CD133, and clone ab19898 targets a non-glycosylated intracellular epitope of CD133 (Table 1). Clone 293C3 was not found suitable for any immunohistochemical staining on paraffin sections and was thus excluded from further analysis.
Table 1.
CD133 Antibody Clones Used in the Present Study, Including Immunogens and Protein Sizes Reported by Studies and Manufacturers as well as Immunohistochemical Methods, Positive Control Tissues, and Prognostic Data Obtained Using Immunohistochemistry
| Antibody Clone | Company | Immunogen | HIER Buffer | Conc. | Detection System | Species and Clonality | Positive Control | Size on Western Blot | Is CD133 Prognostic? |
|---|---|---|---|---|---|---|---|---|---|
| CD133/1 (AC133) | Miltenyi | Second extracellular loop of CD133 (Kemper et al. 2010) | EDTA | 1 + 10 | PV | Mouse monoclonal | Kidney, pancreas | ~120 kDa (Florek et al. 2005; Corbeil et al. 2000) | Yes (Pallini et al. 2008; Thon et al. 2010; Zeppernick et al. 2008) |
| CD133/1 (W6B3C1) | Miltenyi | Glycosylated epitope 1 on the extracellular domain of CD133 | TEG | 1 + 40 | PV | Mouse monoclonal | Kidney, pancreas, placenta | ~120 kDa | No (Christensen et al. 2008) |
| CD133 (C24B9) | Cell Signaling Technology | A non-glycosylated extracellular region surrounding Asp562 of CD133 | EDTA | 1 + 50 | EnV | Rabbit monoclonal | Kidney, pancreas | ~16 kDa (Osmond et al. 2010) ~115 - 130 kDa (Osmond et al. 2010) | NA |
| CD133 (ab19898) | Abcam | Residue 800 to the C-terminus of CD133 | TEG | 1 + 400 | EnV | Rabbit polyclonal | Kidney, pancreas | ~110 kDa | No (Sulman et al. 2008) |
TEG, Tris 10 mM, EGTA, 0.5 mM, pH 9; EDTA, 1 mM, pH 8; HIER, heat-induced epitope retrieval; PV, PowerVision+; EnV, EnVision+; NA, not available.
AC133 was selected because the vast majority of studies use this clone to identify brain tumor stem cells in glioblastomas (Zeppernick et al. 2008; Thon et al. 2010; Pallini et al. 2008; Singh et al. 2004; Singh et al. 2003). It has been used to isolate brain tumor–initiating cells (Singh et al. 2004; Singh et al. 2003) and to show a negative prognostic significance of CD133 in both frozen (Zeppernick et al. 2008; Thon et al. 2010) and paraffin-embedded brain tumor sections (Pallini et al. 2008). W6B3C1 was chosen because it has been used in a study from our group that rejected CD133 as a sole prognostic marker in a series of 114 paraffin-embedded astrocytomas (Christensen et al. 2008). AC133 and W6B3C1 both target a supposed glycosylated epitope 1 of CD133 and should consequently display concordant immunolabeling. A recent study, however, has mapped the AC133 epitope to the extracellular loop 2 of CD133 and suggested that binding of AC133 is not glycosylation dependent (Kemper et al. 2010). Bidlingmaier et al. (2008) have suggested that C24B9 could be a suitable antibody for identifying BTSCs in gliomas as it recognizes a non-glycosylated extracellular epitope of CD133. If tumor cells can change the glycosylation status of their CD133 proteins when they differentiate (Mizrak et al. 2008; Florek et al. 2005; Corbeil et al. 2000), one could bypass problems regarding this matter by using an antibody targeted against non-glycosylated parts of the protein. Clone ab19898 from Abcam (Cambridge, UK) was selected because this or an analogous clone was used in an immunohistochemical study with 70 paraffin-embedded glioblastomas (Sulman et al. 2008), revealing no prognostic significance of CD133. Why these discrepancies exist has yet to be eluded.
To our knowledge, this is the first study comparing CD133 expression profiles of four different and commonly used CD133 antibody clones. The comparison included paraffin-embedded sections of glioblastoma, normal healthy adult brain, kidney, pancreas, and placenta tissues, as well as spheroids and cells derived from the short-term culture SJ-1 and conventional cell lines U87 and Y79. SJ-1, which has been established in our laboratory, is a glioblastoma short-term culture with tumor stem cell properties (Kolenda et al. 2010). U87 is a commonly used commercial cell line for glioblastoma in vitro research with conflicting reports regarding CD133 expression (Christensen et al. 2010; Brehar et al. 2009; Platet et al. 2007). Y79 is a retinoblastoma cell line that has been reported to be CD133+. Studies have confirmed CD133 expression by fluorescence-activated cell sorting (FACS), PCR, and Western blot analysis (Thon et al. 2010; Seigel et al. 2007; Yin et al. 1997).
In the present study, we obtained discordant CD133 expression in glioblastomas, various normal tissues, cultures, and cell lines when using four different CD133 antibody clones. This clearly highlights that choice of CD133 antibody clone most likely is critical for CD133 research and that direct comparison of studies using different CD133 antibody clones for both isolation and identification of CD133 is problematic.
Materials and Methods
Human Subject Tissue
Tissue from 10 glioblastoma patients was chosen (Table 2). All glioblastoma tumor samples were classified by a neuropathologist according to the 2007 World Health Organization (WHO) guidelines (Louis et al. 2007). Patients underwent initial surgical resection for glioblastoma between January and May 2006 at the Department of Neurosurgery, Odense University Hospital, Denmark. Prior to surgical resection, the patients had not received any treatment. To obtain comparable and reproducible results, a series of exclusion criteria for used human subject tissue was defined. Glioblastomas where histological material had been frozen before paraffin embedding were excluded because the morphology of frozen tissue is compromised compared to the morphology of paraffin-embedded tissue (Shi et al. 2008). Because of pronounced tumor heterogeneity, it was not optimal to assess CD133 expression in small biopsy specimens. Tumor biopsies with a diameter of less than 4 mm were thus excluded. Finally, tumors resected only by ultrasonic aspiration were excluded because of varying degrees of tissue degradation (Silverman et al. 1989; Malhotra et al. 1986). The project has been approved by the Data Protection Authority (j.nr. 2009-41-3070) and the Scientific Ethical Committee in Denmark (S-20070021).
Table 2.
Characteristics of the 10 Glioblastoma Patients Included in This Study
| Gender | Age at Surgical Resection, y | Survival Period, mo |
|---|---|---|
| M | 68 | 33 |
| M | 62 | 42 |
| M | 42 | 24 |
| M | 70 | 11 |
| F | 59 | 7 |
| M | 40 | 0 |
| F | 61 | 0 |
| F | 44 | 6 |
| F | 77 | 3 |
| M | 56 | 37 |
| Male-female ratio, 1.5 | Mean age, 58 | Mean survival, 16 |
Culturing of Cells
In the present study, the recently established glioblastoma short-term culture SJ-1 was used as well as the commercial glioblastoma cell line U87 and the commercial retinoblastoma cell line Y79. SJ-1 is a new glioblastoma short-term culture with tumor stem cell properties established in stem cell medium in our laboratory according to the literature (Lee et al. 2006). After several passages, it is capable of spheroid formation at clonal density, expression of stem cell markers in formed spheroids, and expression of neuronal and glial markers upon differentiation (Kolenda et al. 2010). Cells were cultured under normal in vitro culturing conditions (37C, 21% O2, 5% CO2) in a Thermo Steri-cycle CO2 incubator (Thermo Electron Corp., Waltham, MA). Serum-free so-called stem cell medium was chosen for culturing of SJ-1 and U87 cells, whereas serum-containing medium was chosen for Y79 (Seigel et al. 2007; Seigel et al. 2005). Composition of the stem cell medium: Neurobasal A (Sigma-Aldrich, Broendby, Denmark), B27 without vitamin A (Invitrogen A/S, Taastrup, Denmark), N-2 (Invitrogen A/S, Taastrup, Denmark), L-glutamine (Lonza, Verviers, Belgium), Pen/strep (Lonza, Verviers, Belgium), 50 ng/ml epidermal growth factor (EGF; Sigma-Aldrich, Broendby, Denmark), and 50 ng/ml basic fibroblast growth factor (bFGF; Trichem A/S, Skanderborg, Denmark). Composition of serum-containing medium: Dulbecco’s modified Eagle’s medium (DMEM) (D-5671; Sigma-Aldrich), 10% fetal bovine serum (FBS; Fisher Scientific, Roskilde, Denmark), 2 mM NaP (Sigma-Aldrich), 2 mM L-Glutamine (Cambrex, East Rutherford, NJ), 1% non-essential amino acids (NEAA; Cambrex), and 1% penicillin/streptomycin (Cambrex). All cells were cultured in 20 ml medium in 75-cm2 culturing flasks (NUNC, Roskilde, Denmark). Using the given culture conditions, SJ-1 and U87 grew as spheroids and Y79 grew in large clusters.
