New evidence indicates that the conserved plasmid shared among Chlamydial species may be key for understanding and vaccinating against these pathogenic bacteria.
Abstract
Chlamydial plasmids are small, highly conserved, nonconjugative, and nonintegrative DNA molecules that are nearly ubiquitous in many chlamydial species, including Chlamydia trachomatis. There has been significant recent progress in understanding chlamydial plasmid participation in host–microbe interactions, disease, and immune responses. Work in mouse model systems and, very recently, in nonhuman primates demonstrates that plasmid-deficient chlamydial strains function as live attenuated vaccines against genital and ocular infections. Collectively, these studies open new avenues of research into developing vaccines against trachoma and sexually transmitted chlamydial infections.
Human pathogenic chlamydiae are members of a successful and unique lineage of bacteria (Collingro et al., 2011), which infect and cause disease in a wide variety of animals (Longbottom and Coulter, 2003). Although antibiotic chemotherapy is quite effective in treating diagnosed primary uncomplicated ocular and urogenital chlamydial infections, prevention of the serious pathological sequelae, which can develop after infection, and prevention of reinfection are much more difficult tasks (Byrne, 2010). Most chlamydial infections are asymptomatic and thus go undiagnosed and untreated, which can lead to chronic inflammation and irreversible tissue and organ pathology. In some countries and communities, limited access to medical care and infection screening technologies further hampers timely diagnosis and treatment (Centers for Disease Control and Prevention [CDC], 2011). Repeat infections are common, and the risk of developing the serious sequelae of ocular (e.g., blindness) or urogenital infection (e.g., infertility) increases with multiple infections (Darville and Hiltke, 2010).
These obligate intracellular bacteria develop within a membrane-bound vacuole termed the inclusion (Fig. 1), and existence within the inclusion defines much about the biology of the lineage. The challenges to understanding and preventing chlamydial disease are amplified by the difficulties of working with an obligate intracellular bacterial species, in which the development of a practical system of genetic manipulation is only in its infancy. (Binet and Maurelli, 2009; Kari et al., 2011).
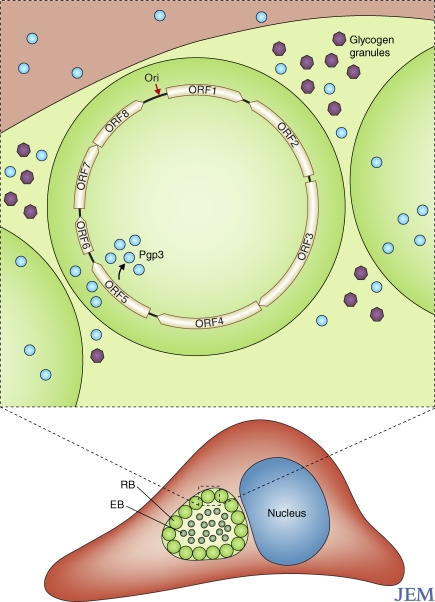
Figure 1.
Chlamydial growth and plasmid structure. The obligately intracellular chlamydiae develop within a host vacuole termed the inclusion. The two developmental forms have different functions in growth. Elementary bodies (EB) are infectious but minimally metabolically active, whereas reticulate bodies (RB) grow and divide, but cannot infect. As shown in a magnified reticulate body, the chlamydial plasmid has eight ORFs, which encode plasmid maintenance and chlamydia-specific functions. ORF5 encodes Pgp3, which is secreted from the bacterium and the inclusion and accumulates in the cytosol of the host cell. Pgp3 is immunogenic in many host species. The chlamydial plasmid is also associated with the accumulation of glycogen granules (purple) in the lumen of the inclusion and, as discussed in the text, functions in virulence in animal model systems.
The chlamydial plasmid
In most microbial systems, autonomously replicating plasmids function as tools of genetic variation and are built with replicative backbones associated with a variety of functional cassettes encoding niche-specific genes or gene sets. Often these plasmids can be horizontally transferred, which leads to rapid distribution or reassociation of phenotypes among strains (Koonin et al., 2001). The spread of antibiotic resistance is often a function of sharing of plasmids (or other laterally transferred genetic elements) among otherwise susceptible strains. Such plasmid transfer is a major and evolving problem in infectious disease worldwide. Many serious diseases are directly the result of virulence factors carried by plasmids, and variation in plasmid structure is often associated with different disease syndromes caused by otherwise related strains and species.
