Many HIV-1 envelope-reactive antibodies shortly after HIV-1 transmission may arise from crow-reactive memory B cells previously stimulated by non-HIV-1 host or microbial antigens
Abstract
The initial antibody response to HIV-1 is targeted to envelope (Env) gp41, and is nonneutralizing and ineffective in controlling viremia. To understand the origins and characteristics of gp41-binding antibodies produced shortly after HIV-1 transmission, we isolated and studied gp41-reactive plasma cells from subjects acutely infected with HIV-1. The frequencies of somatic mutations were relatively high in these gp41-reactive antibodies. Reverted unmutated ancestors of gp41-reactive antibodies derived from subjects acutely infected with HIV-1 frequently did not react with autologous HIV-1 Env; however, these antibodies were polyreactive and frequently bound to host or bacterial antigens. In one large clonal lineage of gp41-reactive antibodies, reactivity to HIV-1 Env was acquired only after somatic mutations. Polyreactive gp41-binding antibodies were also isolated from uninfected individuals. These data suggest that the majority of gp41-binding antibodies produced after acute HIV-1 infection are cross-reactive responses generated by stimulating memory B cells that have previously been activated by non–HIV-1 antigens.
Initial antibody responses to transmitted/founder HIV-1 envelope (Env) do not arise until ∼13 d after the onset of viremia, target gp41, and are nonneutralizing (Tomaras et al., 2008). Whereas early T cell responses to HIV-1 that are coincident with these initial antibody responses drive viral evolution for escape mutants, the early gp41 Env antibody response does not (Bar et al., 2009; Goonetilleke et al., 2009; McMichael et al., 2010). Instead, the first antibodies capable of selecting viral mutants are gp120 autologous neutralizing antibodies that appear only months after transmission (Richman et al., 2003; Wei et al., 2003; Moore et al., 2009).
The progeny of B cells that respond initially to microbial pathogens or vaccines may be sampled in the transient plasmacytosis that appears in the circulation ∼7 d after immunization (Falkoff et al., 1983; Wrammert et al., 2008). To define the clonal nature of early humoral responses to HIV-1 Env, we isolated single CD19+, CD27hi, CD38hi, and CD20lo or CD20− plasmablasts and/or plasma cells (hereafter termed plasma cells) from the blood or bone marrow of acute HIV-1 infection (AHI) subjects and used RT-PCR to amplify rearranged variable regions of Ig heavy and light chain genes (VH and VL, respectively; Wardemann et al., 2003; Tiller et al., 2008; Wrammert et al., 2008; Liao et al., 2009) for Ig gene analysis and for production of recombinant mAbs. AHI subjects were studied at ∼17–30 d after HIV-1 transmission, during a period of plasmacytosis. The levels of antibody mutation frequencies during this period of plasmacytosis were compared with those induced by primary HIV Env immunization in uninfected subjects. Our analysis demonstrated that HIV-1–reactive antibodies in the initial response to HIV-1 in the setting of AHI were more somatically hypermutated than Env antibodies isolated after primary HIV-1 Env vaccination. Analysis of VH sequences of genomic DNA by 454 deep sequencing of four HIV-1 Env-reactive clonal lineages from AHI subjects did not reveal any unmutated lineage members. Similarly rare gp41-reactive mutated mAbs could be isolated from uninfected subjects. These data suggested that many initial Env antibodies in AHI may arise from preexisting mutated Env-cross reactive memory B cells.
RESULTS
The plasma cell response in AHI
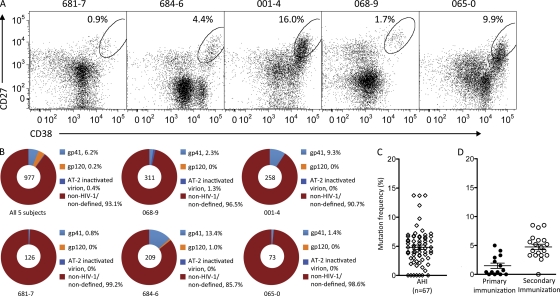
The frequency (mean percentage) of plasma cells in AHI subjects was 6.5 ± 2.8% of total B cells (Fig. 1 A). We produced 977 recombinant mAbs from plasma cells of five AHI subjects (Table I and Fig. 1 B). To ensure that no HIV-1 Env antibodies were missed, autologous HIV-1 gp140 proteins derived from the single transmitted/founder virus from subjects 684–6 and 681–7, as well as a group M consensus Env gp140 and a clade B recombinant (r) gp41, were used to screen antibodies in binding assays (Tomaras et al., 2008).
Figure 1.
Plasma cell response in AHI. (A) Sorting of plasma cells from AHI subjects (681–7, 684–6, 001–4, 0689 and 065–0). Dot plots were gated on CD3−CD14−CD16−CD235a−CD19+ B cells (cells were also gated as CD20lo/− during sorting). Ovals designate the populations that were sorted as plasma cells (CD27hi/+ and CD38hi/+). Numbers indicate percentage of cells in the sort gate. LKP, leukapheresis cells. (B) Specificity of antibodies isolated from plasma cells from each subject, or from all five AHI subjects pooled together. Total numbers of antibodies were indicated at the center of each pie chart. Percentage of antibodies binding to gp41, gp120, AT-2 inactivated HIV-1 clade B virion, and non–HIV-1/nondefined antigens are indicated in different colors. (C and D) Frequency of somatic mutations in VH gene segments of antibodies isolated from all five AHI subjects or from individuals 14 d after the second immunization (42 d after first dose) or 14 d after the fourth immunization (182 d after first dose) with gp120/NefTat/AS01B (D).
Table I.
