Abstract
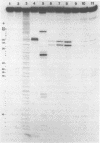
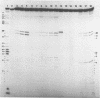
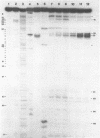
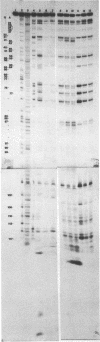
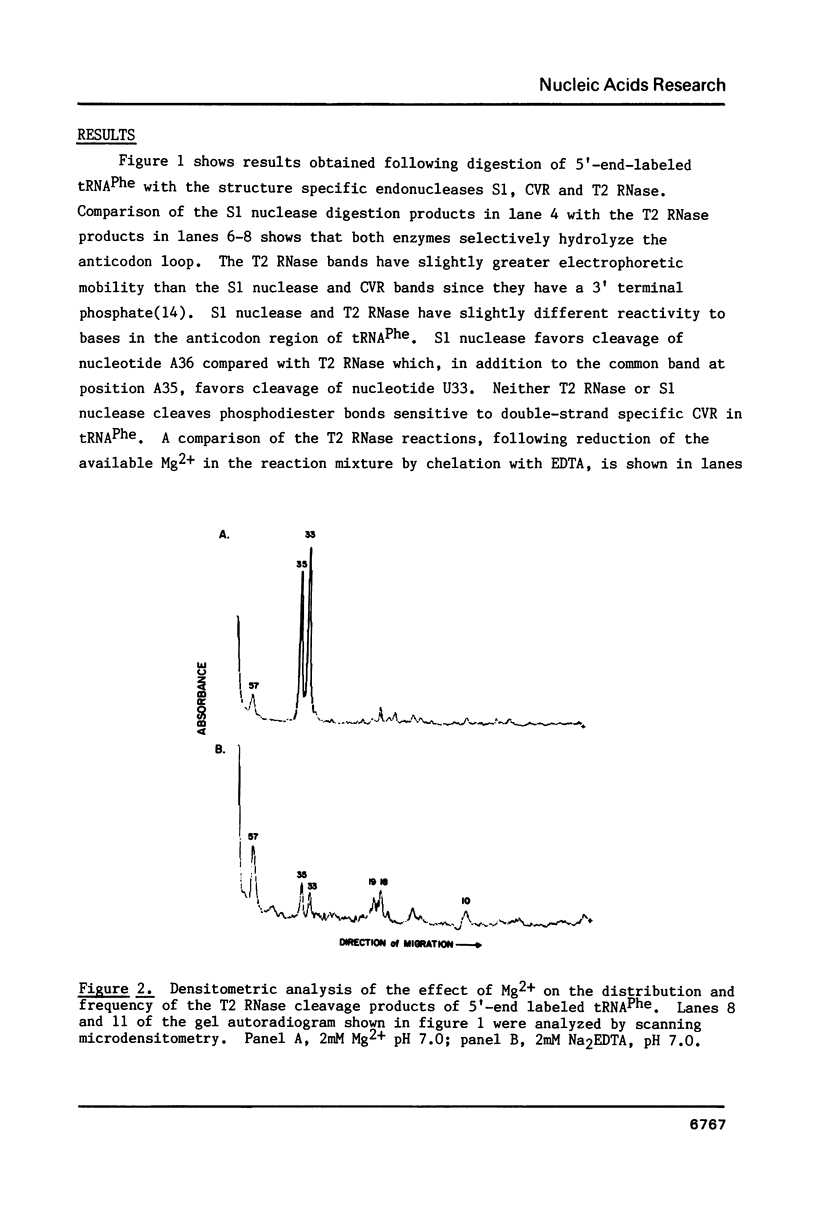
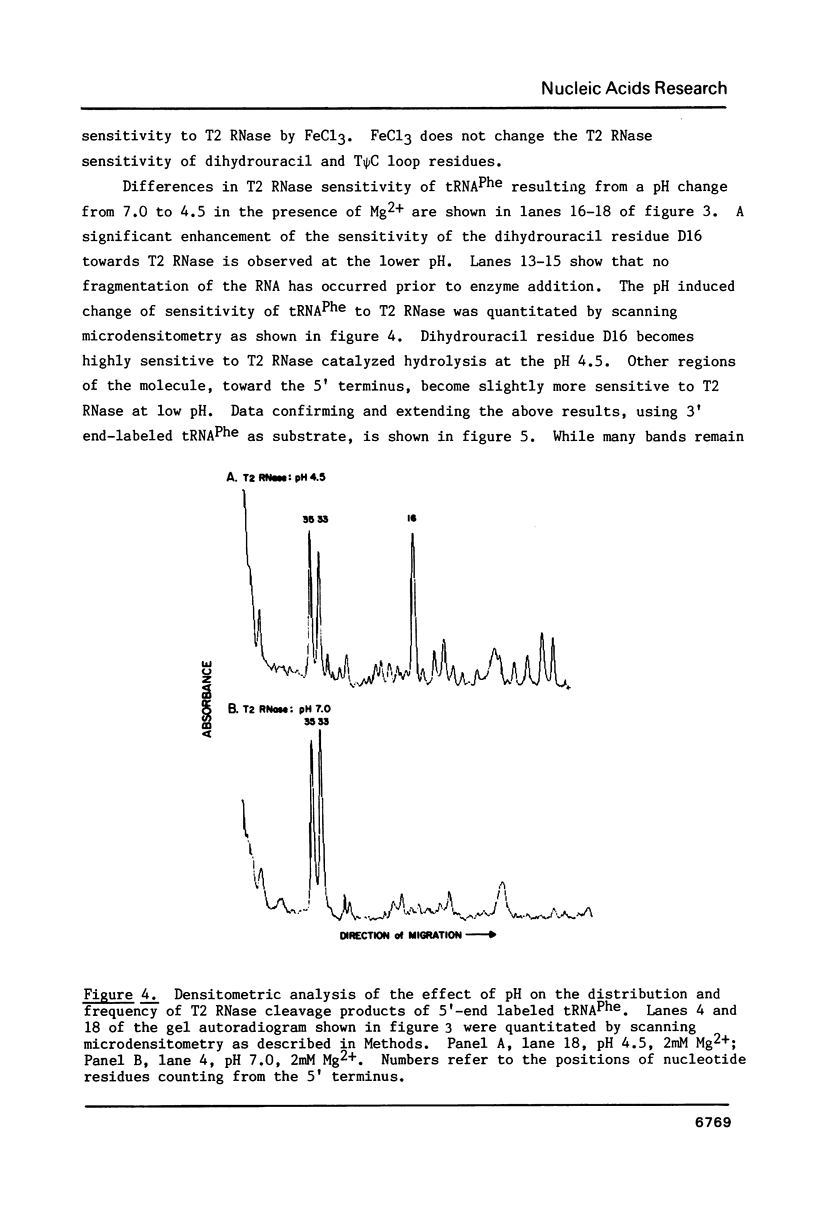
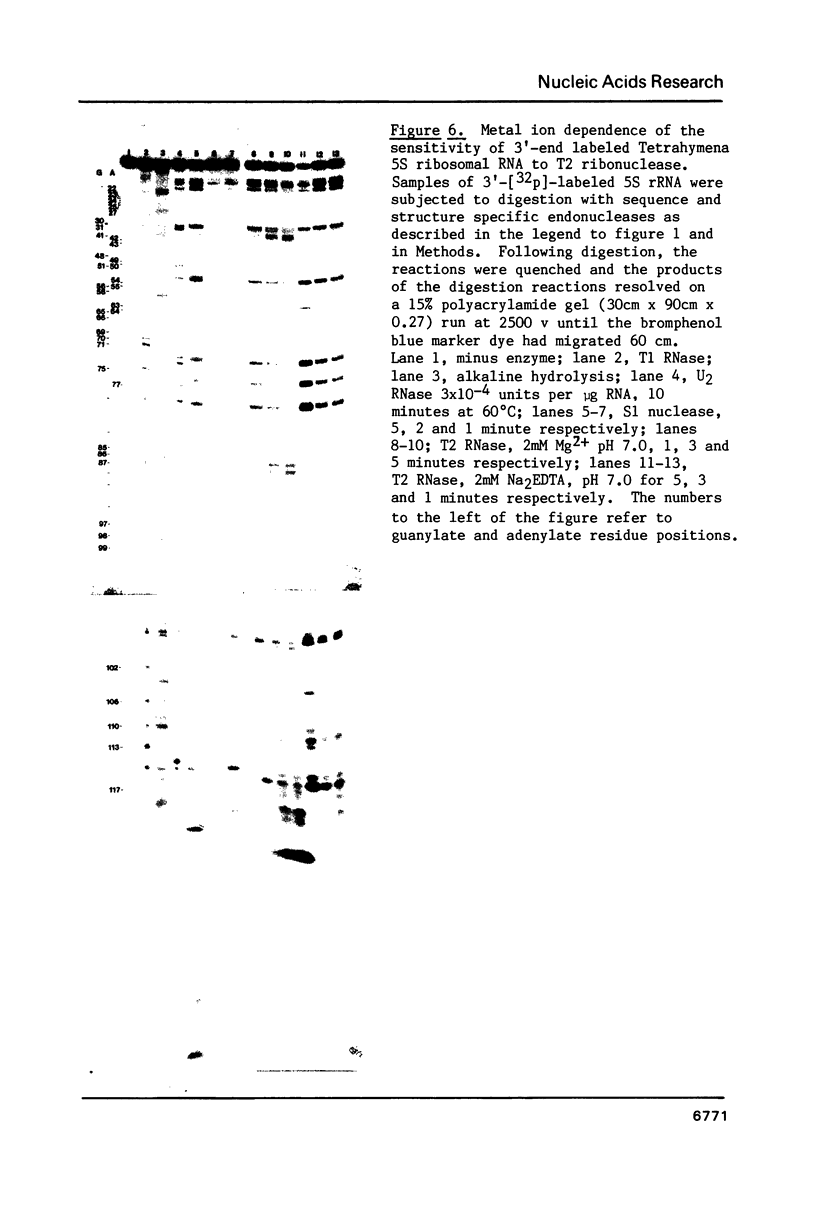
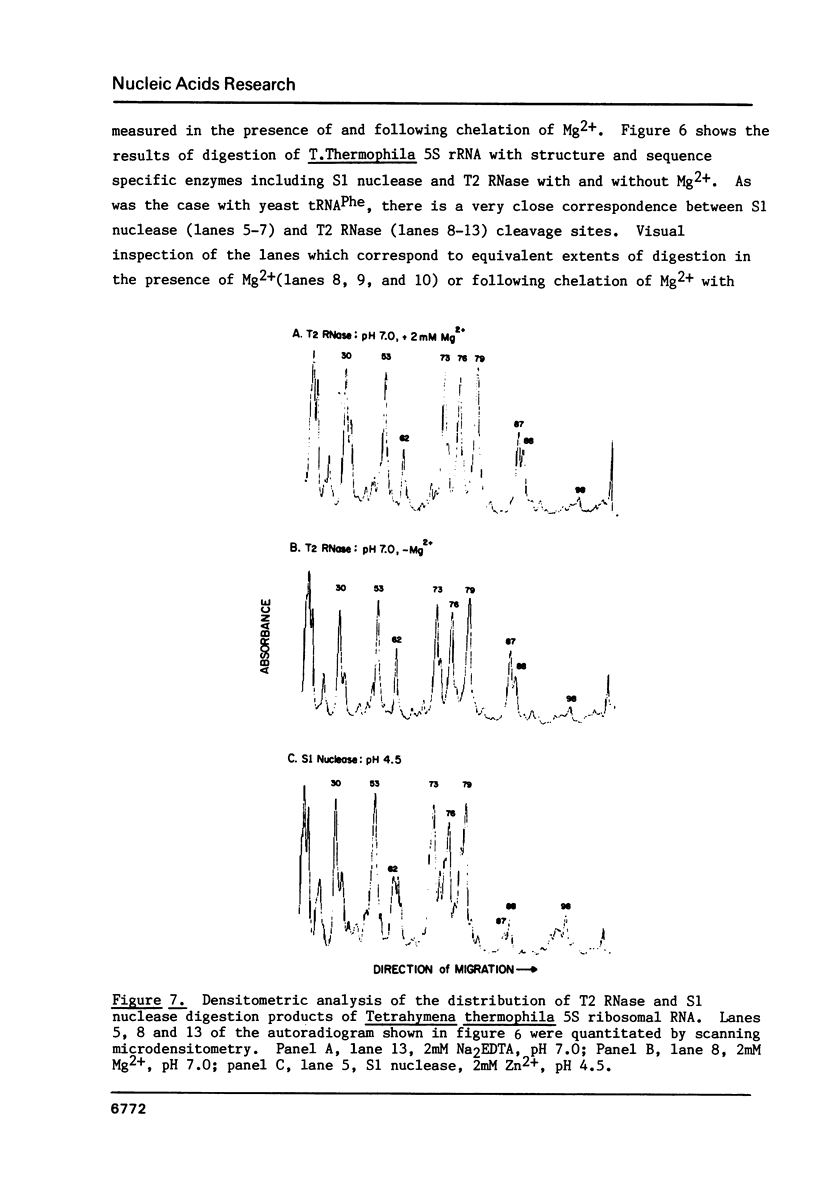
We describe the use of an enzymic probe of RNA structure, T2 ribonuclease, to detect alterations of RNA conformation induced by changes in Mg2+ ion concentration and pH. T2 RNase is shown to possess single-strand specificity similar to S1 nuclease. In contrast to S1 nuclease, T2 RNase does not require divalent cations for activity. We have used this enzyme to investigate the role of Mg2+ ions in the stabilization of RNA conformation. We find that, at neutral pH, drastic reduction of the available divalent metal ions results in a decrease in the ability of T2 RNase to cleave the anticodon loop of tRNAPhe. This change accompanies an increase in the cleavage of the molecule in the T psi C and in the dihydrouracil loops. Similar treatment of Tetrahymena thermophila 5S ribosomal RNA shows that changes in magnesium ion concentration does not have a pronounced effect on the cleavage pattern produced by T2 RNase. T2 RNase activity has a broader pH range than S1 nuclease and can be used to study pH induced conformational shifts in RNA structure. We find that upon lowering the pH from 7.0 to 4.5, nucleotide D16 in the dihydrouracil loop of tRNAPhe becomes highly sensitive to T2 RNase hydrolysis. This change accompanies a decrease in the relative sensitivity of the anticodon loop to the enzyme. The role of metal ion and proton concentrations in maintenance of the functional conformation of tRNAPhe is discussed.