Such variability is not found in chlamydia. Several chlamydial species contain one of a homologous set of ∼7,500 base pair plasmids with a copy number that is approximately fourfold greater than that of the chromosome (Thomas et al., 1997; Pickett et al., 2005; Fig. 1). Within C. trachomatis clinical isolates, the plasmid is virtually ubiquitous. There are occasional studies showing plasmid-negative clinical strains, but little is known about the epidemiology and significance of these relatively rare isolates (Peterson et al., 1990; An et al., 1992; Farencena et al., 1997). Chlamydial plasmids are nonconjugative and nonintegrative. They do not encode antibiotic resistance genes and do not show signs of genetic flexibility. This plasmid and the equally curious chlamydiaphage found in some strains (Storey et al., 1989) represent the entire complement of extrachromosomal elements carried by Chlamydia spp. There is a single example of an integrated plasmid in Chlamydia suis (Dugan et al., 2004), but this is clearly the exception and not the rule.
Nucleotide sequence identity among plasmids from Chlamydia spp. ranges from 69 to 99%, with very high identity within C. trachomatis (Thomas et al., 1997; Seth-Smith et al., 2009). Sequence comparisons suggest that the plasmids evolved in parallel with the different chlamydial species. The C. trachomatis plasmid carries a set of five open reading frames (ORFs), which share identity with episomal maintenance genes common to other plasmids, and a set of three genes encoding proteins of unknown function, each of which is chlamydia specific. Human strains of Chlamydophila pneumoniae lack plasmid, but equine C. pneumoniae strains and other veterinary chlamydial species are plasmid positive.
The chlamydial plasmid also has a practical side. It is a common target for nucleic acid amplification test (NAAT)–based diagnostics of human infections (Fredlund et al., 2004). NAAT-based tests have been, and remain, a valuable aspect of chlamydial disease control. However, clinical isolates with a deletion in the target sequence used in some diagnostic kits (the Swedish Variant) have been identified. This can lead to false negative test results in patients (Unemo and Clarke, 2011). Studies have reported an apparent clonal expansion of C. trachomatis isolates carrying this deletion. It is possible that the inability to detect infections caused by strains carrying the deleted plasmid has contributed to the rapid expansion of the variant strains in patient populations.
As mentioned, the lack of routine genetic tools has been a significant barrier to rapid progress in chlamydial research. Manipulation of the chlamydial plasmid is a logical approach to introduce DNA into these organisms, but such efforts have been frustrating and generally unproductive. However, a very recent study describes successful transformation experiments using hybrid shuttle vectors built with pBR325 and the entire chlamydial plasmid (Wang et al., 2011). The vectors are propagated in both E. coli and C. trachomatis, and the system was used to generate green fluorescent C. trachomatis. This is an exciting and highly significant development in the study of chlamydiae.
The chlamydial plasmid appears to be critical for growth in vivo (Russell et al., 2011), but it is not required for growth in vitro. Multiple laboratories have described phenotypic changes associated with plasmid-deficient strains as compared with closely related wild type strains. Creation of plasmid-deficient isogenic strains began with the careful work of Akira Matsumoto, who developed a plaque-purification technique for chlamydial strains (Matsumoto et al., 1998) and noted that unlike wild-type C. trachomatis strains, three of his plaque-purified clones of C. trachomatis did not accumulate glycogen within the inclusion. Each glycogen-negative clone was then shown to lack the chlamydial plasmid. Subsequent investigations of plasmid-negative strains (O’Connell and Nicks, 2006; Carlson et al., 2008; O’Connell et al., 2011) confirmed the absence of glycogen and examined transcription across the chlamydial genome relative to wild-type strains. Although the precise scope and nature of plasmid-chromosome cross-regulation is not fully understood, transcription of a set of chlamydial chromosomal genes is negatively affected by the absence of plasmid. One C. trachomatis gene that is down-regulated in the plasmid-negative strains is glgA, a glycogen biosynthetic gene. Thus, it is hypothesized that elements encoded by the plasmid regulate, and perhaps directly participate in, the process of glycogen accumulation within chlamydial inclusions.