Clinical information of 5 AHI patients
| Subjectsa | Age (gender) | Days since infectionb | Viral load copies/ml | CD4 count cells /µl | Plasma anti–HIV-1 gp41 | Fiebig stagec | ||||
| EIA | Western blot | IgM | IgG | IgA | ||||||
| 065-0 | 35 (M) | 17 | 391 | 269 | Neg. | Neg. | Neg. | Neg. | Neg. | 2/1 |
| 068-9d | 37 (M) | 30 | 258 | 611 | Pos. | Neg. | Pos. | Neg. | Neg. | 3 |
| 681-7 | 42 (M) | 20 | 5,900,000 | 230 | Neg. | Neg. | Neg. | Neg. | Neg. | 2/1 |
| 684-6 | 32 (M) | 20 | 10,925,000 | 225 | Pos. | Neg. | Pos. | Pos. | Pos. | 3 |
| 001-4 | 26 (M) | 20 | 4,700,000 | 165 | Pos. | Ind. | Pos. | Pos. | Pos. | 4 |
EIA, enzyme immunoassay; Neg., negative; Pos., positive; Ind., indeterminate, as one or more bands are present but the blot did not meet the criteria for positive in that at least one of gp160 and/or gp120, gp41, or gp24 must be present.
Patient no. 068-9 received ART for 7 d after exposure, 21 d before blood and bone marrow samples were obtained from.
Days since infection were determined by clinical information of last contact or by days from onset of AHI symptoms. All patients had admitted to hospitals of recent contact except 684-6, who had AHI symptoms and had negative Western blot analysis with high plasma viral load.
Based on description reported by Fiebig et al (2003).
All five subjects were quickly placed on antiretroviral therapy soon after diagnosis and consequently none developed detectable autologous or heterologous neutralizing antibodies during up to 2 yr of follow up (unpublished data).
Of 977 mAbs from AHI subjects, 67 (6.9%) were HIV-1 Env reactive, and of those 67 mAbs, 61 (6.2%) were reactive with gp41, 2 (0.2%) were reactive with gp120, and 4 (0.4%) were reactive with only aldrithol-2 (AT-2)–inactivated HIV-1 clade B virions (Fig. 1 B and Table S1). Plasma cell–derived IgA1, IgG1, and IgG3 isotype usage predominated for HIV-1–reactive and non-HIV-1–reactive antibodies (Table S1). VH gene family usage in AHI HIV-reactive antibodies were 19.0% VH1, 54.0% VH3, and 12.7% VH4, respectively, and were similar to the distribution of VH families in 34,384 VH sequences collected in the National Center for Biotechnology Information database (Table S2).
The average complementarity-determining region (CDR) H3 length of AHI antibodies in amino acid residues was 15.1 ± 0.4 (Table S3), and mutation frequency in VH gene segments was 5.0 ± 0.4% (Fig. 1 C and Table S4). There were no significant differences in mutation frequencies among Env gp41–, Env gp120–, or AT-2–inactivated virion-reactive antibodies (Fig. S1). The frequency of VH mutations in acute HIV-1 antibodies was high when compared with antibodies from the reported primary response to rotavirus infection (0.3%; Weitkamp et al., 2003), and were more similar to mutation frequencies of antibodies from the previously reported secondary responses to influenza vaccination (6.5%; Wrammert et al., 2008).
Mutation frequency in antibodies from a primary HIV-1 Env immunization
To determine the mutation frequency of a true primary response to HIV-1 Env in the setting of vaccination, we isolated 14 anti-gp120 Env antibodies 14 d after the second immunization (total of 42 d after the first priming immunization) with recombinant HIV-1 W61D gp120 and 20 anti-gp120 Env antibodies 14 d after the fourth immunization (total of 182 d after the first prime) with the same immunogen (Leroux-Roels et al., 2010). We found that 4 of 14 HIV-1–reactive Env antibodies (29%) isolated 14 d after 2 immunizations (42 d after initial immunization) had unmutated VH sequences, and the mean VH mutation frequency of these 14 antibodies was 1.5% (±0.5%; n = 14; Fig. 1 D and Table S5). In contrast, the mean VH mutation frequency of 20 antibodies isolated 182 d after 4 Env immunizations was 4.8% (±0.5%; Fig. 1 D and Table S5). The total number of unmutated HIV-1–reactive antibodies found in AHI was 7 of 67 (10.4%; all gp41 reactive; Fig. 1 C and Fig. S1), and was lower than that found in the unmutated antibodies (28.6%) isolated after primary HIV-1 Env vaccination (P = 0.09; Fisher’s exact test). Furthermore, the mean VH mutation frequency of the plasma cell antibodies 17–30 d after transmission in AHI (5.0 ± 0.4%) was significantly higher than that of antibodies from the day 42 primary response to Env immunization (1.5 ± 0.5%; P < 0.0001, Student’s t test)
Clonality of the Env antibody response in HIV-1 acute infection
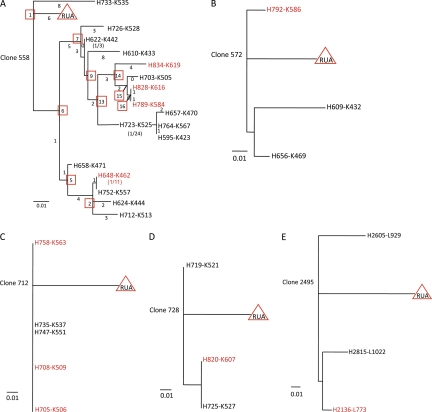
Within 977 AHI plasma cell–derived antibodies, we identified 35 clonal lineages, of which 12 contained members reactive with HIV-1 gp41 Env (Table S6). Antibodies within five HIV-1 gp41-reactive clonal lineages were expressed and studied in this work (Fig. 2, Table II, and Tables S7–S11). One surprisingly large clonal family (clonal lineage 558 from subject 684–6 ∼20 d after transmission) contained 51 members (17 unique; Fig. 2 A and Table S7).