Preparation of Tissues and Cell Cultures for Immunohistochemistry
Fresh tissue biopsies from all glioblastoma patients and control tissues, including brain, kidney, pancreas, and placenta tissue, were all fixed in 4% neutral-buffered formalin with subsequent paraffin embedding. SJ-1, U87, and Y79 cells were likewise prepared for immunohistochemical staining by transference to 4% neutral-buffered formalin overnight followed by paraffin embedding. Then, 3-µm sections were cut on a microtome and placed on Superfrost plus slides (Thermo Fisher Scientific, Copenhagen, Denmark).
Immunohistochemistry
Immunohistochemical staining was carried out on a Dako Autostainer Universal Staining System (Dako Denmark A/S, Glostrup, Denmark). The 3-µm paraffin sections were dewaxed with xylene and rehydrated with ethanol. Endogenous peroxidase activity was quenched by 1.5% hydrogen peroxide. To obtain the optimal staining protocol, various heat-induced epitope retrievals (HIER) and non-HIERs were tested as well as tests of preincubation and different detection systems. The protocol with the highest signal-to-noise ratio was selected as the final, optimized protocol for every CD133 antibody clone (Table 1). To control that changes in procedure only altered the intensity and not the pattern of staining for any given clone, staining of glioblastoma tissues and tissue arrays, including 28 human normal tissues and 12 human tumors, using the same HIER and/or detection system for each antibody in addition to the optimized protocol was compared. This was confirmed for all tissues, but the use of different HIER buffers sometimes introduced or eliminated cross-reactivity with human serum inside blood vessels, a general problem with these antibodies (AC133 in Figs. 1 and 2, ab19898 and C24B9 in Fig. 2). Five epitope retrieval reagents were tested: protease (0.05% protease type XIV; Sigma-Aldrich, St Louis, MO) in TBS (0.05 M, pH 7.4), citrate (10 mM citrate buffer, pH 6), TEG (Tris 10 mM, EGTA 0.5 mM, pH 9), TRS (Target Retrieval Solution, pH 6; Dako Denmark A/S), and EDTA (1 mM, pH 8). Next, preincubation with 0.5% casein, 5% normal serum from secondary antibody species, and 2% bovine serum albumin or Tween-20 detergent was tested. Sections were then incubated for 60 min with primary antibodies diluted in EnVision FLEX Antibody Diluent (Dako Denmark A/S). Visualization of the antigen–antibody complex was carried out with EnVision+ (Dako Denmark A/S), PowerVision+ (Dako Denmark A/S), or CSAII (Dako Denmark A/S) detections systems according to the manufacturer’s manual. DAB was used as chromogen. Finally, sections were counterstained with Mayer’s hematoxylin (Bie & Berntsen, Herlev, Denmark) for 2 min, and coverslips were mounted with Aquatex (Merck, Darmstadt, Germany). Positive and negative controls were included in every run. Kidney, pancreas, and placenta tissues served as CD133 reference tissues as they have been reported to be CD133+ (Immervoll et al. 2008; Christensen et al. 2008; Florek et al. 2005). Omitting primary antibodies served as negative controls as well as controls for nonspecific staining induced by the detection systems alone. Mouse IgG1 and rabbit IgG isotype controls (X0931, X0903, and X0936; Dako Denmark A/S) revealed no relevant staining in glioblastoma, cell culture, or CD133 reference sections. Four CD133 antibody clones were used: CD133/1 AC133 (Miltenyi, Bergisch Gladbach, Germany), CD133/1 W6B3C1 (Miltenyi), ab19898 (Abcam), and CD133 C24B9 (Cell Signaling Technology, Danvers, MA) (Table 1). CD133 antibody clone CD133/2 293C3 (Miltenyi) was also tested but was not found suitable for immunohistochemical detection of CD133 using any combination of primary antibody dilution, HIER, and detection system.
Figure 1.
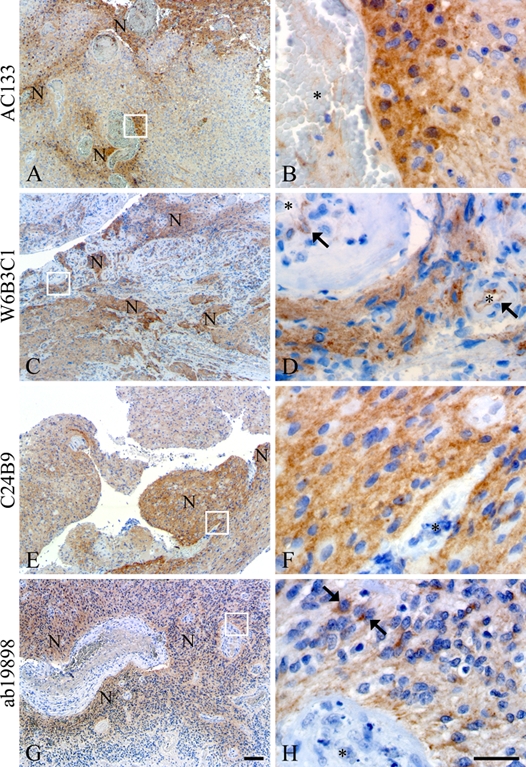
All clones AC133 (A, B), W6B3C1 (C, D), C24B9 (E, F), and ab19898 (G, H) identified niches of perivascular tumor cells in the glioblastomas. W6B3C1 also showed staining of the basal endothelial membrane of most blood vessels (arrows, D). Clone ab19898 intensely stained dispersed cells within the niches (arrows, H). Vessel lumen is indicated by an asterisk (B, D, F, H), and N denotes niches. Scale bars: 50 µm (A, C, E, G); 30 µm (B, D, F, H).
Figure 2.
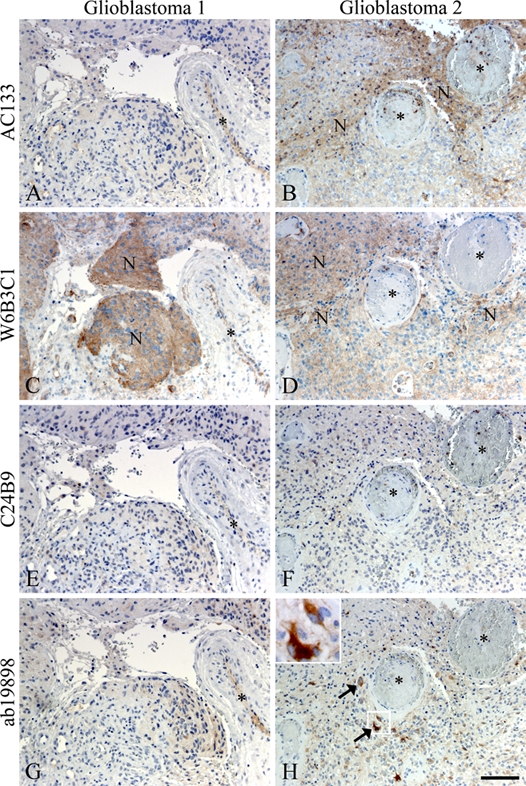
Immunohistochemical staining of adjacent glioblastoma sections with the CD133 antibody clones AC133 (A, B), W6B3C1 (C, D), C24B9 (E, F), and ab19898 (G, H). Differences in intensity and appearance of CD133+ niches were evident. In glioblastoma 1, W6B3C1 identified a clearly demarcated perivascular niche (C), whereas AC133, C24B9, and ab19898 showed little or no reaction in the same area (A, E, G). In glioblastoma 2, both AC133 and W6B3C1 identified niches of CD133+ cells (B, D), whereas C24B9 and ab19898 did not (F, H). Ab19898, however, did identify dispersed single cells with intense cytoplasmatic staining in the same area (arrows and inserts, H). N denotes niches, and vessel lumen is indicated by an asterisk. Scale bar: 100 µm.