Linking disease and vaccines to the chlamydial plasmid
Subsequent in vivo studies using plasmid-deficient strains demonstrated that the plasmid is a virulence factor that significantly impacts clinical disease in mouse model systems. For example, although plasmid-negative strains grew similarly to isogenic wild-type strains, they failed to activate Toll-like receptor 2–dependent immune responses either in vivo or in vitro (O’Connell et al., 2011). Other work indicated that plasmid-negative C. trachomatis strains had significantly higher 50% infective dose values relative to a wild-type matched isolate (Carlson et al., 2008). The plasmid-deficient strains also functioned as successful attenuated live vaccines in mice, wherein prior infection with plasmid-negative strains limited the pathology associated with a subsequent infection by wild-type strains (O’Connell et al., 2007).
The near universal carriage of this single plasmid by C. trachomatis has led to speculation about its importance in human chlamydial infections. Early work demonstrated that chlamydia-infected individuals produce antibodies specific for at least one plasmid-encoded protein, Pgp3, which is encoded by ORF5 (Comanducci et al., 1994). Further work explored the possible utility of Pgp3 as a vaccine candidate (Donati et al., 2003). Recent work has shown that native Pgp3 is a trimeric molecule that is transported to the host cell cytosol during the infectious process (Li et al., 2008; Chen et al., 2010; Fig. 1). Other chlamydial proteins are delivered to the cytosol, where they function to turn the host cell into a “chlamydia factory,” and it is hypothesized that Pgp3 serves a similar function.
Chlamydial vaccine studies are complicated by the possibility that the debilitating pathology associated with chlamydial ocular and urogenital infections represents collateral damage of innate and/or adaptive immune responses (Rockey et al., 2009; Farris and Morrison, 2011). Protection, in contrast, is thought to be serovar or genovar specific, but neither the mechanisms of pathogenesis nor the mechanisms of protection have been clearly elucidated. Several candidate protective antigens have been explored (Rockey et al., 2009), but no molecule has emerged as a clear and practical choice as a single protective antigen. Work in the mouse system suggests that delivery of antigens, or a combination of antigens, is required to generate a protective response. Indeed, a live attenuated veterinary chlamydial vaccine, generated via chemical mutagenesis, is commercially available and routinely used to reduce the incidence of abortion in sheep (Burall et al., 2009).
In this issue of the Journal of Experimental Medicine, Kari et al. describe experiments demonstrating that a plasmid-deficient C. trachomatis strain is both avirulent in ocular infections of macaques and functions as a live attenuated vaccine in that system. These fascinating studies conclusively demonstrate that the plasmid functions as a virulence factor in C. trachomatis ocular infections, consistent with the work conducted in the genital tract in the murine model system. In addition to the colonization studies, Kari et al. demonstrated that three of the six animals infected with the plasmid-negative strain were completely protected against challenge by an isogenic and virulent plasmid-positive strain. Analysis of the major histocompatibility complex genotypes of each animal demonstrated that all three of the solidly protected animals shared a common M1 haplotype, suggesting a potential role for CD4+ T cells in the observed differential protection.
What role does the plasmid play in chlamydial infections and why is it maintained as an extrachromosomal element in this pathogen? Although the latter question remains unaddressed, it appears that some aspect of chlamydial plasmid biology is strongly selected for during successful infection or transmission of the pathogen within a host population (Russell et al., 2011). It is likely that this competitive advantage tips the scale toward pathogenesis in human infections. In addition, as repeated and/or persistent chlamydial infections are strongly associated with severe pathology, it is likely that inflammatory responses elicited by the bacterium are associated with serious disease sequelae. Thus, the plasmid may encode or control expression of proteins that are responsible for successful bacterial growth and/or of the stimulation of a deleterious immune response. The search for deleterious chlamydial antigens has a long history, and genus-common antigens are thought to be major players in this pathogenetic response. Perhaps a major immunopathogenetic determinant is encoded by the plasmid, or the gene products encoded/controlled by the plasmid allow persistence to a level that facilitates deleterious responses to other chlamydial macromolecules. Under the latter model, absence of the plasmid would allow the bacterium to be cleared before an immunopathogenic response can be generated or amplified. Alternatively, glycogen accumulation might be a metabolic equivalent of a virulence factor, providing energy stores as chlamydiae fight to grow and survive in a hostile host environment.