Figure 2.
Phylogenetic trees of antibody clonal lineages derived from AHI. Clonal lineages 558 (A), 572 (B), 712 (C), 728 (D), and 2495 (E) were obtained from AHI subject 684–8. Clonal lineage 2495 was obtained from AHI subject 001–4. Antibodies highlighted in red for each clonal lineage were gp41 reactive and antibodies labeled in black font were gp41-negative in the initial screening. The length of lines in scale indicates extent of nucleotide differences between individual antibodies. Numbers underneath the scale lines indicate frequency of mutation; 0.01 = 1% sites mutated or 0.1 = 10% sites mutated. Inferred unmutated ancestors (RUAs) are indicated by red triangles in each clonal lineage. For clonal lineage 558 (A), numbers under or beside lines denote numbers of nucleotides of VH and VK chain genes of individual antibodies that differ from each other, and numbers in parentheses indicate the number of antibodies that had identical sequences. Inferred reverted intermediate antibodies are indicated by red square boxes for clonal lineage 558.
Table II.
Plasma cell clonal lineages studied from AHI
| Clone ID | Reactivity | Subject | Ig class, isotype (number) | VH | DH | JH | VK or VL | JK or JL | No. of members |
| 558 | HIV-1 gp41 | 684-6 | IgG3(50), G1(1) | 3–7 | 1–26 | 5 | 1–39 | 4 | 51 |
| 572 | HIV-1 gp41 | 684-6 | G1(2), G3(1) | 3–48 | 2–2 | 6 | 3–20 | 4 | 3 |
| 712 | HIV-1 gp41 | 684-6 | A1 | 3–23 | 3–10 | 5 | 1–39 | 4 | 5 |
| 728 | HIV-1 gp41 | 684-6 | G1(2), A1(1) | 1–3 | 2–15 | 6 | 1–39 | 4 | 3 |
| 2495 | HIV-1 gp41 | 001-4 | G3 | 1–2 | 2–21 | 3 | 3–21 | 2 | 3 |
We considered two possible origins for the gp41-reactive, highly mutated B cells recovered from AHI subjects. First, HIV-1 gp41 Env may trigger a naive B cell to proliferate and mutate to this degree, despite reductions of CD4+ T cell numbers and disruption of mucosal germinal centers that occur in AHI (Brenchley, et al., 2004; Levesque et al., 2009). Although it is possible, we thought that this scenario was less plausible because of the low number of mutations that develop in the time from HIV-1 transmission to the time when blood is collected for plasma cell sorting (∼17–30 d; Keele et al., 2008). Second, HIV-1 gp41 may trigger preexisting mutated B cells that cross react with gp41, and after stimulation by gp41Env, Env may become the trigger for further antibody affinity maturation.
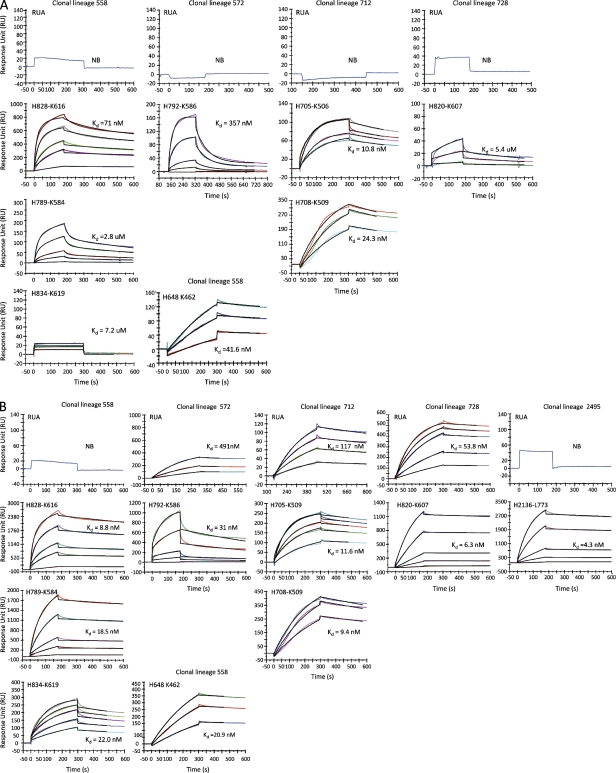
To directly test the hypothesis that autologous Env reactivity in AHI clonal lineages can arise distal to a founder naive B cell, we used somatic diversification analysis-2 (SoDA-2; Volpe et al., 2006; Munshaw and Kepler, 2010) and DNA maximum likelihood (Felsenstein, 1973, 1981, 2005) programs to infer reverted unmutated ancestors (RUAs) of clonal lineages 558, 572, 712, and 728 (Fig. 2, A–D; and Tables S7–11) from subject 684–6 and clonal lineage 2495 from subject 001–4 (Fig. 2 E and Table S11), and also to infer reverted intermediate antibodies in clonal lineage 558 (Fig. 2 A and Table S7). Inferred RUAs and intermediates and observed antibodies from subject 684–6 were assayed against recombinant (r) Env gp140 derived from the autologous transmitted/founder virus (Keele et al., 2008).
