Abstract
Immunofluorescence detection of proteins in growth plate cartilage is often unsuccessful because of innate autofluorescence, fixative-induced fluorescence, and dense cartilage matrix, which can inhibit antibody penetration. To overcome these limitations, the authors have tested various chemical pretreatments, including the autofluorescence quencher sodium borohydride, the antigen retrieval method of boiling sodium citrate, sugar-degrading enzymes (hyaluronidase, heparinase, and chondroitinase), and the proteolytic enzyme protease XXIV. Here the authors show that, in most cases, background fluorescence in cartilage is the primary obstacle to high-quality imaging. Blocking intrinsic fluorescence of the specimen in combination with specific pretreatments allows visualization using antibodies that previously did not generate a robust signal in the growth plate. Each antibody requires a specific combination of chemical pretreatments that must be empirically determined to achieve optimal staining levels. This manuscript contains online supplemental material at http://www.jhc.org. Please visit this article online to view these materials.
Keywords: immunofluorescence, antibody, cartilage, antigen retrieval, autofluorescence, tissue pretreatment
In situ immunodetection of endogenous proteins is a powerful tool in cell and developmental biology. In conjunction with genetic analyses, protein localization has provided important insights into biological mechanisms in many model systems. By contrast, immunological techniques have been used sparingly in cartilage research because these methods show low sensitivity and inconsistent results. When performed, protein localization is often detected using precipitating chromogenic substrates (Kvist et al. 2008) that do not provide quantitative data or the single-cell or subcellular resolution required to simultaneously determine the localization of multiple proteins. For these reasons, our understanding of the cell biological processes that underlie the development and maintenance of cartilage is predominately based on the analysis of in vitro systems.
The developing endochondral skeleton, which uses a cartilage template intermediate to generate mature mineralized bone, is an excellent system for studies of cartilage cell biology because the complete range of cell types found during differentiation is present simultaneously (reviewed by Kronenberg 2003). The growth plate cartilage of long bones is composed of a continuum of maturing chondrocytes with stem cell–like resting chondrocytes (RZ) residing at each end of the bone followed by proliferative chondrocytes that are flattened and stacked in columns, which mature into prehypertrophic and ultimately hypertrophic chondrocytes. Growth plate chondrocytes are embedded in dense, region-specific extracellular matrix, including collagen type II and IX (immature chondrocytes) or type X (hypertrophic chondrocytes) (von der Mark et al. 1976; Irwin et al. 1985; Schmid and Linsenmayer 1985a, 1985b; Nishimura et al. 1990). However, surrounding individual chondrocytes is a pericellular matrix containing collagen type IV, fibronectin, and laminin (Kvist et al. 2008). The properties of these matrices are modified by associated proteoglycans (reviewed in Gentili and Cancedda 2009).
These specific properties of the extracellular matrix also contribute to artifacts in immunofluorescence studies by producing innate and fixation-induced autofluorescence and by inhibiting antibody penetration. Various methods have been described to improve antigen detection. In most cases, individual approaches are described in relation to a specific protein, leaving uncertainty as to whether these methods can be applied broadly to different types of proteins or different tissues. In addition, much of the effort to improve protein detection has focused on increasing the available immunoreactive epitopes using antigen retrieval methods. In cartilage, these methods often produce variable results, and only epitopes present at high concentrations are readily observed. More sensitive methods are required to detect lower abundance proteins or to obtain quantitative protein expression data in cartilage.
Here we present a systematic analysis of chemical pretreatments, individually and in combination, which decrease autofluorescence and remove interfering molecules from the extracellular matrix. The pretreatments tested included sodium borohydride (NaBH4) (Weber et al. 1978; Baschong et al. 2001; Langelier et al. 2000), boiling sodium citrate (Na-citrate) (Imam et al. 1995; Dreier, Gunther, et al. 2008), hyaluronidase (Dreier, Gunther, et al. 2008; Kluppel et al. 2005; Blumbach et al. 2008), heparinase II (Melrose et al. 2003), chondroitinase (Kluppel et al. 2005; Blanc et al. 2005), or protease XXIV (Rheinhardt and Finkbeiner 2001; Dreier, Opolka, et al. 2008). The results demonstrate that each of these methods can increase the sensitivity of antibody staining in the cartilage growth plate; however, each antibody/antigen requires a unique combination of the aforementioned pretreatments to obtain optimal fluorescence signal.
Materials and Methods
Mouse Strains and Animal Care
Mouse (Swiss Webster; Jackson Laboratories, Bar Harbor, ME) husbandry and use were in accordance with National Institutes of Health (NIH) guidelines and approved by the Animal Care and Use Committee of Northwestern University.
Tissue Preparation, Embedding, and Sectioning
All tissue was harvested from newborn (P0) to postnatal day 3 (P3) mice. Hindlimbs and forelimbs were skinned and fixed in 4% paraformaldehyde (PFA; Sigma-Aldrich, St Louis, MO) overnight at 4C before preparing tissue for frozen sections or paraffin embedding. All steps were performed with gentle rocking. For frozen sections, half of the limbs harvested were incubated with 5 mg/ml sodium borohydride (NaBH4; Sigma-Aldrich) for 2 × 30 min at room temperature and washed several times with phosphate-buffered saline (PBS). All samples were then sequentially incubated at 4C in 30% sucrose/PBS for 3 hr, a 1:1 solution of optimal cutting temperature compound (OCT) and 30% sucrose for 1 hr, and OCT compound (Sakura Finetek, Torrance, CA) for 1 hr. The tissue was frozen in OCT compound using a mixture of dry ice and ethanol. Blocks were cut into 10- to 20-µm sections using a cryostat (Leica, Heidelberg, Germany) and collected on Superfrost Plus slides (VWR International, Radnor, PA).