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appel B., Erdmann V. A., Stulz J., Ackerman T. Determination of base pairing in Escherichia coli and Bacillus stearothermophilus 5S RNAs by infrared spectroscopy. Nucleic Acids Res. 1979 Oct 25;7(4):1043–1057. doi: 10.1093/nar/7.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Göringer U., Wagner R. Structural investigation of Phe-tRNAPhe from E.coli bound to the ribosomal A-site. Nucleic Acids Res. 1983 Feb 11;11(3):575–589. doi: 10.1093/nar/11.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina-Stein M., Crothers D. M. Conformational changes of transfer ribonucleic acid. The pH phase diagram under acidic conditions. Biochemistry. 1974 Jun 18;13(13):2771–2775. doi: 10.1021/bi00710a017. [DOI] [PubMed] [Google Scholar]
- Butzow J. J., Eichhorn G. L. Different susceptibility of DNA and RNA to cleavage by metal ions. Nature. 1975 Mar 27;254(5498):358–359. doi: 10.1038/254358a0. [DOI] [PubMed] [Google Scholar]
- Cole P. E., Yang S. K., Crothers D. M. Conformational changes of transfer ribonucleic acid. Equilibrium phase diagrams. Biochemistry. 1972 Nov 7;11(23):4358–4368. doi: 10.1021/bi00773a024. [DOI] [PubMed] [Google Scholar]
- Johnston P. D., Redfield A. G. Study of transfer ribonucleic acid unfolding by dynamic nuclear magnetic resonance. Biochemistry. 1981 Jul 7;20(14):3996–4006. doi: 10.1021/bi00517a008. [DOI] [PubMed] [Google Scholar]