As naive B cells responding to antigens often have low affinity binding in vivo (Dal Porto et al., 2002; Shih et al., 2002), we used Luminex assays and surface plasmon resonance (SPR) to assess the Env reactivity of RUAs of HIV-1 Env-reactive clonal lineages from AHI, RUAs, and observed antibodies. We found that none of the four RUAs from subject 684–6 reacted with autologous rgp140 in SPR assays (Fig. 3 A), whereas observed antibodies from each clone did react with autologous rgp140 in SPR assays (Fig. 3 A). Thus, detectable reactivity of all 4 clones from subject 684–6 with autologous transmitted/founder rgp140 appeared to be acquired after somatic hypermutation of the RUA. Interestingly, 3 of 4 subject 684–6 RUAs from lineages 572, 712, and 728 did cross react with heterologous clade B rgp41 in both SPR (Fig. 3 B) and Luminex assays (Fig. S2 A). This finding suggested either that cross-reactivity of the RUA with rgp41 reflected epitope similarity of rgp41 with the non–HIV-1 antigen that triggered the gp41 antibodies, or suggested that autologous recombinant 684–6 rgp140 protein does not express the gp41 epitopes that were expressed on native 684–6 transmitted/founder Env in virions in vivo. An additional gp41 clonal lineage from subject 001–4 (clone 2495; Fig. 2 E and Table S11) was also studied, and the 2495 RUA, like the RUA of clonal lineage 558, was rgp41 nonreactive, whereas the lineage 2495 observed antibody (H2136-L773) was strongly rgp41 reactive in SPR assay (dissociation constants [Kd] = 4.3 nM; Fig. 3 B), as well as in Luminex assays (Fig. S2 A).
Figure 3.
Binding of RUA antibodies and observed antibodies to 684–6 transmitted/founder gp140 and to HIV-1 MN rgp41 was analyzed by SPR. RUAs and observed antibodies were derived from HIV-1 gp41-reactive clonal lineages 558, 572, 712, and 728 as indicated at the top of each graph. Approximately 1,000–3,000 Response Units of 684–6 gp140 (A) or HIV-1 MN rgp41 (B) were immobilized on adjacent flow cells of the same sensor chip. Each antibody was injected in a dose range starting at 100 µg/ml for 3 min at 30 µl/min on a BIAcore 3000 instrument (GE Healthcare) using PBS, pH 7.4, as running buffer. For binding Kd measurements, each antibody that bound to 684–6 gp140 (A) or to HIV-1 MN rgp41 (B) was injected at concentrations ranging from 80 to 1µg/ml and at a flow rate of 50 µl/min for 3min. Data are representative of three separate experiments.
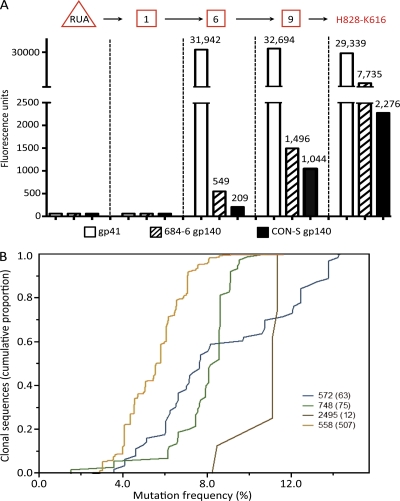
Next, observed antibodies, the RUA, and intermediate antibodies of clonal lineage 558 (Fig. 2 A and Table S7) were assayed against HIV-1 clade B rgp41, autologous rgp140, and group M consensus (CON-S) rgp140 in Luminex assays (Tomaras et al., 2008) to determine where the HIV-1 Env reactivity of antibodies in clone 558 was acquired (Fig. 4). Although neither clone 558 RUA nor the earliest intermediate antibody (no. 1) reacted with rgp41, autologous rgp140, or CON-S rgp140, the reverted intermediates 6 and 9 and observed antibody H828-K616 all bound well to the three Env proteins (Fig. 4 A). Thus, in clone 558, autologous Env and rgp41 did not react with the inferred RUA or with intermediate no. 1, suggesting that Env reactivity was indeed acquired after somatic mutations had been induced in the RUA and inferred intermediate no. 1 by an as yet unknown antigen.
Figure 4.
Characteristics of anti–HIV-1 gp41 Env clonal lineages. (A) Analysis of the reactivity of inferred RUA, intermediates, and observed antibodies in clonal lineage 558 with HIV-1 rgp41, autologous Env gp140, and CON-S Env gp140. The RUA (triangle) and reverted intermediates 1, 6, and 9 (square boxes), as well as observed antibody H828-K616, were assayed for reactivity with rgp41, autologous 684–6 gp140, and the group M consensus gp140 CON-S by Luminex assays. (B) VH sequences of clonal lineages identified by 454 deep sequencing. Shown are the numbers of clonal sequences (on the y axis) with the percentage of nucleotide change from germline sequences (on the x axis) for each of HIV-1 Env-reactive clonal lineage m (indicated by clonal lineage number with number of sequences found in parentheses) identified by 454 deep sequencing from genomic DNA of B cells of acute HIV-1 subjects. Sequences with identical V, D, and J segment usage and identical V-D and D-J junctions were considered to be members of an individual B cell clone. Data are representative of three separate experiments.
It was important to determine evidence of antigen drive in clonal lineage 558. Selection for replacement mutations was reflected in synonymous and nonsynonymous mutations in clone 558 VH and VL CDR and framework regions (Table S12). Whereas 85.7% of Ig CDR mutations were nonsynonymous, only 68.8% of framework mutations were nonsynonymous (χ2 = 15.3; P < 10−6).
We also asked if the affinity of antibody binding to rgp41 increased as somatic mutations accumulated. We produced additional reverted inferred intermediate antibodies of lineage 558 (Fig. 2 A), evaluated the binding of these antibodies to HIV-1 Env rgp41 using Luminex assays (Tomaras et al., 2008), and determined their Kd of binding to HIV-1 Env rgp41. With the accumulation of mutations, the individual apparent affinities (Kd) of clone members for gp41 binding in general increased, e.g., from 63.3 nM at inferred intermediate no. 6 to 4.7 nM at antibody H789-K584, and to 0.6 nM at observed antibody H828-K616 (Table III).
Table III.