For paraffin embedding, fixed limbs were dehydrated through an ethanol series into xylenes and then embedded in paraffin (Paraplast X-tra; Tyco-Healthcare, Mansfield, MA). Samples were cut in 8-µm sections and floated onto Superfrost Plus (VWR International) glass slides, air dried overnight, and then baked at 50C for 30 min. Slides were dewaxed in xylenes, rehydrated through an ethanol series, and washed in PBS. Half of the samples were incubated in 5 mg/ml NaBH4 for 2 × 30 min in slide mailers and washed several times in PBS before proceeding to immunodetection.
Antibody Staining and Pretreatments
Tissue sections were incubated in 4% PFA for 10 min at room temperature followed by washing in PBST (PBS + 0.1% Tween 20). Next, samples were treated with one or more optional pretreatments. All incubation steps occurred in a humidified chamber. For a detailed protocol and list of reagents, see SF1, ST1, ST2, and ST3. Pretreatment 1: Slides were placed in boiling 0.01 M sodium citrate (pH 6.4) for 10 min (without additional heating) and then washed in PBS. Pretreatment 2: Samples with or without antigen retrieval were first incubated in 0.25% hyaluronidase (H6254; Sigma-Aldrich) in PBS for 1 hr rocking gently at a shallow angle (100 µl/slide), then washed in PBST before incubating in a combination of 200 mU/ml of Chondroitinase ABC (C3667; Sigma-Aldrich) and 2.5 mU/ml Heparinase II (H6512; Sigma-Aldrich) in HEPES buffer plus 3 mM CaCl2 at 37C for 1 hr (~200 µl/slide). Pretreatment 3: Sections were treated with proteinase XXIV (P8038; Sigma-Aldrich) 25 µg/ml in PBS for 10 min at 37C. All pretreatments were then washed in Tris-buffered saline + 0.01% Tween 20 (TBST; pH 7.4) blocked with 10% heat-inactivated sheep serum (HISS) in TBST for at least 1 hr at room temperature prior to incubating with 100 µl of primary antibody in 2% HISS/TBST overnight at 4C (this and subsequent incubation steps were performed in a humidified chamber with gentle rocking). Following primary antibody incubation, sections were washed in TBST, incubated with secondary antibody in 2% HISS/TBST for ~2 to 4 hr at room temperature, and washed in TBST. Tissue was stained with 200 ng/ml 4′,6-diamidino-2-pheylindole (DAPI; Sigma-Aldrich) in PBS for 1 min, washed in TBST, and mounted in gelvatol or Prolong Gold anti-fade mounting medium (Invitrogen, Carlsbad, CA).
Results
Antigen Retrieval Modestly Improves Sensitivity
Antigen retrieval using boiling sodium citrate (Na-citrate) is a common method employed to increase sensitivity in immunofluorescence studies (Imam et al. 1995). However, when this method is applied to growth plate cartilage, without any additional pretreatments, the results are often variable. Some antibodies fail to localize proteins in situ even when using antigen retrieval methods, and antibodies that can be visualized often show low signal-to-noise ratio (signal:noise) or poor specificity for subcellular localization (Fig. 1). For example, pSmad, cyclinD1, and collagen IV (Col IV) do not show specific signal without pretreatment. After antigen retrieval, the nuclear proteins phospho-Smad1/5/8 (pSmad) and cyclinD1 (ST1; Baldin et al. 1993; Sewing et al. 1993) show detectable levels of immunofluorescence; however, the signal:noise ratio is very low, and specific localization to the nucleus is not consistently observed (Fig. 1). Similarly, antigen retrieval does not increase signal for Col IV and only marginally improves the signal for Col X (Fig. 1). These data suggest that antigen masking by fixation and other manipulations during tissue processing are not the primary causes of low signal:noise for immunofluorescence detection of proteins in cartilage.
Figure 1.
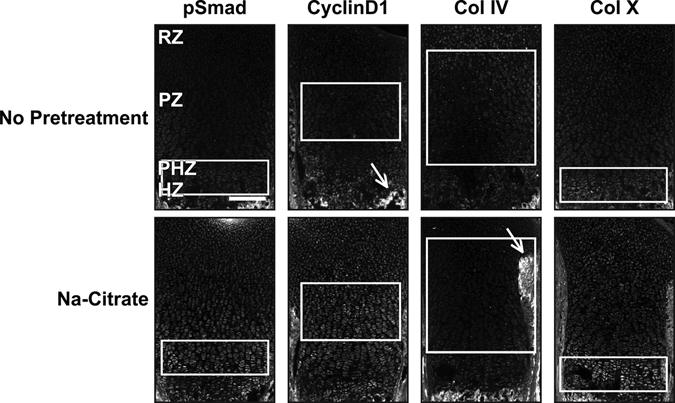
Antigen retrieval modestly enhances immunofluorescence signal. Sections from wild-type P0 mouse growth plates do not have appreciable immunofluorescence without pretreatment for the following antibodies: phospho-Smad1/5/8 (pSmad, 1:100, 1:200), cyclinD1 (1:200), collagen IV (Col IV, 1:200), and collagen X (Col X, 1:200). Sodium citrate (Na-citrate) treatment increases the signal for pSmad in the prehypertrophic zone (PHZ) and for cyclinD1 in the proliferative zone (PZ). By contrast, Col IV was not visualized in the cartilage matrix either with or without Na-citrate pretreatment (white arrows point to the perichondrium and the mineralized matrix). Col X has weak expression in the hypertrophic zone (HZ) without pretreatment and a stronger signal:noise ratio with Na-citrate pretreatment. White boxes indicate location where signal is present or expected. Scale bar equals 200 µm in all panels.