Assessment Guidelines
Adjacent sections of 10 glioblastomas were simultaneously stained with the four antibody clones and subsequently evaluated using Computer Assisted Stereology Toolbox (CAST) 2.1.6.0. (Visiopharm, Hoersholm, Denmark) coupled to an Olympus BX50 light microscope equipped with a motorized XY stage (Prior ProScan, Rockland, MA). The immunostainings were evaluated by a trained PhD student (S.K.H.) in close collaboration with an experienced neuropathologist (B.W.K.). The evaluation was blinded (i.e., the observer had no knowledge of which CD133 antibody clone was evaluated). Adjacent sections stained with Mayer’s hematoxylin were used to identify areas with vital tumor tissue and exclude any invasion zone and necrotic or damaged areas. First, the region of interest was outlined at low magnification (2× objective). The CD133+ niches were quantified with a 4× objective using a point grid, whereas a higher magnification (20× objective) was used for evaluation of single cells and blood vessels. On each section, 500 to 800 points were evaluated to ensure that 200 valid points were counted per section (Gundersen 1992; Gundersen et al. 1988; Gundersen and Jensen 1987). A niche was defined as a clearly demarcated entity of cells corresponding to a minimum of five cells (Gilbertson and Rich 2007). In case of large CD133+ niches with staining that gradually decreased toward the periphery, the niche had to be distinct from the surrounding background. Both CD133+ and negative blood vessels were counted.
Counted CD133+ single cells were dispersed, had a positive membrane and/or cytoplasm together with visible nuclei, were clearly distinct from the background, and did not belong to a niche. Single cells were subclassified into angular single cells, including tumor cells, with ramifications, protrusions, and spindle-shaped morphologies or smooth cells without any cellular protrusions. All CD133+ neutrophils in and around the tumor tissue were excluded. Tumor volume fractions of total CD133+ cells, CD133+ niches, and CD133+ angular and smooth single cells were calculated as well as CD133+ blood vessel volume fractions (Gundersen 1986). Intraobserver variance was determined by sampling sections from each antibody group again and comparing data from this run with data from the initial run. Intraobserver variance was always less than 10%.
Statistical Analysis
Statistical comparison of CD133+ niches, CD133+ single cells, and CD133+ blood vessel volume fractions was carried out using one-way analysis of variance (ANOVA) with Bonferroni’s multiple comparison test. To test whether significant correlation existed between volume fractions, Pearson’s linear correlation coefficient (r) was computed with two-tailed p values and r > 0.8 indicating strong correlation. The statistical tests were performed using GraphPad Prism 5.01 software (GraphPad Software, La Jolla, CA), and an overall significance level of p<0.05 was defined.
Results
Identification of CD133+ Niches in Glioblastomas
In glioblastoma tissues, all antibody clones identified CD133+ tumor cells in niches of diverse size, localization, and intensity (Table 3). The size of the niches varied from large positive areas to small perivascular niches comprising only a few cells (Fig. 1). All antibody clones displayed intense cytoplasmatic staining of a small fraction of cells within the CD133+ niches, and this phenomenon was prevalent with ab19898 (Fig. 1H, arrows). When comparing the distribution of CD133 staining, using the different clones in adjacent sections of the same glioblastomas, differences became apparent, as the niches were rarely located in the same areas (Fig. 2). In one glioblastoma, W6B3C1 identified a clearly demarcated perivascular niche (Fig. 2C), whereas AC133, C24B9, and ab19898 showed little or no reaction in the same area (Fig. 2A,E,G). In another glioblastoma, AC133 and W6B3C1 identified distinct niches in the same area (Fig. 2B,D), whereas C24B9 and ab19898 did not (Fig. 2F,H). Dispersed single cells with intense cytoplasmatic staining were, however, observed in this area with ab19898 (Fig. 2H, arrows and insert).
Table 3.
Overview Table Illustrating the Presence of Inconsistent Staining Characteristics
| AC133 | W6B3C1 | C24B9 | ab19898 | ||
|---|---|---|---|---|---|
| Glioblastomas | |||||
| CD133+ niches | + | + | + | (+) | |
| CD133+ single cells | (+) | + | + | + | |
| CD133+ single-cell phenotype | Smooth | Smooth (angular) | Smooth | Angular | |
| CD133+ blood vessels | − | + | − | − | |
| Stem cell zones | |||||
| CD133+ SVZ ependymal cells | − | + | − | + | |
| CD133+ SVZ subependymal cells | + | + | + | + | |
| CD133+ SGZ cells | − | − | + | + | |
| Cell cultures | |||||
| CD133+ SJ-1 cells | (+) | + | + | (+) | |
| CD133+ U87 cells | − | (+) | (+) | + | |
| CD133+ Y79 cells | (+) | + | + | (+) | |
| Positive control tissues | |||||
| Kidney | |||||
| CD133+ Bowman’s capsule | + | + | + | − | |
| CD133+ tubuli | (+) | (+) | + | + | |
| Pancreas | |||||
| CD133+ islets of Langerhans | − | − | + | − | |
| CD133+ pancreatic acini/tubules | + | + | + | + | |
| Placenta | |||||
| CD133+ trophoblasts/syncytiotrophoblasts | − | + | − | + | |
+, positive; −, negative; (+), few positive; SVZ, subventricular zone; SGZ, subgranular zone.
Quantitative analysis revealed that W6B3C1 identified most CD133 (max = 2.8%, mean = 1.3%) and that significantly higher total CD133+ volume fractions were obtained using W6B3C1 compared to AC133 (p<0.01; Fig. 3A). CD133+ niche volume fractions obtained for the different clones were not significantly different from each other (Fig. 3B). Figure 4 shows the volume fractions obtained when using the four different clones in each of the 10 glioblastomas. Looking at total CD133+, a strong correlation existed between AC133 and ab19898 (r = 0.856, p=0.002) as well as W6B3C1 and C24B9 (r = 0.803, p=0.005). Regarding niches, a strong correlation (r > 0.800) was also identified between AC133 and ab19898 (r = 0.831, p=0.003; Table 4).
Figure 3.
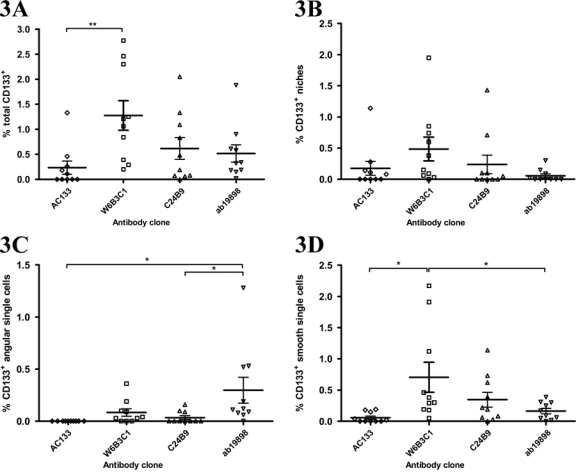
Scatter plots showing the tumor volume fractions of total CD133 positivity (A), CD133+ niches (B), CD133+ angular single cells (C), and CD133+ smooth single cells (D) obtained by staining adjacent sections with CD133 antibody clones AC133, W6B3C1, C24B9, and ab19898 in 10 glioblastomas. Total CD133+ volume fraction (A) includes volume fractions of CD133+ niches (B), angular single cells (C), and smooth single cells (D). Significantly higher total CD133+ volume fractions were identified using W6B3C1 than when using AC133 (**). The volume fraction of CD133+ angular single cells identified using ab19898 was significantly higher than the volume fraction identified using AC133 and C24B9 (C, *). Tumor volume fraction of CD133+ smooth single cells was significantly higher when identified by W6B3C1 than when identified with AC133 and ab19898 (D, *). Horizontal lines denote mean and standard error of mean. *p<0.05. **p<0.01.
Figure 4.
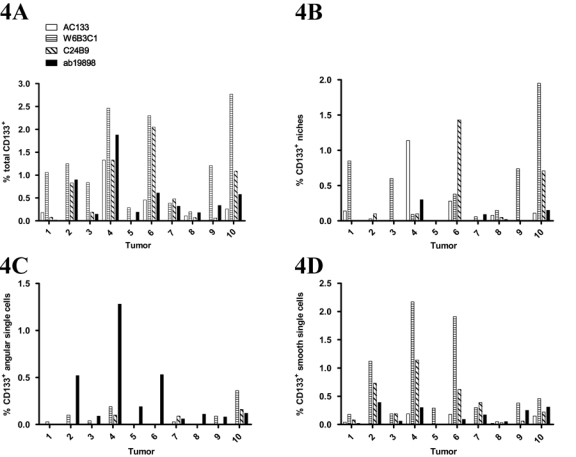
Graphs showing tumor volume fraction variation of total CD133 positivity (A), CD133+ niches (B), CD133+ angular single cells (C), and CD133+ smooth single cells (D) obtained by staining adjacent sections from 10 glioblastomas with CD133 antibody clones AC133, W6B3C1, C24B9, and ab19898.
Table 4.