Apparent binding affinity of observed, RUA, and intermediate antibodies in clonal lineage 558 to HIV-1 gp41
| Antibody ID | EC50 | Apparent Kd |
| µg/ml | nM | |
| H789-K584 | 0.7 | 4.7 |
| H684-K462 | 56.3 | 375.3 |
| H828-K616 | 90 | 0.6 |
| CL558_RUA | NB | NA |
| CL558_1 | NB | NA |
| CL558_2 | 26.7 | 0.2 |
| CL558_5 | 22.8 | 152 |
| CL558_6 | 9.5 | 65.3 |
| CL558_7 | 7.8 | 52 |
| CL558_9 | 25.7 | 171.3 |
| CL558_13 | 18.2 | 121.3 |
| CL558_14 | 21.9 | 146 |
| CL558_15 | 5.3 | 35.3 |
| CL558_16 | 2.1 | 14 |
Binding of antibodies to HIV-1 gp41 was determined in Luminex assays at antibody concentrations ranging from 50 to 0.2 µg/ml. NB, No detectable binding; NA, not applicable.
We next asked if deep sequencing could reveal examples of unmutated or less extensively hypermutated VH sequences of antibodies in gp41 clonal lineages. Thus, we performed 454 deep sequencing on V(D)J gene segments (Boyd et al., 2009) of genomic DNAs derived from PBMCs of all 5 acute HIV-1 subjects and obtained an average depth of total of 3,300 Ig VH sequences per subject. We found the VH sequences of four HIV-1 gp41 clonal lineages (Fig. 4 B) that had been initially identified among antibodies derived from the sorted single plasma cells (Table S6). These included 63 VH sequences with mutation frequencies ranging from 3.5 to 13.5% for clone 572, 75 VH sequences with mutation frequencies from 1.5 to 10.8% (median, 11.1%) for clone 748, 12 VH sequences with mutation frequencies from 8.2 to 11.4% (median, 11.1%) for clone 2495, and, finally, 507 VH sequences with mutation frequencies ranging from 2.5 to 11.6% (median, 5.6%) for clonal lineage 558 (Fig. 4 B and Table S6). Importantly, we found no unmutated VH sequences, or even any sequences with fewer than three mutations, in any of these four clonal lineages, a finding compatible with the hypothesis that many of the responding antibodies to gp41 stimulation are derived from preexisting memory B cells.
Polyreactive nature of the initial antibody response to Env
We determined polyreactivity of AHI gp41 antibodies with gut flora whole-cell lysate antigens by Luminex assays (Tomaras et al., 2008). Reactivity of the RUAs and/or observed antibodies in clonal lineages 558, 572, and 728 with gut flora was present but weak (Fig. S2 A), whereas the RUAs of clonal lineage 2495 did not react with gut flora antigens (Fig. S2 A). In contrast, the three observed gp41 antibodies of lineage 712 reacted strongly with gut flora (Fig. S2 A), while the RUA of clonal lineage 712 did not react with gut flora antigens (Fig. S2 A).
We also tested RUAs and observed antibodies from clones 558, 572, 712, 728, and 2495 for polyreactivity by assaying for reactivity with HEp-2 epithelial cells. Although neither the clonal lineage 558 RUA nor inferred intermediate 1 reacted with HEp-2 cells, inferred intermediate 6 did—a change in specificity that coincided with the acquisition of reactivity for rgp41. In contrast to the increase in affinity for rgp41 in the 558 lineage, reactivity to HEp-2 cells in the clonal lineage waxed and waned (Fig. 5).
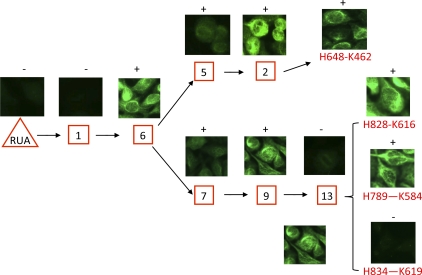
Figure 5.
Binding of the RUA, inferred reverted intermediate antibodies, and observed antibodies in clonal lineage 558 to HEp-2 epithelial cells in the indirect immunofluorescence staining assays. Numbers in square boxes are inferred intermediate antibodies. The observed antibody members are highlighted in red font. Positive or negative immunofluorescence staining of antibodies on HEp-2 epithelial cells is indicated above the pictures for the indicated antibodies. Data are representative of three separate experiments.
In the other lineages, several patterns of HEp-2 reactivity were seen (Fig. S2 B). In lineage 572, the RUA and observed mAb H656-K469 were HEp-2 reactive, whereas observed mAb H792-K586 was nonreactive. Similarly, the RUA and observed antibodies H705-K506, H708-K509, and H758-K563 in clonal lineage 712 were HEp-2 reactive. The RUA of clonal lineage 728 was also HEp-2 positive, but observed mAb H820-K607 was negative. Finally, the RUA of clonal lineage 2495 was similar to lineage 558 and was not polyreactive for HEp-2 cells (Fig. S2 B), whereas observed gp41 antibody H2136-L773 in lineage 2495 was polyreactive (Fig. S2 B).
The specificity of gp41-reactive antibodies in clone 558 was to a conformational determinant on rgp41 recognized best on blue native gels (Fig. S3 A). Interestingly, although weakly reactive with gut flora in Luminex (Fig. S2 A), lineage 558 gp41 antibody CL558_2 cross reacted with a 520-kD molecule present in both aerobic and anaerobic gut flora lysates, providing candidates for one source of non–HIV-1 antigens that could prime for cross-reactive antibody responses with HIV-1 Env (Fig. S3 B).
Regarding AHI gp41 antibody neutralization of HIV-1, as expected (Tomaras et al., 2008), none of the observed Env antibodies neutralized the autologous transmitted/founder viruses of subject 684–6 in pseudovirus neutralization assays (not depicted).