Sugar-Degrading Enzymes Alone Do Not Enhance Immunofluorescence Detection of Proteins
Because antigen retrieval alone does not provide a robust, general method for immunofluorescence in growth plate cartilage, we sought to identify additional treatments that would improve detection of endogenous proteins in fixed sections of cartilage. We initially chose to focus on proteoglycans of the extracellular matrix that have been proposed to reduce antibody penetration into tissue. We tested several published methods to remove the carbohydrate polymers that decorate many extracellular matrix–associated proteins. We first determined the efficacy of hyaluronidase, an enzyme that degrades the glycosaminoglycan hyaluronan found in the extracellular matrix (Quintarelli et al. 1978). Hyaluronidase is commonly used as a pretreatment of cartilage prior to immunodetection of proteins (Blumbach et al. 2008; Dreier, Opolka, et al. 2008; Kvist et al. 2008). In addition, we tested heparinase and chondroitinase, which degrade heparin and glycosaminoglycans, respectively (reviewed in Langelier et al. 2000; Gentili and Cancedda 2009). In the absence of antigen retrieval, these three enzymes individually did not substantially improve detection of these proteins (Fig. 2A,B).
Figure 2.
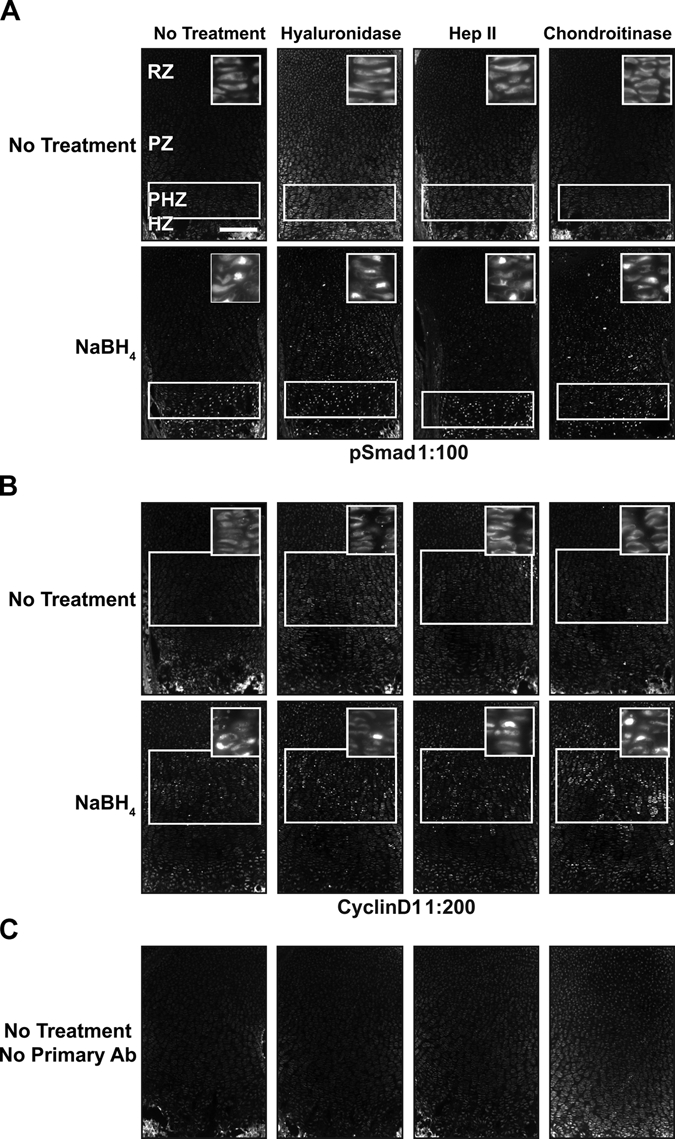
NaBH4 and hyaluronidase or chondroitinase increases sensitivity. (A) pSmad cannot be localized without pretreatment, but it is nuclear localized in the prehypertrophic zone (PHZ) and hypertrophic zone (HZ) when treated with NaBH4 alone. Adding hyaluronidase or chondroitinase alone does not produce a visible signal. NaBH4 treatment in combination with hyaluronidase or chondroitinase reveals nuclear localized pSmad in the resting zone (RZ) and proliferative zone (PZ). Heparinase II does not increase sensitivity over NaBH4 alone. (B) Cyclin D1 sensitivity is also increased by NaBH4 alone or in combination with hyaluronidase or chondroitinase. It is localized to the nuclei of RZ and PZ chondrocytes. (C) Only background fluorescence is visualized without primary antibody. White boxes indicate location where signal is present or expected. Scale bar: in A–C, 200 µm.
NaBH4 Significantly Increases the Sensitivity of Immunofluorescence Detection
While performing the above studies, we noted that background fluorescence is a general problem in the cartilage growth plate (Fig. 3A). This could be due to fixative-induced fluorescence or to innate autofluorescence found in the cartilage growth plate. Innate fluorescence in untreated sections of fixed cartilage is sufficiently intense to permit the analysis of growth plate architecture (Retting et al. 2009). To overcome the limitations imposed by high levels of background fluorescence, we tested the reducing agent NaBH4, which quenches both tissue autofluorescence and fixative-induced fluorescence (Weber et al. 1978; Baschong et al. 2001). This method proved highly effective as NaBH4 greatly reduced autofluorescence in fixed cartilage for both frozen and paraffin-embedded samples, and it blocked the often substantial increase in tissue fluorescence associated with pretreatments such as antigen retrieval or sugar-degrading enzymes (Figs. 3B, 4A). Decreasing autofluorescence revealed distinct nuclear localization of both pSmad and cyclinD1 in growth plate cartilage (Figs. 2A,B, 4B). These results demonstrate that autofluorescence is the primary cause of low signal:noise ratios and poor subcellular localization of antigens in samples of fixed cartilage. Similarly, NaBH4 pretreatment greatly reduces autofluorescence in tissues such as the heart, liver, lung, and kidney (Fig. 5). Although effective on many tissues, NaBH4 pretreatment does not substantially reduce autofluorescence in blood cells (Fig. 5). Together, these data show that blocking autofluorescence by NaBH4 treatment is an essential first step for most antibodies in multiple tissues.