Correlation Matrix of Identified Volume Fractions Listing Pearson Correlation Coefficients (r) at the Top and Significances (p) at the Bottom
| % Total CD133+ | % CD133+ Niches | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AC133 | W6B3C1 | C24B9 | ab19898 | AC133 | W6B3C1 | C24B9 | ab19898 | ||
| AC133 | 1.000 | AC133 | 1.000 | ||||||
| — | — | ||||||||
| W6B3C1 | 0.654 | 1.000 | W6B3C1 | −0.155 | 1.000 | ||||
| 0.040 | — | 0.670 | — | ||||||
| C24B9 | 0.609 | 0.803 | 1.000 | C24B9 | 0.120 | 0.309 | 1.000 | ||
| 0.062 | 0.005 | — | 0.742 | 0.385 | — | ||||
| ab19898 | 0.856 | 0.639 | 0.632 | 1.000 | ab19898 | 0.831 | 0.104 | 0.009 | 1.000 |
| 0.002 |
0.047 |
0.050 |
— |
0.003 |
0.775 |
0.981 |
— |
||
| % CD133+ Angular Single Cells | % CD133+ Smooth Single Cells | ||||||||
| AC133 |
W6B3C1 |
C24B9 |
ab19898 |
AC133 |
W6B3C1 |
C24B9 |
ab19898 |
||
| AC133 | 1.000 | AC133 | 1.000 | ||||||
| — | — | ||||||||
| W6B3C1 | 1.000 | W6B3C1 | 0.761 | 1.000 | |||||
| — | 0.011 | — | |||||||
| C24B9 | 0.824 | 1.000 | C24B9 | 0.599 | 0.904 | 1.000 | |||
| 0.003 | — | 0.067 | 0.000 | — | |||||
| ab19898 | 0.251 | 0.232 | 1.000 | ab19898 | 0.259 | 0.427 | 0.592 | 1.000 | |
| 0.485 | 0.520 | — | 0.471 | 0.218 | 0.071 | — | |||
Identification of CD133+ Single Cells in Glioblastomas
In the glioblastomas, variations in morphology, distribution, and intensity of dispersed single cells were observed. All antibody clones identified a range of dispersed single cells with sizes varying from small to larger and multinucleated types as well as cells with ramifications, protrusions, and spindle-shaped and smooth morphologies (Fig. 5, Table 3). Clone AC133 identified few single cells, and staining of identified cells was cytoplasmatic and/or membranous. Identified cells predominantly displayed a round, smooth phenotype, but angular cells with protrusions were also observed (Fig. 5A, arrowhead). With W6B3C1, the cell membrane typically stained stronger than the cytoplasm (Fig. 5B, arrows). The morphology of identified single cells was often round and smooth (Fig. 5B, arrows), but angular cells with protrusions and/or processes were also observed (Fig. 5B, arrowhead). Clone C24B9 identified round and smooth cells with a staining that was constricted to the cytoplasm (Fig. 5C, arrow). Clone ab19898 identified smaller single cells with a round morphology as well as large multinucleated cells with eccentric nuclei (Fig. 5D) and angular cells with protrusions and processes (Fig. 2H). The staining was always cytoplasmatic, and the larger cells were often intensely stained (Fig. 5D).
Figure 5.
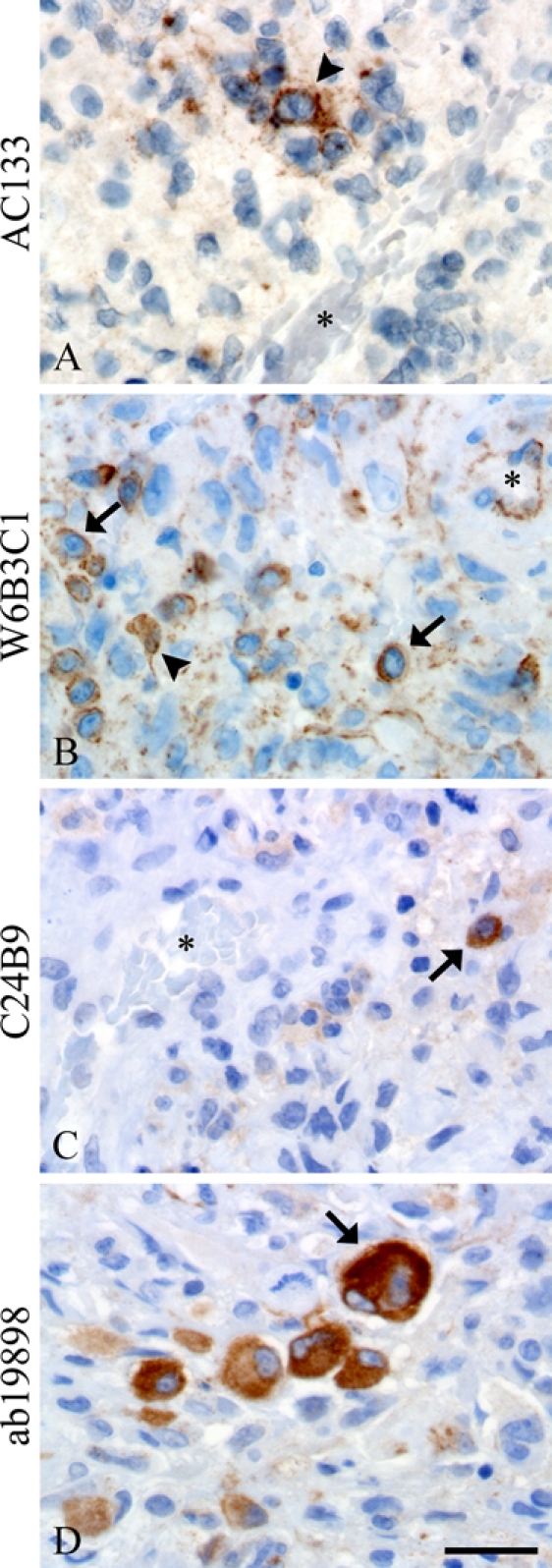
All clones identified CD133+ dispersed single cells in glioblastomas, but their morphology ranged from cells having ramifications and protrusions (arrowheads, A, B) to small (arrows, B, C) or bigger multinucleated cells (arrow, D). W6B3C1 had a tendency to stain tumor cell plasma membranes more than the cytoplasm (arrows, B), whereas AC133, C24B9, and ab19898 mostly stained the cytoplasm (arrowhead, A; arrows, C, D). Asterisks denote blood vessel lumen. Scale bar: 30 µm.
The identified volume fraction of CD133+ angular single cells was significantly higher when using ab19898 than when using AC133 and C24B9 (Fig. 3C; p<0.05). In contrast, the identified volume fraction of CD133+ smooth single cells was significantly higher when using W6B3C1 than when using AC133 and ab19898 (Fig. 3D; p<0.05). AC133 identified almost no single cells with mean volume fractions only ranging between 0% and 0.1%. Regarding angular single-cell volume fractions, a strong correlation was found between W6B3C1 and C24B9 (r = 0.824, p=0.003; Table 4). Likewise, a dominating pattern of simultaneous high smooth single-cell volume fractions was observed using these clones (Fig. 4D, Table 4; r = 0.904, p=0.000).
Only Clone W6B3C1 Identified CD133+ Blood Vessels in Glioblastoma
W6B3C1 was the only clone to stain tumor blood vessels in the investigated glioblastomas (Table 3). Staining of blood vessels was mostly located to the basal or luminal endothelial membrane, but some vessels also showed distinct staining of the outer vessel border near the tunica adventitia, resulting in a railway-like staining pattern (Fig. 6A). Clone W6B3C1 labeled 82.8% (mean) of all tumor blood vessels (Fig. 6B).
Figure 6.
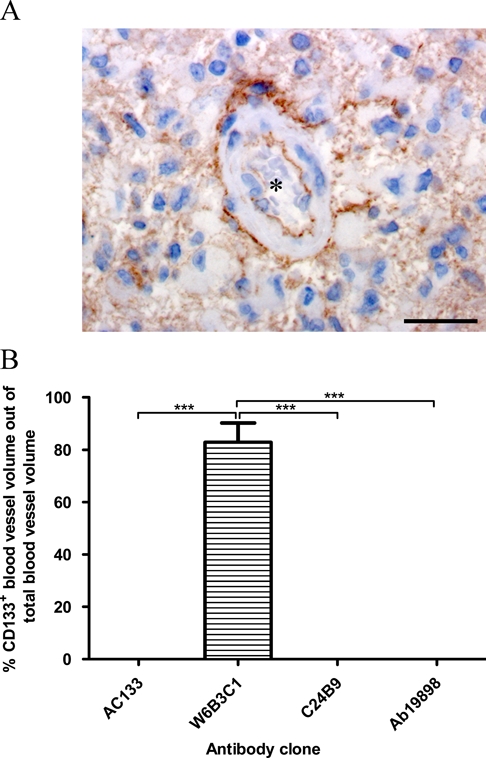
W6B3C1 displayed a railway-like staining pattern of some blood vessels (A), whereas no stained blood vessels were identified using AC133, ab19898, or C24B9. The percentage of CD133+ blood vessel volume out of total blood vessel volume obtained by each antibody clone was estimated (B). Scalebar: 30 µm, ***p<0.001.