One explanation for the polyreactivity seen in HIV-1 gp41 RUAs and their progeny is that HIV-1 may be activating a polyreactive pool of B cells. Tsuiji et al. (2006) have demonstrated that a subset of the human IgM+, CD27+ memory (mutated) B cell compartment is reactive with bacterial antigens. We found that 8 of 11 (72.7%) of the observed gp41 antibodies in the clonal lineages tested reacted with antigens present in lysates of gut flora (Fig. S2 A). Thus, we hypothesized that, if a subset of the IgM memory pool of B cells were being preferentially activated in AHI, then rgp41+ mutated IgM B cells should also be present in acute infection.
In this regard, 4 of 61 rgp41 mAbs isolated from blood plasma cells were IgMs. Although 1 IgM was unmutated, the VH mutations in 3 IgM gp41 mAbs were 4.0, 4.8, and 5.9%. In addition, using Epstein-Barr virus (EBV) transformation (Levesque et al., 2009), we isolated a mutated human VH4-4 IgM mAb 1E7 (VH mutation frequency, 1.9%) from bone marrow of a subject with recent HIV-1 infection that reacted with HIV-1 gp41 Env and cross reacted with gut flora antigens (Fig. S4), but did not neutralize HIV-1 (unpublished data). Thus, polyreactive nonneutralizing mutated IgM antibodies are being activated in AHI that react with HIV-1 Env rgp41.
Mutated HIV-1 Env-reactive antibodies are present in HIV-1 uninfected subjects
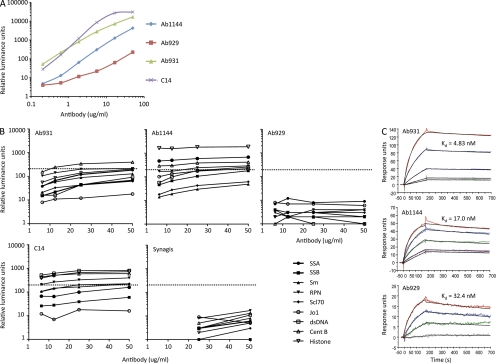
If the notion is plausible that the initial HIV-1 Env responses can be generated, at least in part, by HIV-1 Env gp41 interacting with memory B cells that express a B cell receptor that has been mutated as a result of stimulation by a non–HIV-1 antigen, then such antibodies should exist in HIV-1–uninfected subjects. We have identified 3 IgG1 antibodies (Ab1144, Ab929, and Ab931) from a total of 270 antibodies generated from plasma cells sorted from PBMC of 2 uninfected subjects using the same plasma cell sorting strategy for isolation of AHI, and one IgM antibody generated by EBV transformation of B cells from an uninfected terminal ileum (C14; Table IV and Fig. 6 A). All these antibodies reacted with 4 HIV-1 Env rgp41 and were mutated with a mean VH mutation frequency of 7.0 ± 3.0%. Two of three IgG and one IgM gp41-reactive antibodies were polyreactive with reactivity to multiple autoantigens (Fig. 6 B), although not to our gut flora whole-cell lysate (not depicted). In SPR assays, three IgG1 antibodies (Ab1144, Ab929, and Ab931) isolated from an uninfected subject (022–3) had HIV-1 gp41-binding affinity of 4.8 , 17, and 32.4 nM (Fig. 6 C), respectively, which are similar in affinity to gp41-positive antibodies isolated from acute HIV-1 subjects (Fig. 3 B).
Table IV.
HIV-1 Env-reactive antibodies isolated from uninfected subjects
| VH | VK/L | |||||||||
| PTID | ID | Isotype | Family | Mutation frequency | CDR3 length | ID | Family | Mutation frequency | CDR3 length | Specificity |
| % | aa | % | aa | |||||||
| 022-3 | H0929 | G3 | 3–23 | 7.7 | 18 | λ0308 | VL1–47 | 0.035 | 11 | gp41 |
| 022-3 | H0931 | G3 | 4–61 | 10.4 | 24 | κ0692 | VK3–15 | 8.6 | 11 | gp41 |
| 022-3 | H1141 | G1 | 3–48 | 7.5 | 17 | λ0378 | VL3–10 | 3.6 | 11 | gp41 |
| C14 | HC14-2 | M | 3–74 | 3.0 | 8 | λC14-2 | VL1–44 | 0.7 | 12 | gp41 |
Figure 6.
Reactivity of antibodies isolated from uninfected subjects. Three IgG antibodies (Ab1144, Ab929, and Ab931) and one IgM antibody (C14) in threefold dilutions ranging from 50 to 0.2 µg/ml (x axis) were evaluated for reactivity with HIV-1 rgp41 by Luminex assays (A) and for reactivity with a panel of autoantigens by ANA assays (B). The dotted lines indicate the cutoff values ≥120 luminance units used to denote positivity. Binding affinity of Ab1144, Ab929, and Ab931 in dose ranges (starting concentration at 40 µg/ml) to HIV-1 MN gp41 was measure by SPR (C). Data are representative of at least two separate experiments.
DISCUSSION
In this study, we have demonstrated that the HIV-1 Env gp41 plasma cell repertoire of subjects early in AHI is frequently polyreactive with host cell and gut flora antigens and that polyreactive antibodies that cross react with HIV-1 Env can be isolated from HIV-1 uninfected subjects.