Figure 3.
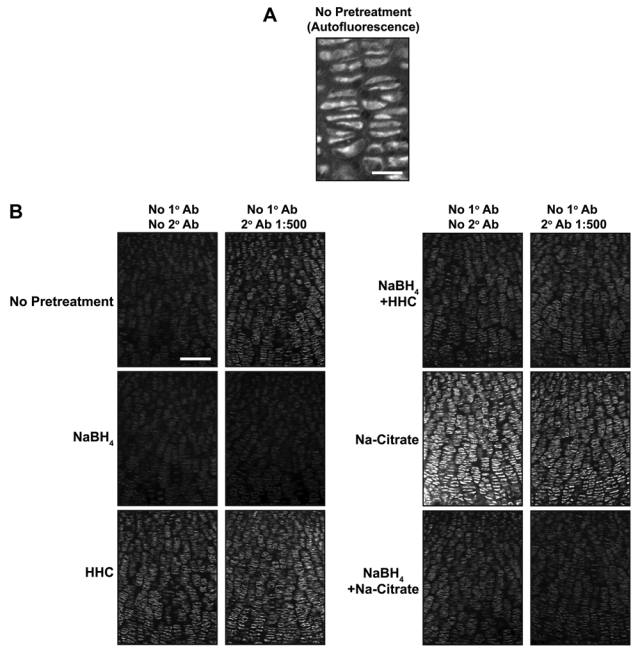
NaBH4 quenches autofluorescence in growth plate cartilage. (A) Innate immunofluorescence is readily detected in untreated sections of fixed cartilage. (B) Nonspecific fluorescence was detected in sections of P0 mouse growth plate subjected to various pretreatments (left) and then incubated with (right column) or without (left column) secondary antibody. NaBH4 pretreatment quenches innate tissue autofluorescence and substantially reduces the background fluorescence induced by tissue pretreatments. HHC, hyaluronidase + heparinase II + chondroitinase. Scale bars: in A, 25 µm; in B, 100 µm.
Figure 4.
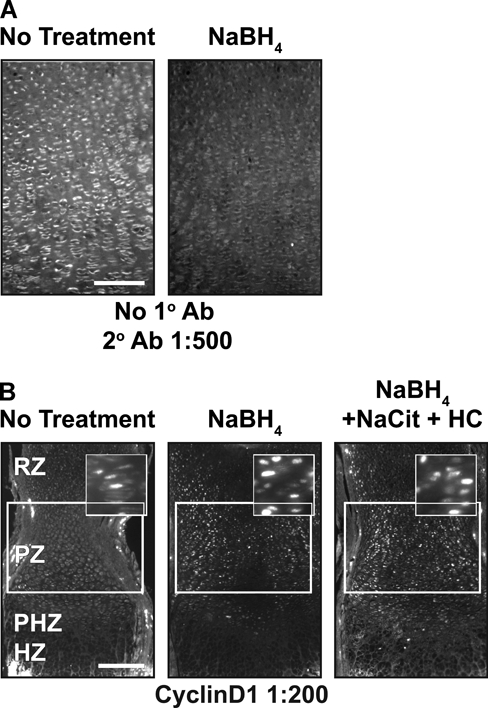
NaBH4 quenches autofluorescence and in combination with hyaluronidase or chondroitinase increases sensitivity in paraffin sections. (A) Autofluorescence is quenched when paraffin sections are treated with NaBH4. (B) Cyclin D1 sensitivity is increased by NaBH4 alone or in combination with boiling sodium citrate, hyaluronidase, and chondroitinase in paraffin sections of tissue. Scale bars: in A, 100 µm; in B, 200 µm.
Figure 5.
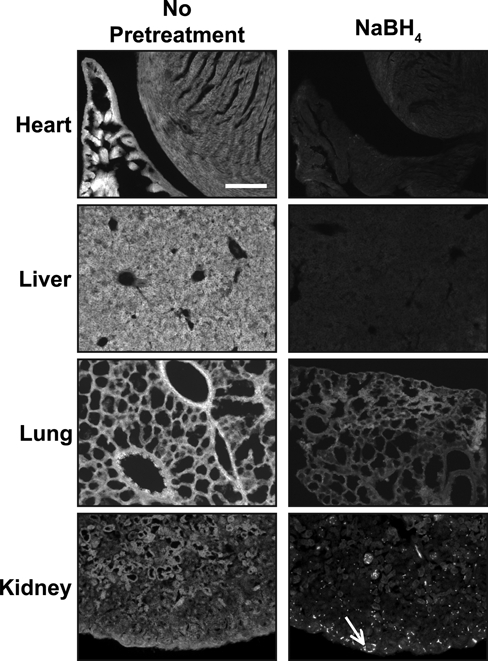
NaBH4 quenches autofluorescence in multiple tissues. Autofluorescence is detected in heart, liver, lung, and kidney. In all tissues tested, NaBH4 pretreatment quenches innate tissue autofluorescence. In the kidney, reduction of background fluorescence by NaBH4 reveals blood cells (white arrow) and does not significantly reduce the autofluorescence from the blood cells. Scale bar equals 100 µm in all panels.