CD133 Staining in Stem Cell Zones
Comparative staining of adjacent paraffin sections of stem cell regions in adult healthy brain tissue revealed considerable differences (Fig. 7, Table 3). All antibody clones did, however, identify a subpopulation of cells in the subventricular zone of the lateral ventricle with a juxtanuclear staining that seemed to be constricted to one pole of the cell. The majority of these cells were clustered in subependymal bands (Fig. 7A,C,E,G). Clones W6B3C1 (Fig. 7C) and ab19898 (Fig. 7G) revealed staining of ependymal cells, whereas no ependymal positivity was seen using AC133 (Fig. 7A) and C24B9 (Fig. 7E). Clones C24B9 and ab19898 displayed weak to moderate cytoplasmatic staining of cells in the hippocampal subgranular zone (Fig. 7F,H), whereas AC133 and W6B3C1 did not (Fig. 7B,D). Moreover, all clones, except AC133, showed diffuse staining of the entire hippocampal subgranular zone. Only W6B3C1 identified blood vessels (arrows and inserts, Fig. 7D).
Figure 7.
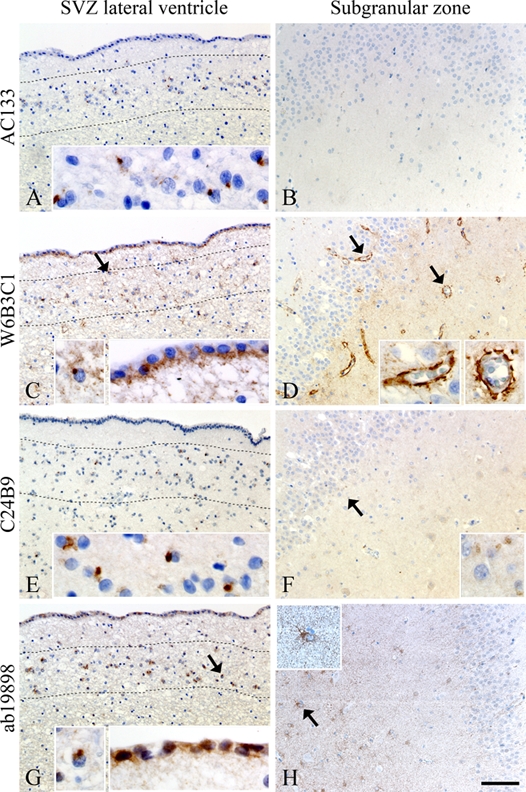
Comparative staining on sections of stem cell regions in healthy adult brain tissue using antibody clones AC133 (A, B), W6B3C1 (C, D), C24B9 (E, F), and ab19898 (G, H). A subependymal cell population with juxtanuclear staining was identified in the subventricular zone (SVZ) of the lateral ventricle by all antibody clones. The juxtanuclear staining was constricted to one pole of the cells, and the majority of positive cells were clustered in subependymal bands delineated by dotted lines (arrows and inserts, A, C, E, G). W6B3C1 and ab19898 also stained ependymal cells in SVZ (C, G), whereas no ependymal positivity was seen using AC133 and C24B9 (A, E). C24B9 and ab19898 stained the cytoplasm of few cells in the hippocampal subgranular zone (arrows and inserts, F, H), whereas AC133 was blank negative (B). Clone W6B3C1 was the only antibody to identify brain blood vessels (arrows and inserts, D). Scale bar: 100 µm.
CD133 Staining of Cell Cultures
Comparative staining of three different cell lines also revealed differences (Table 3). Intense CD133 expression was seen in SJ-1 spheroids with W6B3C1 and C24B9 (Fig. 8D,G), whereas sparse staining was seen with AC133 and ab19898 (Fig. 8A,J). Very little staining was observed in spheroids derived from the commercial glioblastoma cell line U87 using clones AC133, W6B3C1, and C24B9 (Fig. 8B,E,H), but clone ab19898 intensely stained numerous cells (Fig. 8K). The staining of SJ-1 and U87 spheroids was mostly cytoplasmatic, but membranous staining was also seen. All clones showed some immunoreactivity in the retinoblastoma cell line Y79. The staining generally had a dotted membranous and/or cytoplasmatic localization, but dispersed juxtanuclear staining was also observed (Fig. 8C,F,I,L). Staining was more widespread and distinct with clones W6B3C1 and C24B9 (Fig. 8F,I) than with clones AC133 and ab19898 (Fig. 8C,L).
Figure 8.
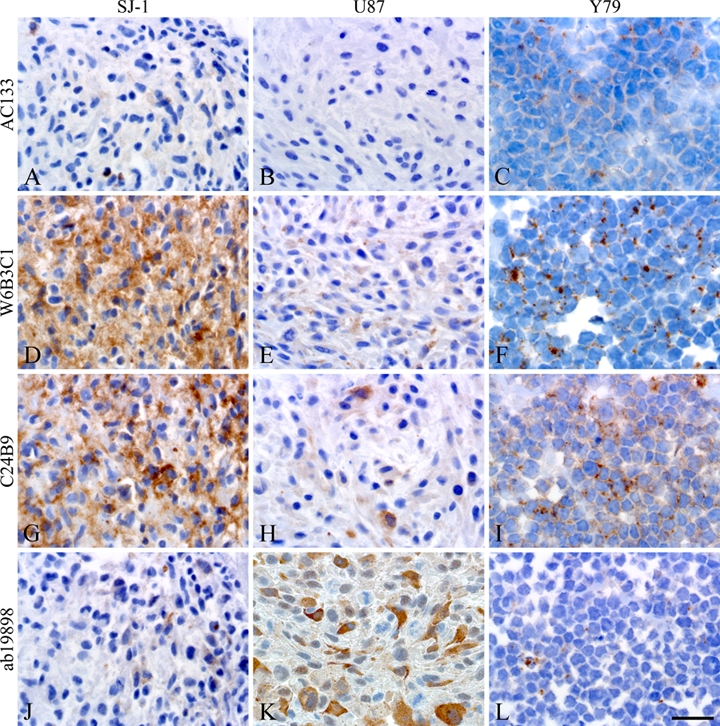
Comparative staining on paraffin sections of glioblastoma short-term culture SJ-1 spheroids, glioblastoma cell line U87 spheroids, and retinoblastoma cell line Y79 clusters. W6B3C1 and C24B9 identified intense expression of CD133 in SJ-1 (D, G), whereas AC133 and ab19898 did not (A, J). Little immunoreactivity was seen in U87 using AC133, W6B3C1, and C24B9 (B, E, H), but ab19898 displayed intense cytoplasmatic staining of numerous cells (K). All clones stained cells from the Y79 cell line, and the staining was mostly dotted and membranous and/or cytoplasmatic with the appearance of dispersed juxtanuclear staining as well (C, F, I, L). Images of SJ-1 and U87 were from adjacent sections of the same spheroid. Scale bar: 30 µm.
CD133 Staining in Kidney, Pancreas, and Placenta Tissues
Staining of tissues reported to be CD133+ in the literature was also inconsistent among the clones (Fig. 9, Table 3). In kidney tissue, only three out of four clones showed staining of parietal layer cells in the Bowman’s capsule (Fig. 9A,D,G), and kidney duct cells were stained in varying degrees. Duct cell staining with AC133 (arrow, Fig. 9A) and W6B3C1 (not shown) was apical and membranous, whereas staining with C24B9 and ab19898 was mostly cytoplasmatic (Fig. 9G,J). In pancreas tissue, the four antibodies showed a varying degree of apical/endoluminal staining in the majority of acini and ducts, but the overall signature remained consistent (Fig. 9B,E,H,K). Still, some diffuse staining of the islets of Langerhans was only observed with C24B9 (Fig. 9, arrow in H), and cytoplasmatic staining of pancreatic duct cells was mainly observed with ab19898 (Fig. 9, arrow in K). In placental tissue, only two out of four clones stained trophoblasts/syncytiotrophoblasts in chorionic villi (Fig. 9C,F,I,L).
Figure 9.