Although the mechanisms of attenuated virulence and/or of immune protection are not clearly defined by Kari et al., this study is a seminal contribution to the field of chlamydia pathogenesis and forms the basis for continued exploration of the role of the plasmid in the chlamydial infectious process. Future studies, perhaps encouraged by this work, will need to address the challenging issues of antigenic variation and host genetic variation in designing vaccine strategies.
Acknowledgments
Richard Morrison of the University of Arkansas for Medical Sciences is acknowledged for critical reading of this manuscript. Editorial comments were also provided by members of my laboratory.
The Rockey laboratory is supported by Public Health Service awards AI069214 and AI086469.
References
- An Q., Radcliffe G., Vassallo R., Buxton D., O’Brien W.J., Pelletier D.A., Weisburg W.G., Klinger J.D., Olive D.M. 1992. Infection with a plasmid-free variant Chlamydia related to Chlamydia trachomatis identified by using multiple assays for nucleic acid detection. J. Clin. Microbiol. 30:2814–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R., Maurelli A.T. 2009. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc. Natl. Acad. Sci. USA. 106:292–297 10.1073/pnas.0806768106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burall L.S., Rodolakis A., Rekiki A., Myers G.S., Bavoil P.M. 2009. Genomic analysis of an attenuated Chlamydia abortus live vaccine strain reveals defects in central metabolism and surface proteins. Infect. Immun. 77:4161–4167 10.1128/IAI.00189-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne G.I. 2010. Chlamydia trachomatis strains and virulence: rethinking links to infection prevalence and disease severity. J. Infect. Dis. 201:S126–S133 10.1086/652398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J.H., Whitmire W.M., Crane D.D., Wicke L., Virtaneva K., Sturdevant D.E., Kupko J.J., III, Porcella S.F., Martinez-Orengo N., Heinzen R.A., et al. 2008. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect. Immun. 76:2273–2283 10.1128/IAI.00102-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) 2011. CDC Grand Rounds: Chlamydia prevention: challenges and strategies for reducing disease burden and sequelae. MMWR Morb. Mortal. Wkly. Rep. 60:370–373 [PubMed] [Google Scholar]
- Chen D., Lei L., Lu C., Galaleldeen A., Hart P.J., Zhong G. 2010. Characterization of Pgp3, a Chlamydia trachomatis plasmid-encoded immunodominant antigen. J. Bacteriol. 192:6017–6024 10.1128/JB.00847-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A., Tischler P., Weinmaier T., Penz T., Heinz E., Brunham R.C., Read T.D., Bavoil P.M., Sachse K., Kahane S., et al. 2011. Unity in variety - the pan-genome of the Chlamydiae. Mol. Biol. Evol. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comanducci M., Manetti R., Bini L., Santucci A., Pallini V., Cevenini R., Sueur J.M., Orfila J., Ratti G. 1994. Humoral immune response to plasmid protein pgp3 in patients with Chlamydia trachomatis infection. Infect. Immun. 62:5491–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darville T., Hiltke T.J. 2010. Pathogenesis of genital tract disease due to Chlamydia trachomatis. J. Infect. Dis. 201:S114–S125 10.1086/652397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati M., Sambri V., Comanducci M., Di Leo K., Storni E., Giacani L., Ratti G., Cevenini R. 2003. DNA immunization with pgp3 gene of Chlamydia trachomatis inhibits the spread of chlamydial infection from the lower to the upper genital tract in C3H/HeN mice. Vaccine. 21:1089–1093 10.1016/S0264-410X(02)00631-X [DOI] [PubMed] [Google Scholar]
- Dugan J., Rockey D.D., Jones L., Andersen A.A. 2004. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob. Agents Chemother. 48:3989–3995 10.1128/AAC.48.10.3989-3995.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farencena A., Comanducci M., Donati M., Ratti G., Cevenini R. 1997. Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect. Immun. 65:2965–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris C.M., Morrison R.P. 2011. Vaccination against Chlamydia genital infection utilizing the murine C. muridarum model. Infect. Immun. 79:986–996 10.1128/IAI.00881-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredlund H., Falk L., Jurstrand M., Unemo M. 2004. Molecular genetic methods for diagnosis and characterisation of Chlamydia trachomatis and Neisseria gonorrhoeae: impact on epidemiological surveillance and interventions. APMIS. 112:771–784 10.1111/j.1600-0463.2004.apm11211-1205.x [DOI] [PubMed] [Google Scholar]