- Jones C. R., Kearns D. R. Identification of a unique ethidium bromide binding site on yeast tRNAPhe by high resolution (300 MHz) nuclear magnetic resonance. Biochemistry. 1975 Jun 17;14(12):2660–2665. doi: 10.1021/bi00683a016. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Quigley G. J., Suddath F. L., McPherson A., Sneden D., Kim J. J., Weinzierl J., Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973 Jan 19;179(4070):285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Sussman J. L., Suddath F. L., Quigley G. J., McPherson A., Wang A. H., Seeman N. C., RICH A. The general structure of transfer RNA molecules. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4970–4974. doi: 10.1073/pnas.71.12.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazaki T., Hori H., Osawa S., Mita T., Higashinakagawa T. The nucleotide sequences of 5S rRNAs from three ciliated protozoa. Nucleic Acids Res. 1982 Jul 24;10(14):4409–4412. doi: 10.1093/nar/10.14.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard R. E., Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981 Oct 10;9(19):5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan T., Silverman S., Kohli J., Söll D. Nucleotide sequence of phenylalanine transfer RNA from Schizosaccharomyces pombe: implications for transfer RNA recognition by yeast phenylalanyl-tRNA synthetase. Biochemistry. 1978 May 2;17(9):1622–1628. doi: 10.1021/bi00602a007. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Lockard R. E., Vamvakopoulos N., Rieser L., RajBhandary U. L., Vournakis J. N. Secondary structure of mouse and rabbit alpha- and beta-globin mRNAs: differential accessibility of alpha and beta initiator AUG codons towards nucleases. Cell. 1980 Jan;19(1):91–102. doi: 10.1016/0092-8674(80)90391-8. [DOI] [PubMed] [Google Scholar]
- Peattie D. A., Herr W. Chemical probing of the tRNA--ribosome complex. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2273–2277. doi: 10.1073/pnas.78.4.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Teeter M. M., Rich A. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):64–68. doi: 10.1073/pnas.75.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. Initial stages of the thermal unfolding of yeast phenylalanine transfer RNA as studied by chemical modification: the effect of magnesium. Eur J Biochem. 1977 Nov 15;81(1):91–101. doi: 10.1111/j.1432-1033.1977.tb11930.x. [DOI] [PubMed] [Google Scholar]
- Schreier A. A., Schimmel P. R. Interaction of manganese with fragments, complementary fragment recombinations, and whole molecules of yeast phenylalanine specific transfer RNA. J Mol Biol. 1974 Jul 5;86(3):601–620. doi: 10.1016/0022-2836(74)90183-1. [DOI] [PubMed] [Google Scholar]
- Stulz J., Ackermann T., Appel B., Erdmann V. A. Determination of base pairing in yeast 5S and 5.8S RNA infrared spectroscopy. Nucleic Acids Res. 1981 Aug 11;9(15):3851–3861. doi: 10.1093/nar/9.15.