Although ∼30% of antibodies isolated 42 d after HIV-1 Env priming with 2 immunizations were unmutated, only ∼10.4% of plasma cell–derived AHI HIV-1 antibodies were unmutated. Moreover, in the Env clonal response in AHI, VH deep sequencing did not reveal any unmutated members of four of the Env clonal lineages in this study. In a primary vaccination response to HIV-1 Env, we found the VH mutation frequency to be 1.5% (Fig. 1 D). Even after a secondary boosting in an Env vaccine setting, we were able to find 1 of 20 Env antibodies that were unmutated. Similarly, in a primary antibody response to rotavirus infection, Weitkamp et al. found a VH mutation rate of 0.3% (Weitkamp et al., 2003). In contrast, Wrammert et al. (2008) found the mutation rate in a secondary response to influenza vaccine vaccination to be 6.5%. Thus, HIV-1–reactive antibodies in the initial response to HIV-1 Env in AHI have mutation frequencies that are more consistent with that of antibodies in a secondary antibody response than that of antibodies in an Env primary response. These analyses raised the hypothesis that at the time of AHI, HIV-1 preferentially activated preexisting memory B cells that had been originally triggered by non–HIV-1 antigens, but whose BCRs cross react with HIV-1 gp41, and upon contact with gp41 undergo antigen drive and clonal expansion. This hypothesis has been tested in three ways.
First, we isolated one clonal lineage (558) from AHI that was sufficiently large to infer an RUA and reverted intermediates, and demonstrated that the gp41 reactivity of this clonal lineage with the autologous Env was acquired after the RUA and one intermediate. Second, we asked if mutated Env-reactive B cells could be isolated from the plasma cell repertoires of uninfected subjects, and found that, indeed, uninfected subjects have gp41 Env-reactive B cells with B cell receptors that both bind Env and cross react with non-HIV-1 antigens. Third, we asked if mutated IgM mAbs could be generated from HIV-1 infection that bind gp41 and gut flora, and we found one such IgM mAb 1E7 (Fig. S4). This reactivity with gut flora could indicate that it is primarily gut flora–specific B cells that are triggered in AHI by HIV-1 Env gp41, or it could mean only that the initial responsive B cells to HIV-1 Env are polyreactive and gut flora antigens are simply one such common cross reaction.
It should be noted that although we used very sensitive methods (SPR) for studying the reactivity of unmutated ancestor antibodies with gp41 and autologous gp140 Env, a physiologically relevant interaction of gp41 with a membrane-bound naive B cell receptor may have occurred in vivo, leading to B cell triggering. Our studies cannot rule out this possibility.
It is not known why many of the initial responding Env antibodies in AHI are polyreactive. We have previously shown that some of the rare broadly neutralizing mAbs are polyreactive (Haynes et al., 2005), and for some of these antibodies, such as those that are directed to the gp41 membrane proximal neutralizing determinants, their polyreactivity appears to induce tolerance control of production of these types of antibodies (Verkoczy et al., 2009). Mouquet et al. (2010) have demonstrated that, in HIV-1 chronically infected subjects who have plasma broad neutralizing activity, ∼75% of all Env antibodies were polyreactive and that polyreactivity facilitates high-affinity binding to virions with few Env spikes. Thus, one possibility is that HIV-1 Env is processed in the spleen, lymph nodes, and MALT in such a manner as to preferentially activate polyreactive B cells. If this is the case, then the antibodies that are induced in Env vaccination would have the same degree of polyreactivity as seen in HIV-1 infection. Studies are underway to address this issue.
The degree of polyclonality in the AHI response was striking, particularly in contrast to the highly clonal response seen in influenza vaccination (Wrammert et al., 2008) and in pandemic H1N1 swine influenza infection (Wrammert et al., 2010). However, a similar polyclonal response has been seen in mice in γ herpes virus infections (Sangster et al., 2000) and in humans with seasonal influenza infections (unpublished data). Thus, this degree of polyclonal activation is not unique to AHI.
We and others have previously reported Env-reactive B cells from HIV-1–uninfected subjects (Berberian et al., 1993; Chin et al., 1995; Chen et al., 2009; Levesque et al., 2009). The importance of our studies here is that with the same methods as used in AHI subjects, we can isolate rare mutated IgG and IgM antibodies with binding properties similar to those isolated from AHI (Fig. 6 and Table III), a finding that is critical to the hypothesis that HIV-1 Env can activate previously activated, mutated memory B cells that have Env cross reactivity. Moreover, we have found a range of reactivity of uninfected human sera to gp41 Env recombinant proteins (unpublished data). Several studies, including studies presented here, have demonstrated with a variety of techniques that HIV-1–reactive B cells are present before infection in humans. Our present studies suggest that a subset of these B cells is involved in the initial response to HIV-1 Env gp41 in AHI.
Why gp120 antibodies that neutralize the autologous transmitted/founder virus only develop 3 mo or longer after transmission—too late to prevent or control infection—is not clear (Wei et al., 2003; Richman et al., 2003; Keele et al., 2008). This initial gp41 antibody response in AHI does not select HIV-1 escape mutants (unpublished data). That a nonneutralizing, nonprotective gp41 response occurs first, and the potentially protective gp120 response occurs late, suggests some type of immune modulation or direction by gp41 away from an initial gp120 response. It will be of great interest to probe the plasma cell and memory B cell repertoires of HIV-1 subjects as they evolve autologous neutralizing antibody responses, and to determine the fates of gp41 clonal lineages as gp120 clonal lineages arise.
In summary, our study provides considerable new insight into the dysfunctional nature of the initial antibody response to HIV-1 in AHI. That the initial antibodies are polyreactive and more mutated than expected provides evidence consistent with the hypothesis that HIV-1 diverts the initial antibody response to Env by stimulating mutated B cells that have been previously activated by non-HIV-1 antigens, thus rendering the earliest antibody responses to HIV-1 nonprotective.
MATERIALS AND METHODS
Study subjects.