- Kari L., Goheen M.M., Randall L.B., Taylor L.D., Carlson J.H., Whitmire W.M., Virok D., Rajaram K., Endresz V., McClarty G., et al. 2011. Generation of targeted Chlamydia trachomatis null mutants. Proc. Natl. Acad. Sci. USA. 108:7189–7193 10.1073/pnas.1102229108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Makarova K.S., Aravind L. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709–742 10.1146/annurev.micro.55.1.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chen D., Zhong Y., Wang S., Zhong G. 2008. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect. Immun. 76:3415–3428 10.1128/IAI.01377-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longbottom D., Coulter L.J. 2003. Animal chlamydioses and zoonotic implications. J. Comp. Pathol. 128:217–244 10.1053/jcpa.2002.0629 [DOI] [PubMed] [Google Scholar]
- Matsumoto A., Izutsu H., Miyashita N., Ohuchi M. 1998. Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J. Clin. Microbiol. 36:3013–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell C.M., Nicks K.M. 2006. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology. 152:1601–1607 10.1099/mic.0.28658-0 [DOI] [PubMed] [Google Scholar]
- O’Connell C.M., Ingalls R.R., Andrews C.W., Jr, Scurlock A.M., Darville T. 2007. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J. Immunol. 179:4027–4034 [DOI] [PubMed] [Google Scholar]
- O’Connell C.M., AbdelRahman Y.M., Green E., Darville H.K., Saira K., Smith B., Darville T., Scurlock A.M., Meyer C.R., Belland R.J. 2011. Toll-like receptor 2 activation by Chlamydia trachomatis is plasmid dependent, and plasmid-responsive chromosomal loci are coordinately regulated in response to glucose limitation by C. trachomatis but not by C. muridarum. Infect. Immun. 79:1044–1056 10.1128/IAI.01118-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E.M., Markoff B.A., Schachter J., de la Maza L.M. 1990. The 7.5-kb plasmid present in Chlamydia trachomatis is not essential for the growth of this microorganism. Plasmid. 23:144–148 10.1016/0147-619X(90)90033-9 [DOI] [PubMed] [Google Scholar]
- Pickett M.A., Everson J.S., Pead P.J., Clarke I.N. 2005. The plasmids of Chlamydia trachomatis and Chlamydophila pneumoniae (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology. 151:893–903 10.1099/mic.0.27625-0 [DOI] [PubMed] [Google Scholar]
- Rockey D.D., Wang J., Lei L., Zhong G. 2009. Chlamydia vaccine candidates and tools for chlamydial antigen discovery. Expert Rev. Vaccines. 8:1365–1377 10.1586/erv.09.98 [DOI] [PubMed] [Google Scholar]
- Russell M., Darville T., Chandra-Kuntal K., Smith B., Andrews C.W., Jr, O’Connell C.M. 2011. Infectivity acts as in vivo selection for maintenance of the chlamydial cryptic plasmid. Infect. Immun. 79:98–107 10.1128/IAI.01105-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith H.M., Harris S.R., Persson K., Marsh P., Barron A., Bignell A., Bjartling C., Clark L., Cutcliffe L.T., Lambden P.R., et al. 2009. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 10:239 10.1186/1471-2164-10-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey C.C., Lusher M., Richmond S.J. 1989. Analysis of the complete nucleotide sequence of Chp1, a phage which infects avian Chlamydia psittaci. J. Gen. Virol. 70:3381–3390 10.1099/0022-1317-70-12-3381 [DOI] [PubMed] [Google Scholar]
- Thomas N.S., Lusher M., Storey C.C., Clarke I.N. 1997. Plasmid diversity in Chlamydia. Microbiology. 143:1847–1854 10.1099/00221287-143-6-1847 [DOI] [PubMed] [Google Scholar]
- Unemo M., Clarke I.N. 2011. The Swedish new variant of Chlamydia trachomatis. Curr. Opin. Infect. Dis. 24:62–69 10.1097/QCO.0b013e32834204d5 [DOI] [PubMed] [Google Scholar]
- Wang Y., Kahane S., Cutcliffe L., Skilton R., Lambden P., Clarke I. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 7:e1002258 10.1371/journal.ppat.1002258 [DOI] [PMC free article] [PubMed] [Google Scholar]