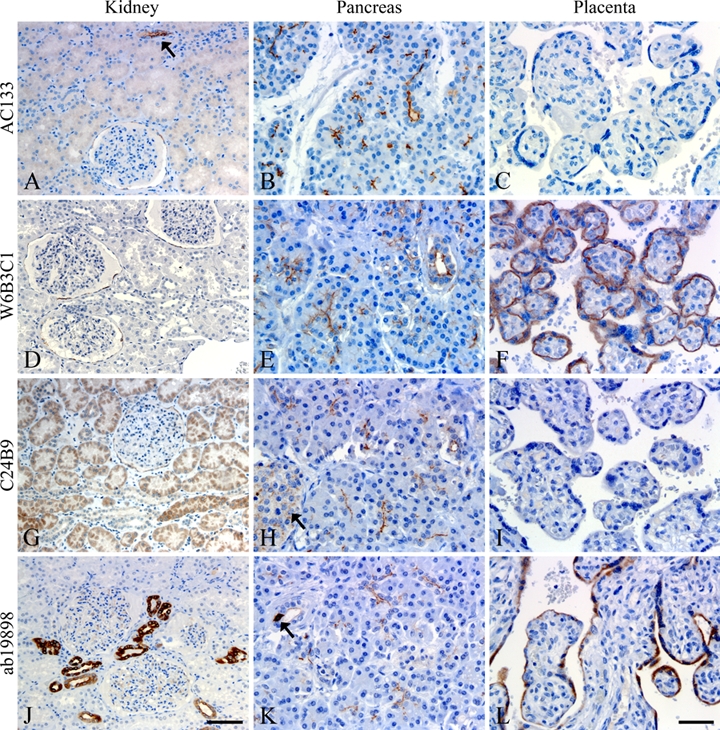
Comparative staining of kidney (A, D, G, J), pancreas (B, E, H, K), and placenta (C, F, I, L) tissues. A varying degree of CD133 positivity was observed in kidney glomeruli, tubuli, and collecting ducts between the clones (A, D, G, J). Arrow in A shows a stained collecting duct. Positive reaction in pancreas was mainly constricted to the apical/endoluminal surface of glandular epithelial and ductal cells (B, E, H, K), but some diffuse staining of islets of Langerhans (arrow, H) and cytoplasmatic staining of pancreatic ductal cells could be observed (arrow, K). Placental tissue was negative using AC133 and C24B9 (C, I) and positive in trophoblasts/syncytiotrophoblasts of chorionic villi using W6B3C1 and ab19898 (F, L). Scale bars: 100 µm (A, D, G, J); 50 µm (B, C, E, F, H, I, K, L).
Discussion
CD133 Staining in Glioblastomas Varies among the Clones
The present study shows that the use of different CD133 antibody clones for immunohistochemical identification of CD133+ cells in glioblastomas caused different results (Table 3). To our surprise, the identified cells rarely seemed to be identical (Fig. 2), and the quantitative analysis revealed a clear difference between the clones regarding fractions of total CD133+, CD133+ niches, and single cells (Figs. 3 and 4).
We obtained most CD133 expression using clone W6B3C1 (Fig. 3A), further highlighted by this being the only clone to identify tumor blood vessels (Fig. 6A). In contrast to previous quantitative CD133 studies, we never found that CD133 volume fractions amounted between 30% and 60% of the brain tumor sections (Son et al. 2009; Zeppernick et al. 2008; Zhang M et al. 2008; Singh et al. 2004). Our results are more in agreement with quantitative results reported by flow cytometric studies (Clément et al. 2009; Joo et al. 2008; Shu et al. 2008) and a recent study using clone AC133 on snap-frozen brain tumor sections (Thon et al. 2010), thus supporting the hypothesis that tumor stem cells are a rare population of cells in solid tumors.
Regarding total CD133+, a correlation existed between AC133 and ab19898 as well as between W6B3C1 and C24B9 (Fig. 4A, Table 4). However, qualitative data strongly suggested that cells identified with these clones were not identical (Figs. 2 and 5). Mean CD133+ niche volume fractions were not significantly different (Fig. 3B), but regarding the clones’ individual niche identification in each tumor, a correlation only existed between AC133 and ab19898 (Fig. 4B, Table 4). As above, a qualitative comparison of this correlation strongly indicated that cells identified with AC133 and ab19898 were not identical (Fig. 2).
Evidence suggests that neural stem cells and tumor stem cells lie within a protective niche in close proximity to the vasculature (Calabrese et al. 2007; Scheres 2007). We have previously shown that CD133 can be used to identify perivascular niches in glioblastomas (Christensen et al. 2008), and to our knowledge, we are the only group who has reported quantitative data of CD133+ niches in glioblastomas. When investigating dispersed single cells, it quickly became apparent that cell populations differed so much in form and size that cells should be divided into angular and smooth single cells to minimize sampling bias. Clone ab19898 identified most angular single cells, whereas W6B3C1 identified most smooth single cells (Fig. 3C,D). Angular and smooth single-cell volume fractions obtained using W6B3C1 and C24B9 were strongly correlated (Table 4), and qualitative data in glioblastomas and cell lines suggested the same (Fig. 8). Thus, these two clones may identify some of the same tumor single cells, but regarding tumor blood vessels (Fig. 6), stem cell zones in healthy brain tissue (Fig. 7), and control tissues (Fig. 9), differences were still evident.
Thus, it is very likely that the inconsistent CD133 immunolabeling obtained in CD133 studies is a result of using different antibody clones binding to different poorly characterized epitopes. Other contributors could be protocol differences with use of different detection systems such as the ultra-sensitive detection system CSA II (Christensen et al. 2008), overnight incubation times (Zhang M et al. 2008; Singh et al. 2004; Singh et al. 2003), paraffin (Pallini et al. 2008; Christensen et al. 2008; Zhang M et al. 2008; Singh et al. 2004; Singh et al. 2003) or cryostat sections (Zeppernick et al. 2008; Thon et al. 2010), semi-quantitative scoring systems (Zeppernick et al. 2008; Pallini et al. 2008; Zhang M et al. 2008), and inter- and intraobserver variability.
The Use of CD133 as a Marker for Tumor Stem Cells Is Controversial
CD133 is a glycoprotein (Corbeil et al. 2000; Miraglia et al. 1997), and tumor stem cells may change the glycosylation status of CD133 when they differentiate (Mizrak et al. 2008; Florek et al. 2005; Corbeil et al. 2000). Using antibodies that only identify the glycosylated epitopes may therefore cause identification of only a subset of CD133+ cells. According to the manufacturers, two of the antibodies used in the present study (AC133 and W6B3C1) target a glycosylated extracellular epitope of CD133, and they may therefore not identify all CD133+ cells. By using the CD133 antibody clone C24B9, an antibody targeted against a non-glycosylated epitope, one could theoretically be able to bypass problems concerning glycosylation and overcome problems with false-positive identification of irrelevant glycosylated epitopes (Bidlingmaier et al. 2008). If CD133 glycosylation was the only variable, then C24B9 staining would also be present in areas stained by the other clones. However, this is not the case, as seen in Figure 2, suggesting that the picture may be more complex than first assumed.
A recent publication has shown that glioblastoma cells negative for AC133 are positive for C24B9 and that the protein identified by C24B9 is truncated (16 kDa; Osmond et al. 2010). This seems to indicate that, in addition to problems regarding glycosylation status, there are problems with antibodies not identifying all variants of CD133. Interestingly, at least 28 alternatively spliced CD133 isoforms have been shown to exist (Fargeas et al. 2007; Jaszai et al. 2007; Fargeas et al. 2004), primarily showing C-terminal domain variations (Fargeas et al. 2007; Fargeas et al. 2004), which could be problematic for antibodies targeted to this area. According to the manufacturer, ab19898 is targeted to this area, and it seems to identify a slightly truncated protein (Table 1), which, according to the present study, mostly is present in cells with an angular morphology (Figs. 2H and 3C). As different CD133 antibodies, most likely, do not cover all splice variants, this could be why inconsistent immunolabeling was obtained in the present study and why different protein sizes were identified in various Western blots (Table 1).
Previous studies have shown that the AC133 epitope is downregulated upon tumor cell differentiation, which the authors explained to be due to loss of glycosylation (Mizrak et al. 2008; Florek et al. 2005; Corbeil et al. 2000). To add to the complexity, a recent colon cancer study has shown that the binding of AC133 is not dependent on glycosylated epitopes (Kemper et al. 2010). Instead, they suggest that the observed downregulation is due to inaccessibility of the AC133 epitope, which may depend on changing of tertiary structure or binding of another protein (Kemper et al. 2010).
The controversy is further substantiated by several studies reporting the existence of CD133– tumor stem cells derived from gliomas or glioma cell lines (Son et al. 2009; Joo et al. 2008; Gunther et al. 2008; Wang et al. 2008; Beier et al. 2007). An interesting quandary is whether these CD133– cells really are CD133– or if CD133 status is concluded on the basis of using CD133 antibodies that are not detecting CD133.