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troutt A., Savin T. J., Curtiss W. C., Celentano J., Vournakis J. N. Secondary structure of Bombyx mori and Dictyostelium discoideum 5S rRNA from S1 nuclease and cobra venom ribonuclease susceptibility, and computer assisted analysis. Nucleic Acids Res. 1982 Jan 22;10(2):653–664. doi: 10.1093/nar/10.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T. Purification and properties of RNase T2. J Biochem. 1966 Aug;60(2):115–132. doi: 10.1093/oxfordjournals.jbchem.a128410. [DOI] [PubMed] [Google Scholar]
- Vary C. P., Vournakis J. N. RNase H-catalyzed site-specific deadenylylation of rabbit alpha- and beta- globin mRNAs. Secondary structure of 3'-noncoding regions. J Biol Chem. 1984 Mar 10;259(5):3299–3307. [PubMed] [Google Scholar]
- Vary C. P., Vournakis J. N. Secondary structure of eukaryotic messenger RNA. Biochem Soc Symp. 1982;47:61–78. [PubMed] [Google Scholar]
- Vournakis J. N., Celantano J., Finn M., Lockard R. E., Mitra T., Pavlakis G., Troutt A., van den Berg M., Wurst R. M. Sequence and structure analysis of end-labeled RNA with nucleases. Gene Amplif Anal. 1981;2:267–298. [PubMed] [Google Scholar]
- Wells B. D. The conformation of the tRNAPhe anticodon loop monitored by fluorescence. Nucleic Acids Res. 1984 Feb 24;12(4):2157–2170. doi: 10.1093/nar/12.4.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede P., Woo N. H., Rich A. Initiator tRNAs have a unique anticodon loop conformation. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3289–3293. doi: 10.1073/pnas.76.7.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]