Antibodies Require Specific Combinations of Pretreatments for Optimal Signal
To increase robustness of protein detection and decrease variability between samples, we tested combinations of pretreatments. The combination of Na-citrate antigen retrieval with sugar-degrading enzymes had modest effects on immunofluorescence levels (Fig. 6). Not surprisingly, the combination of NaBH4 pretreatment of tissue with hyaluronidase and/or chondroitinase also improved signal specificity (Fig. 2A,B). Combining three pretreatments—NaBH4, antigen retrieval, and sugar-degrading enzymes—resulted in the highest signal and subcellular specificity for the nuclear proteins cyclinD1 and pSmad and the cilia-associated protein pericentrin (Jurczyk et al. 2004) (Figs. 4B, 6, 7). In combination with NaBH4, both hyaluronidase and chondroitinase produced a stronger, more specific signal than heparinase (Fig. 2A,B). Together, NaBH4 and all three enzymes (hyaluronidase + heparinase II + chondroitinase [HHC]) combined also produced a high signal:noise ratio (Figs. 6, 7). Thus, sugar-degrading enzymes can enhance immunofluorescence detection in cartilage if a signal plateau has not already been reached with NaBH4 alone.
Figure 6.
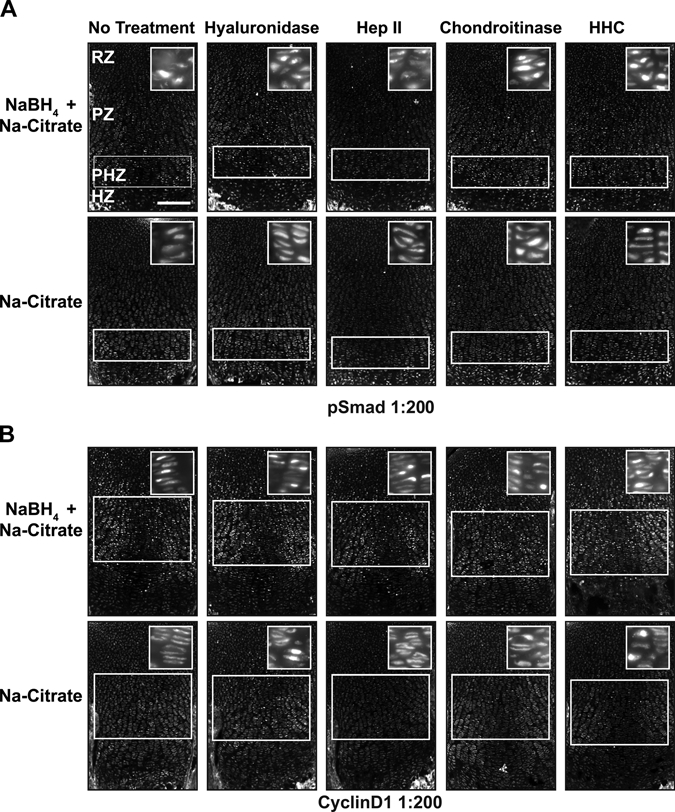
Combining pretreatments optimizes signal detection. Immunofluorescence detection of the stated antigens was performed on sections of P0 mouse growth plate cartilage subjected to the designated combinations of pretreatments (header and left of rows). Antigen retrieval (Na-citrate) alone results in a high level of cytosolic signal for anti-pSmad (A) and anti-cyclinD1 (B), as demonstrated by the clearly discernable structure of the columnar chondrocytes of the proliferative zone. (A) Detection of pSmad in the prehypertrophic zone (PHZ) is improved with Na-citrate pretreatment in combination with NaBH4 (note loss of visible columns). Hyaluronidase, chondroitinase, or a combination of three sugar-degrading enzymes (hyaluronidase + heparinase II + chondroitinase [HHC]) reveals nuclear staining in the proliferative zone (PZ) and resting zone (RZ) when used in combination with Na-citrate and NaBH4. (B) Similarly, NaBH4 and Na-citrate pretreatment significantly increased the signal:noise ratio for cyclinD1 in the PZ and RZ. The addition of hyaluronidase and chondroitinase or HHC further increased the signal and the specific localization of the antigen. Multiple pretreatments reveal higher levels of signal in the proliferative chondrocytes relative to the resting chondrocytes. White boxes indicate location where signal is present or expected. Scale bar in A–C, 200 µm.
Figure 7.
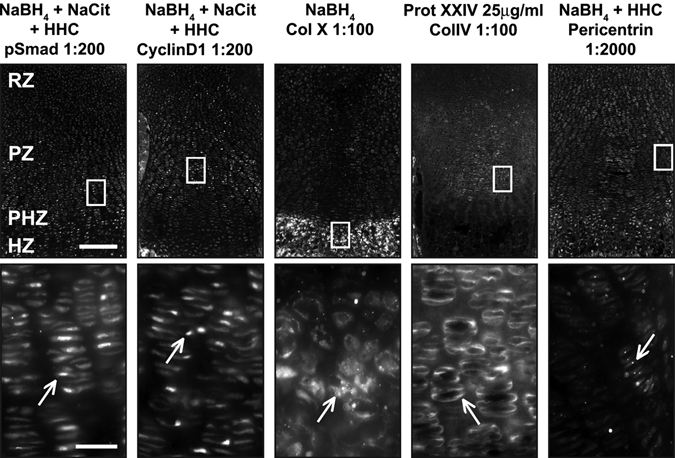
Optimal conditions reveal proper localization of antibodies. For each pretreatment condition (header), a low-magnification immunofluorescence image (top) and a high-magnification image (bottom) corresponding to the white box (top) are presented. (A) Optimal conditions for pSmad, cyclinD1, and pericentrin include NaBH4, Na-citrate, and hyaluronidase + heparinase II + chondroitinase (HHC). High-magnification images of boxed regions show proper localization of antibodies (white arrows). pSmad is nuclear localized through the growth plate. CyclinD1 is nuclear localized in dividing cells in the resting zone (RZ) and proliferative zone (PZ). Pericentrin is found throughout the growth plate at the base of primary cilia. The optimal condition for the matrix protein collagen X (Col X) is NaBH4 alone, but for collagen IV (Col IV), treatment with proteinase XXIV (Prot XXIV) produces optimal results. Col X is found in the matrix of the HZ, whereas Col IV is found in the matrix surrounding resting and proliferative chondrocytes (RZ and PZ). Scale bars: in A, 200 µm for 10× images and 25 µm for 40× images.