CD133 and Clinical Outcome
It is hard to ignore CD133 as a marker of immature stem-like tumor cells with tumor-initiating capabilities, but its detection is problematic. No prognostic significance has been reported with other CD133 antibodies than AC133 (Zeppernick et al. 2008; Thon et al. 2010; Pallini et al. 2008) and 293C3 (Beier et al. 2008), suggesting that cells expressing these antigens are unique and different from cells expressing other CD133 antigens. Thus, it is most likely the distribution of AC133 and 293C3 antigen and not the distribution of CD133 protein itself that is prognostic. AC133 has been used to show a negative prognostic significance of CD133 in both frozen (Zeppernick et al. 2008; Thon et al. 2010) and paraffin-embedded brain tumor sections (Pallini et al. 2008), and 293C3 has been used to correlate the presence of CD133+ cells in high-grade brain tumors with a poor clinical outcome (Beier et al. 2008). The latter used flow cytometric isolation of the CD133+ cells, which subsequently formed neurospheres, differentiated into all neural lineages, and were tumorigenic in nude mice (Beier et al. 2008)—all the hallmarks of true tumor stem cells. An important question is whether CD133+ cells identified by immunohistochemistry and flow cytometry are identical and whether the use of different CD133 antibody clones and protocols interfere with the results.
Neural Stem Cells, Cell Lines, and Control Tissues
The CD133 antibody clones all identified a subpopulation of cells in the subventricular zone of the lateral ventricle with a juxtanuclear staining that seemed to be constricted to one pole of the cells. The majority of these cells were clustered in subependymal bands (Fig. 7) as previously reported (Sakariassen et al. 2007). This is in accordance with the general idea of neural stem cells residing in discrete stem cell niches in the adult subventricular zone (Riquelme et al. 2008; Zhu et al. 2005). Juxtanuclear accumulation of CD133 has previously been reported (Sakariassen et al. 2007), but why this is seen has yet to be elucidated. However, CD133 has previously been proposed to have a role in membrane protrusions and to be associated with cholesterol-based membrane micro domains (Corbeil et al. 2001). It could be speculated that the CD133 juxtanuclear accumulation could be related to plasma membrane organization. Another possibility could be that the stained dots represent CD133 protein undergoing posttranslational modification in the Golgi apparatus because shaping of the stained dots has a close resemblance to shaping of the Golgi apparatus.
When investigating CD133 expression in SJ-1, U87, and Y79 cells, inconsistent results were apparent (Fig. 8, Table 3). In spheroids from SJ-1, a glioblastoma short-term culture with tumor stem cell properties established in the Department of Pathology, Odense University Hospital, intense CD133 expression was only obtained when using W6B3C1 and C24B9 (Fig. 8D,G). U87, which is a commonly used commercial cell line for glioblastoma research, has previously been shown not to express CD133 using AC133, 293C3 (Platet et al. 2007), or W6B3C1 (Christensen et al. 2010) and to express CD133 using 293C3 (Brehar et al. 2009). We also obtained low CD133 levels in U87, except when using ab19898, as this antibody displayed a distinct and ubiquitous cytoplasmatic staining of cells (Fig. 8K). CD133 expression in Y79 has previously been confirmed by PCR and Western blot analysis (Thon et al. 2010; Seigel et al. 2007), and in Y79 sections, we obtained most positive cells with W6B3C1 and C24B9, suggesting that their antigens are more widely distributed in this cell line (Fig. 8F,I). It is somewhat problematic that anything from no CD133 expression to high CD133 expression can be obtained depending on which antibody is being used. These findings make direct comparison of studies using different CD133 antibody clones for both isolation and identification of CD133 problematic.
Our results on kidney, pancreas, and placenta tissue (Fig. 9, Table 3) supported many previous observations (Immervoll et al. 2008; Christensen et al. 2008; Florek et al. 2005). However, qualitative comparison of the four antibodies revealed a dissimilar overall CD133 distribution. Furthermore, some of the tissues found positive for the AC133 antigen in the present study have previously been reported as negative in other immunohistochemical studies using the same clone (AC133) (Thon et al. 2010; Miraglia et al. 1997). This discrepancy could be due to our optimized protocol for formalin-fixed, paraffin-embedded tissues using detection systems with enhanced sensitivity, specific HIER protocols, and high antibody concentrations.
In conclusion, we obtained qualitative and quantitative discordant immunohistochemical CD133 expression in all investigated tissues using the four different CD133 antibody clones (AC133, W6B3C1, C24B9, and ab19898). We conclude that the use of different CD133 antibody clones for immunohistochemical identification of CD133 in paraffin sections will most likely cause different results. It is a problem in this field of research that anything from no CD133 expression to high CD133 expression can be demonstrated depending on which antibody is being used. The present study emphasizes that choice of CD133 antibody clone is critical and that conclusions based on the use of CD133 primary antibodies should be drawn with caution.
Acknowledgments
We are very grateful for the technical support and advice from laboratory technician Ole Nielsen regarding optimization of immunohistochemical protocols. The excellent immunohistochemical work carried out by medical laboratory technicians Helle Wohlleben and Tanja Dreehsen is thankfully acknowledged.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
The author(s) disclosed receipt of the following financial support for the research and/or authorship of this article: This work was supported by the Danish Cancer Society, Danish Medical Research Council, Hørslev Foundation, Danish Cancer Research Foundation, Kathrine and Vigo Skovgaard’s Foundation, Johs. Clemmesen’s Cancer Foundation, Eva and Henry Frænkel’s Memorial Foundation, Hede Nielsen Foundation, Dagmar Marshall Foundation, Einar Willumsen’s Memorial Foundation, Harboe Foundation, and Merchant Sven Hansen and wife Ina Hansen’s Foundation.
References
- Al-Hajj M, Wicha MS, Ito-Hernandez A, Morrison SJ, Clarke MF. 2003. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 100:3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. 2007. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 67:4010–4015 [DOI] [PubMed] [Google Scholar]
- Beier D, Wischhusen J, Dietmaier W, Hau P, Proescholdt M, Brawanski A, Bogdahn U, Beier CP. 2008. CD133 expression and cancer stem cells predict prognosis in high-grade oligodendroglial tumors. Brain Pathol. 18:370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidlingmaier S, Zhu X, Liu B. 2008. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J Mol Med. 86:1025–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehar FM, Bleotu C, Stefan LM, Buzgariu W, Chivu M, Utoiu E, Matei L, Ciurea AV, Tascu A. 2009. Isolation and partial characterization of a new human glioblastoma cell line. Chirurgia. 104:453–461 [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, et al. 2007. A perivascular niche for brain tumor stem cells. Cancer Cell. 11:69–82 [DOI] [PubMed] [Google Scholar]
- Christensen K, Aaberg-Jessen C, Andersen C, Goplen D, Bjerkvig R, Kristensen BW. 2010. Immunohistochemical expression of stem cell, endothelial cell, and chemosensitivity markers in primary glioma spheroids cultured in serum-containing and serum-free medium. Neurosurgery. 66:933–947 [DOI] [PubMed] [Google Scholar]
- Christensen K, Schrøder HD, Kristensen BW. 2008. CD133 identifies perivascular niches in grade II–IV astrocytomas. J Neurooncol. 90:157–170 [DOI] [PubMed] [Google Scholar]
- Clément V, Dutoit V, Marino D, Dietrich PY, Radovanovic I. 2009. Limits of CD133 as a marker of glioma self-renewing cells. Int J Cancer. 125:244–248 [DOI] [PubMed] [Google Scholar]
- Clément V, Marino D, Cudalbu C, Hamou MF, de Mlynarik VTN, Dietrich PY, Gruetter R, Hegi ME, Radovanovic I. 2010. Marker-independent identification of glioma-initiating cells. Nat Methods. 7:224–228 [DOI] [PubMed] [Google Scholar]
- Corbeil D, Roper K, Fargeas CA, Joester A, Huttner WB. 2001. Prominin: a story of cholesterol, plasma membrane protrusions and human pathology. Traffic. 2:82–91 [DOI] [PubMed] [Google Scholar]
- Corbeil D, Roper K, Hellwig A, Tavian M, Miraglia S, Watt SM, Simmons PJ, Peault B, Buck DW, Huttner WB. 2000. The human AC133 hematopoietic stem cell antigen is also expressed in epithelial cells and targeted to plasma membrane protrusions. J Biol Chem. 275:5512–5520 [DOI] [PubMed] [Google Scholar]
- Fargeas CA, Huttner WB, Corbeil D. 2007. Nomenclature of prominin-1 (CD133) splice variants: an update. Tissue Antigens. 69:602–606 [DOI] [PubMed] [Google Scholar]
- Fargeas CA, Joester A, Missol-Kolka E, Hellwig A, Huttner WB, Corbeil D. 2004. Identification of novel prominin-1/CD133 splice variants with alternative C-termini and their expression in epididymis and testis. J Cell Sci. 117(Pt 18):4301–4311 [DOI] [PubMed] [Google Scholar]
- Florek M, Haase M, Marzesco AM, Freund D, Ehninger G, Huttner WB, Corbeil D. 2005. Prominin-1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res. 319:15–26 [DOI] [PubMed] [Google Scholar]
- Gilbertson RJ, Rich JN. 2007. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 7:733–736 [DOI] [PubMed] [Google Scholar]
- Gundersen HJ. 1986. Stereology of arbitrary particles: a review of unbiased number and size estimators and the presentation of some new ones, in memory of William R. Thompson. J Microsc. 143(Pt 1):3–45 [PubMed] [Google Scholar]
- Gundersen HJ. 1992. Stereology: the fast lane between neuroanatomy and brain function—or still only a tightrope? Acta Neurol Scand Suppl. 137:8–13 [DOI] [PubMed] [Google Scholar]
- Gundersen HJ, Bendtsen TF, Korbo L, Marcussen N, Moller A, Nielsen K, Nyengaard JR, Pakkenberg B, Sorensen FB, Vesterby A. 1988. Some new, simple and efficient stereological methods and their use in pathological research and diagnosis, APMIS. 96:379–394 [DOI] [PubMed] [Google Scholar]
- Gundersen HJ, Jensen EB. 1987. The efficiency of systematic sampling in stereology and its prediction. J Microsc. 147(Pt 3):229–263 [DOI] [PubMed] [Google Scholar]
- Gunther HS, Schmidt NO, Phillips HS, Kemming D, Kharbanda S, Soriano R, Modrusan Z, Meissner H, Westphal M, Lamszus K. 2008. Glioblastoma-derived stem cell–enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 27:2897–2909 [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. 2007. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 1:313–323 [DOI] [PubMed] [Google Scholar]
- Immervoll H, Hoem D, Sakariassen PO, Steffensen OJ, Molven A. 2008. Expression of the stem cell marker CD133 in pancreas and pancreatic ductal adenocarcinomas. BMC Cancer. 8:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaszai J, Fargeas CA, Florek M, Huttner WB, Corbeil D. 2007. Focus on molecules: prominin-1 (CD133). Exp Eye Res. 85:585–586 [DOI] [PubMed] [Google Scholar]
- Joo KM, Kim SY, Jin X, Song SY, Kong DS, Lee JI, Jeon JW, Kim MH, Kang BG, Jung Y, et al. 2008. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 88:808–815 [DOI] [PubMed] [Google Scholar]
- Kemper K, Sprick MR, de Scopelliti BMA, Vermeulen L, Hoek M, Zeilstra J, Pals ST, Mehmet H, Stassi G, Medema JP. 2010. The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 70:719–729 [DOI] [PubMed] [Google Scholar]
- Kolenda J, Jensen SS, Aaberg-Jessen C, Christensen K, Andersen C, Brunner N, Kristensen BW. 2010. Effects of hypoxia on expression of a panel of stem cell and chemoresistance markers in glioblastoma-derived spheroids. J Neurooncol. September 11 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. 2006. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403 [DOI] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. 2007. Identification of pancreatic cancer stem cells. Cancer Res. 67:1030–1037 [DOI] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. 2007. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114:97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitland NJ, Collins AT. 2008. Prostate cancer stem cells: a new target for therapy. J Clin Oncol. 26:2862–2870 [DOI] [PubMed] [Google Scholar]
- Malhotra V, Malik R, Gondal R, Beohar PC, Parkash B. 1986. Evaluation of histological appearance of tissues removed by cavitron ultrasonic surgical aspirator (CUSA). Acta Neurochir (Wien). 81(3–4):132–134 [DOI] [PubMed] [Google Scholar]
- Miraglia S, Godfrey W, Yin AH, Atkins K, Warnke R, Holden JT, Bray RA, Waller EK, Buck DW. 1997. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 90:5013–5021 [PubMed] [Google Scholar]
- Mizrak D, Brittan M, Alison MR. 2008. CD133: molecule of the moment. J Pathol. 214:3–9 [DOI] [PubMed] [Google Scholar]
- O’Brien CA, Pollett A, Gallinger S, Dick JE. 2007. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 445:106–110 [DOI] [PubMed] [Google Scholar]
- Osmond TL, Broadley KW, McConnell MJ. 2010. Glioblastoma cells negative for the anti-CD133 antibody AC133 express a truncated variant of the CD133 protein. Int J Mol Med. 25:883–888 [DOI] [PubMed] [Google Scholar]
- Pallini R, Ricci-Vitiani L, Banna GL, Signore M, Lombardi D, Todaro M, Stassi G, Martini M, Maira G, Larocca LM, et al. 2008. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 14:8205–8212 [DOI] [PubMed] [Google Scholar]
- Platet N, Liu SY, Atifi ME, Oliver L, Vallette FM, Berger F, Wion D. 2007. Influence of oxygen tension on CD133 phenotype in human glioma cell cultures. Cancer Lett. 258:286–290 [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. 2007. Identification and expansion of human colon-cancer-initiating cells. Nature. 445:111–115 [DOI] [PubMed] [Google Scholar]
- Riquelme PA, Drapeau E, Doetsch F. 2008. Brain micro-ecologies: neural stem cell niches in the adult mammalian brain. Philos Trans R Soc Lond B Biol Sci. 363:123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakariassen PO, Immervoll H, Chekenya M. 2007. Cancer stem cells as mediators of treatment resistance in brain tumors: status and controversies. Neoplasia. 9:882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. 2007. Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol. 8:345–354 [DOI] [PubMed] [Google Scholar]
- Seigel GM, Campbell LM, Narayan M, Gonzalez-Fernandez F. 2005. Cancer stem cell characteristics in retinoblastoma. Mol Vis. 11:729–737 [PubMed] [Google Scholar]
- Seigel GM, Hackam AS, Ganguly A, Mandell LM, Gonzalez-Fernandez F. 2007. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol Vis. 13:823–832 [PMC free article] [PubMed] [Google Scholar]
- Shi SR, Liu C, Pootrakul L, Tang L, Young A, Chen R, Cote RJ, Taylor CR. 2008. Evaluation of the value of frozen tissue section used as gold standard for immunohistochemistry. Am J Clin Pathol. 129:358–366 [DOI] [PubMed] [Google Scholar]
- Shu Q, Wong KK, Su JM, Adesina AM, Yu LT, Tsang YT, Antalffy BC, Baxter P, Perlaky L, Yang J, et al. 2008. Direct orthotopic transplantation of fresh surgical specimen preserves CD133+ tumor cells in clinically relevant mouse models of medulloblastoma and glioma. Stem Cells. 26:1414–1424 [DOI] [PubMed] [Google Scholar]
- Silverman JF, Jones FD, Unverferth M, Berns L. 1989. Cytopathology of neoplasms of the central nervous system in specimens obtained by the Cavitron Ultrasonic Surgical Aspirator. Acta Cytol. 33:576–582 [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. 2003. Identification of a cancer stem cell in human brain tumors. Cancer Res. 63:5821–5828 [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. 2004. Identification of human brain tumour initiating cells. Nature. 432:396–401 [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam DH, Lee J, Fine HA. 2009. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 4:440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulman EP, Goodman LD, Le TT, Pelloski CE, Bhat K, Gumin J, Lang FF, Colman H, Aldape KD. 2008. A novel marker of glioma stem cells that is prognostic for treatment response and patient outcome. Presented at the 99th Annual Meeting of the American Association for Cancer Research, April 12–16, San Diego, CA Abstract 3777 [Google Scholar]
- Thon N, Damianoff K, Hegermann J, Grau S, Krebs B, Schnell O, Tonn JC, Goldbrunner R. 2010. Presence of pluripotent CD133(+) cells correlates with malignancy of gliomas. Mol Cell Neurosci. 43:51–59 [DOI] [PubMed] [Google Scholar]
- Wang J, Sakariassen PO, Tsinkalovsky O, Immervoll H, Boe SO, Svendsen A, Prestegarden L, Rosland G, Thorsen F, Stuhr L, et al. 2008. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer. 122:761–768 [DOI] [PubMed] [Google Scholar]
- Yin AH, Miraglia S, Zanjani ED, Meida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. 1997. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 90:5002–5012 [PubMed] [Google Scholar]
- Zeppernick F, Ahmadi R, Campos B, Dictus C, Helmke BM, Becker N, Lichter P, Unterberg A, Radlwimmer B, Herold-Mende CC. 2008. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res. 14:123–129 [DOI] [PubMed] [Google Scholar]
- Zhang M, Song T, Yang L, Chen R, Wu L, Yang Z, Fang J. 2008. Nestin and CD133: valuable stem cell-specific markers for determining clinical outcome of glioma patients. J Exp Clin Cancer Res. 27:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, Yan PS, Huang TH, Nephew KP. 2008. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 68:4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF. 2005. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 8:119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]