The optimal combination of pretreatments that increased sensitivity for detection of pSmad and cyclinD1 did not yield strong signal with antibodies against extracellular matrix proteins. No appreciable signal was observed for the matrix protein Col X following HHC treatment (data not shown). Instead, NaBH4 alone produced the most robust signal for Col X (Fig. 7). In contrast, the matrix protein Col IV could not be visualized with NaBH4, Na-citrate antigen retrieval, or sugar-degrading enzymes. In this case, we chose to treat with proteinase XXIV, a proteolytic enzyme used for antigen retrieval (Rheinhardt and Finkbeiner 2001). Robust and specific signal for Col IV was detected after limited proteolytic digestion of the tissue sections (Fig. 7).
Antibody Dilution Can Be Increased with Cartilage Pretreatment
We next asked if the increase in signal:noise translates into increased sensitivity for detection of endogenous antigens. When used at high concentrations on pretreated tissue sections, anti-cyclinD1 shows strong nuclear signal along with high signal in the cytosol (Fig. 8). Diluting the antibody preparation substantially reduces the cytosolic fluorescence while retaining the bona fide nuclear signal (Fig. 8). By analyzing an antibody dilution series, regions of the growth plate with higher levels of signaling (proliferative zone) are readily distinguished from those with lower levels of signal (resting zone). The region-specific differences in cyclinD1 concentration (Fig. 8) parallel known differences in cell cycle activity as evaluated by BrdU labeling (Yang et al. 2003; Ahrens et al. 2009). Similarly, judging from the maximal dilution of antibody that produces zone-specific signal under optimal immunodetection conditions, pSmad is present at higher levels in nuclei of the prehypertrophic zone compared to the resting and the proliferative zones. Pretreatments also increase sensitivity for detection of extracellular proteins. With optimal pretreatment conditions, higher concentrations of antibody result in diffuse signal throughout the cartilage matrix for anti–Col IV, whereas lower concentrations of antibody yield a more specific pericellular signal (Fig. 8). In addition, although Col X signal is visualized at a 1:100 dilution of antibody without NaBH4 pretreatment, detection of signal at a 1:500 dilution requires NaBH4 pretreatment (Fig. 8). Altogether, this demonstrates that a specific combination of pretreatments increases the sensitivity of immunofluorescence detection and provides for greater signal specificity with lower antibody consumption (SF1, ST2, ST3).
Figure 8.
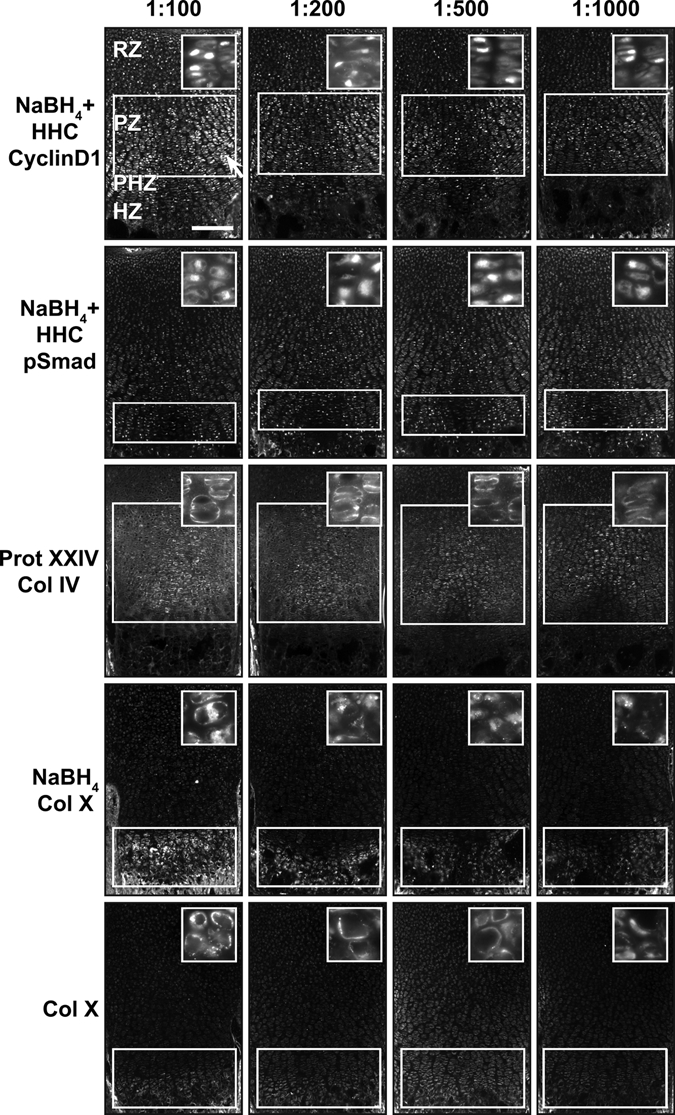
Antibody dilution can be increased with cartilage pretreatment. (A) Optimal pretreatment conditions of cyclinD1, pSmad, collagen IV (Col IV), and collagen X (Col X) allow visualization of antibodies at lower dilutions, up to 1:1000. CyclinD1 has nonspecific cytoplasmic staining (white arrow) when the concentration is too high (1:100). Note how the contrast between signal in the resting and the proliferative zone increases with antibody dilution. Optimal conditions increase the sensitivity of immunofluorescence for Col X. If there is no pretreatment, Col X has only weak signal at a dilution of 1:100. However, the signal:noise ratio is much higher for Col X at 1:100 when it is pretreated with NaBH4, and signal can be visualized even at a dilution of 1:1000. White bars indicate highest levels of expression. White boxes indicate location where signal is present or expected. Scale bar: in A, 200 µm.
Discussion
Localization of endogenous proteins is an essential tool for elucidating the molecular underpinnings of cell biology. Whereas many antibody preparations are effective for immunolocalization of proteins in cultured cells, only a subset of these reagents has proven useful in the analysis of complex tissues such as cartilage. Anecdotally, failure to obtain an immunofluorescence signal is often attributed to antibody quality or concentration of the target protein. Perhaps more problematic is the potential for false-positive signals where protein localization (Campos-Xavier et al. 2009) does not correlate to known gene expression domains (Ahrens et al. 2009). Our work here suggests that suboptimal tissue preparation is a leading factor that limits the utility of and confidence in immunofluorescence results in cartilage.
The different optimal pretreatments identified in this study suggest that the interplay of multiple assay parameters determines the effectiveness of a given antibody in immunofluorescence for the cartilage growth plate. Given the limited number of antibodies tested to date, we cannot be certain from the results of these experiments whether the sensitivity of signal to the presence of proteoglycans depends on the subcellular localization of the protein, the properties of the antigen, or the specific composition of the antibody preparation. However, as a starting point, it appears that both autofluorescence and carbohydrate polymers strongly interfere with visualizing the subcellular localization of proteins by immunofluorescence in the cartilage growth plate. By contrast, autofluorescence and interference by proteoglycans may be secondary to recovering native protein structure or exposing buried epitopes for detection of matrix proteins. Accordingly, pretreatment with carbohydrate-degrading enzymes decreases specific signal from antibodies against the matrix proteins tested. Regardless of the reasons for these differences, our results demonstrate that testing multiple pretreatment conditions is necessary to optimize signal strength and spatial resolution for a given antibody preparation.
Determining optimal pretreatment conditions for a particular antibody confers distinct advantages over traditional immunofluorescence methods. First, these improved methods allow visualization of endogenous proteins using antibodies that previously did not produce sufficient signal for visualization. Second, higher antibody dilutions conserve antibody and save money. Third, and perhaps most important, the ability to use a lower concentration of antibodies permits the assessment of protein expression levels within a given tissue. Using a dilution series, the threshold antibody concentration for protein detection in different regions of the same tissue or in different subcellular compartments can be determined, allowing for the possibility of obtaining semi-quantitative data that can enhance modeling of pathway dynamics in situ.
Altogether, this work demonstrates that the limitations of immunofluorescence in growth plate cartilage can be overcome through appropriate pretreatment of the tissue. Although this work primarily involves analysis of frozen sections, we show that these methods, specifically NaBH4 pretreatment, are applicable to previously sectioned paraffin-embedded tissue and therefore could be used to substantially increase the information obtained from archival samples of healthy and diseased human cartilage. Moreover, these techniques may also be applied to other tissues to increase the sensitivity of protein signal using immunofluorescence. In other tissues such as bone marrow, myocardium, cartilage, and spinal cord, NaBH4 alone (Baschong et al. 2001; Langelier et al. 2000; Casella et al. 2004) or antigen retrieval alone has been used for optimal staining, but combinations of these techniques have not been applied to these or other tissue types. In addition, tissues that are rich in matrix proteins are prime candidates for using hyaluronidase, chondroitinase, or heparinase pretreatments to increase the signal:noise ratio in immunofluorescence studies. Altogether, this work demonstrates that a unique cocktail of pretreatments can increase sensitivity for particular antibodies in immunofluorescence studies. In our experience, these methods can turn non-performing antibodies into useful antibodies and thereby increase the repertoire of unique, commercially available antibodies that can be applied to study cell biology in vivo.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
The author(s) disclosed receipt of the following financial support for the research and/or authorship of this article: This work was supported by the Cellular Molecular Basis of Disease Training Grant (MJA), the National Institutes of Health/NIAMS (AR054857), and the National Center for Research Resources (NCRR). Grant sponsors: National Institutes of Health/NIAMS; AR054857.
References
- Ahrens MJ, Li Y, Jiang H, Dudley AT. 2009. Convergent extension movements in growth plate chondrocytes require gpi-anchored cell surface proteins. Development. 136:3463–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. 1993. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 7:812–821 [DOI] [PubMed] [Google Scholar]
- Baschong W, Suetterlin R, Laeng RH. 2001. Control of autofluorescence of archival formaldehyde-fixed, paraffin-embedded tissue in confocal laser scanning microscopy (CLSM). J Histochem Cytochem. 49:1565–1572 [DOI] [PubMed] [Google Scholar]
- Blanc A, Tran-Khanh N, Filion D, Buschmann MD. 2005. Optimal processing method to obtain four-color confocal fluorescent images of the cytoskeleton and nucleus in three-dimensional chondrocyte cultures. J Histochem Cytochem. 53:1171–1175 [DOI] [PubMed] [Google Scholar]
- Blumbach K, Niehoff A, Paulsson M, Zaucke F. 2008. Ablation of collagen IX and COMP disrupts epiphyseal cartilage architecture. Matrix Biol. 27:306–318 [DOI] [PubMed] [Google Scholar]
- Campos-Xavier AB, Martinet D, Bateman J, Belluoccio D, Rowley L, Tan TY, Baxová A, Gustavson KH, Borochowitz ZU, Innes AM, et al. 2009. Mutations in the haparan-sulfate proteoglycan glypican 6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. Am J Hum Genet. 84:760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella GT, Bunge MB, Wood PM. 2004. Improved immunocytochemical identification of neural, endothelial, and inflammatory cell types in paraffin-embedded injured adult rat spinal cord. J Neurosci Methods. 139:1–11 [DOI] [PubMed] [Google Scholar]
- Dreier R, Gunther BK, Mainz T, Nemere I, Bruckner P. 2008. Terminal differentiation of chick embryo chondrocytes requires shedding of a cell surface protein that binds 1,25-dihydroxy-vitamin D3. J Biol Chem. 283:1104–1112 [DOI] [PubMed] [Google Scholar]
- Dreier R, Opolka A, Grifka J, Bruckner P, Grassel S. 2008. Collagen IX–deficiency seriously compromises growth cartilage development in mice. Matrix Biol. 27:319–329 [DOI] [PubMed] [Google Scholar]
- Gentili C, Cancedda R. 2009. Cartilage and bone extracellular matrix. Curr Pharm Des. 15:1334–1348 [DOI] [PubMed] [Google Scholar]
- Imam SA, Young L, Chaiwun B, Taylor CR. 1995. Comparison of two microwave based antigen-retrieval solutions in unmasking epitopes in formalin-fixed tissue for immunostaining. Anticancer Res. 15:1153–1158 [PubMed] [Google Scholar]
- Irwin MH, Silvers SH, Mayne R. 1985. Monoclonal antibody against chicken type IX collagen: preparation, characterization, and recognition of the intact form of type IX collagen secreted by chondrocytes. J Cell Biol. 101:814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyk A, Gromley A, Redick S, San Agustin J, Witman G, Pazour GJ, Peters DJ, Doxsey S. 2004. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J Cell Biol. 166:637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluppel M, Wight TN, Chan C, Hinek A, Wrana JL. 2005. Maintenance of chondroitin sulfation balance by chondroitin-4-sulfotransferase 1 is required for chondrocyte development and growth factor signaling during cartilage morphogenesis. Development. 132:3989–4003 [DOI] [PubMed] [Google Scholar]
- Kronenberg HM. 2003. Developmental regulation of the growth plate. Nature. 423:332–336 [DOI] [PubMed] [Google Scholar]
- Kvist AJ, Nystrom A, Hultenby K, Sasaki T, Talts JF, Aspberg A. 2008. The major basement membrane components localize to the chondrocyte pericellular matrix: a cartilage basement membrane equivalent? Matrix Biol. 27:22–33 [DOI] [PubMed] [Google Scholar]
- Langelier E, Suetterlin R, Hoemann CD, Aebi U, Buschmann MD. 2000. The chondrocyte cytoskeleton in mature articular cartilage: structure and distribution of actin, tubulin, and vimentin filaments. J Histochem Cytochem. 48:1307–1320 [DOI] [PubMed] [Google Scholar]
- Melrose J, Smith S, Ghosh P, Whitelock J. 2003. Perlecan, the multidomain heparan sulfate proteoglycan of basement membranes, is also a prominent component of the cartilaginous primordia in the developing human fetal spine. J Histochem Cytochem. 51:1331–1341 [DOI] [PubMed] [Google Scholar]
- Nishimura I, Muragaki Y, Hayashi M, Ninomiya Y, Olsen BR. 1990. Tissue-specific expression of type IX collagen. Ann N Y Acad Sci. 580:112–119 [DOI] [PubMed] [Google Scholar]
- Quintarelli G, Vocaturo A, Roden L, Bellocci M, Vassallo LM. 1978. Role of hyaluronic acid in the in vivo aggregation of cartilage proteoglycans. Connect Tissue Res. 5:237–248 [DOI] [PubMed] [Google Scholar]
- Retting KN, Song B, Yoon BS, Lyons KM. 2009. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 136:1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinhardt JM, Finkbeiner WE. 2001. Protease XXIV increases detection of mucin gene expression during in situ hybridization in archival tissue. J Histochem Cytochem. 49:923–924 [DOI] [PubMed] [Google Scholar]
- Schmid TM, Linsenmayer TF. 1985a. Developmental acquisition of type X collagen in the embryonic chick tibiotarsus. Dev Biol. 107:373–381 [DOI] [PubMed] [Google Scholar]
- Schmid TM, Linsenmayer TF. 1985b. Immunohistochemical localization of short chain cartilage collagen (type X) in avian tissues. J Cell Biol. 100:598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewing A, Burger C, Brusselbach S, Schalk C, Lucibello FC, Muller R. 1993. Human cyclin D1 encodes a labile nuclear protein whose synthesis is directly induced by growth factors and suppressed by cyclic AMP. J Cell Sci. 104(Pt 2):545–555 [DOI] [PubMed] [Google Scholar]
- von der Mark K, von der Mark H, Gay S. 1976. Study of differential collagen synthesis during development of the chick embryo by immunofluorescence: II. Localization of type I and type II collagen during long bone development. Dev Biol. 53:153–170 [DOI] [PubMed] [Google Scholar]
- Weber K, Rathke PC, Osborn M. 1978. Cytoplasmic microtubular images in glutaraldehyde-fixed tissue culture cells by electron microscopy and by immunofluorescence microscopy. Proc Natl Acad Sci U S A. 75:1820–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Topol L, Lee H, Wu J. 2003. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development. 130:1003–1015 [DOI] [PubMed] [Google Scholar]