Blood and bone marrow samples were collected from 5 AHI subjects at 17 to 30 d after transmission, as estimated from patient history and Fiebig classification (Table I) for plasma cell sorting and for isolation of genomic DNA for sequence analysis of V(D)J segments of Ig heavy chain genes by the 454 platform with Titanium chemistry (Roche). Table I shows the characteristics of the subjects studied. Subjects 065–0 and 641–7 were at Feibig Stage 1 according to the Fiebig classification (Fiebig et al., 2003), whereas subjects 068–9, 684–6, and 001–4 were at Feibig stage 2. Five subjects immunized with gp120/NefTat/AS01B from the GSK trial PRO-HIV-002 (Leroux-Roels, et al., 2007) were studied. PBMCs were obtained 14 d after the second immunization (total of 42 d after the first dose, and 14 d after fourth immunization (total of 182 d after the first dose) with the same immunogen (Leroux-Roels, et al., 2007). PBMCs were also obtained from two HIV-1 negative healthy donors and used for single plasma cell sorts for isolating VH and VL gens for generating recombinant antibodies. Additionally, PBMCs were obtained from another HIV-1–negative healthy donor and used for generation of EBV-transformed human cell cultures for production human mAbs. All work related to human subjects was in compliance with Institutional Review Board protocols approved by the Duke University Health System Institutional Review Board, Rockefeller University Hospital Institutional Review Board; Institutional Review Board of the University of Alabama at Birmingham; University of North Carolina at Chapel Hill, Office of Human Research Ethic Biomedical Institutional Review Board; and Ethics Comité, UZ Ghent, Ghent, Belgium.
Single-cell flow cytometry sorting strategy.
PBMCs from blood or bone marrow samples were labeled with panels of fluorochrome antibody conjugates specific for human IgG-(PE), CD3 (PE-Cy5), CD16 (PE-Cy5), CD19 (APC-Cy7), CD20 (PE-Cy7), CD27 (Pacific Blue), CD235a (PE-Cy5), IgD (PE), IgM (FITC; BD), and CD14 (PE-Cy5) and CD38 (APC-Cy5.5; both from Invitrogen) as previously described (Wrammert et al., 2008; Liao et al., 2009). All antibody reagents were titered and used at optimal concentrations for flow cytometry. As Wrammert et al. (2008) have shown that the antibody secreting cells (ASCs) present in human PBMC populations express a distinctive CD19+, CD38hi, IgD−, CD20lo, or CD20− cell phenotype. To characterize single human ASCs, we sorted single plasmablasts/cytes (CD3−CD14−CD16−CD235a−CD19+CD20−CD27hiCD38hi) from human PBMC samples cells recovered from AHI and control subjects into the individual wells of 96-well plates containing 20 µl/well of RT reaction buffer (Invitrogen) as previously described (Tomaras et al., 2008; Bar et al., 2009). Antigen-specific memory B cell sorts were performed for isolation of HIV-1 gp120 Env-reactive antibodies from subjects immunized with gp120/NefTat/AS01B from the GSK trial PRO-HIV-002 (Leroux-Roels, et al., 2007) using AF647 and Pacific blue dual color labeled-B.W61D gp120 using the method as previously described (Scheid et al., 2009).
PCR amplification of plasmablast/plasma cell immunoglobulin VH and VL genes.
The Ig VH and VL genes of the sorted plasmablast were amplified by RT and nested PCR using the method as previously reported (Wardemann et al., 2003; Tiller et al., 2008; Wrammert et al., 2008; Liao et al., 2009). The PCR products amplified by this method contain enough coding region sequences for the constant regions of either heavy- or light-chain genes for allowing the identification of IgH subclass and types of light chains (Liao et al., 2009). Isolated VH and VL genes were used for assembling full-length Ig IgG1 heavy- and light-chain expression cassette by overlapping PCR to express recombinant IgG1 antibodies (Liao et al., 2009).
Sequencing, sequence annotation, quality control, and data management of Ig VH and VL sequences.
All PCR products of Ig VH and VL genes were purified using a PCR purification kit (QIAGEN) and sequenced in forward and reverse directions using an ABI 3700 instrument and BigDye sequencing kit (Applied Biosystems). Base calling for each sample was performed using Phred (Ewing and Green, 1998; Ewing et al., 1998). The forward and reverse strands of the antibody genes were assembled into one final nucleotide sequence using a novel assembly algorithm based on the quality scores at each base position (Munshaw and Kepler, 2010). The estimated PCR artifact rate was 0.28 or ∼1 PCR artifact per 5 genes amplified. The isotype and subclasses of Ig heavy-chain and types of light-chain were determined by comparing the constant region sequences of the isolated Ig VH and VL PCR products with the constant region sequences of the known Ig isotypes and subclass, κ, and λ genes using a computer alignment algorithm (Smith and Waterman, 1981). The unmutated rearrangement of each quality-assured antibody sequence was determined using SoDA (Volpe et al., 2006). Genetic information inferred by using SoDA, such as gene segment usage, somatic mutations, n-nucleotides, and CDR3 length is stored in a relational database to facilitate subsequent statistical analysis.
Antibodies from a given subject were regarded as clonally related if they met the following criteria for both the heavy and light chains: (a) They were inferred to use the same V and J gene segments. D-segment identification is subject to substantial uncertainty, so inferred differences in D-segment use were not sufficient to rule out clonal relatedness of two antibodies. (b) Identical CDR3 length. (c) 70% or greater nucleotide identity with CDR3. Phylogenetic analysis using the DNA Maximum Likelihood method of the PHYLIP 3.63 package (Felsenstein, 2005) was conducted using the inferred unmutated common ancestor from SoDA as the root.
High-throughput DNA sequencing of Ig V(D)J gene segments.
Genomic DNA samples were isolated from aliquots of PBMCs of all five acute HIV-1 subjects using QIAamp DNA Mini kit (QIAGEN). Multiplexed primers targeting the V and J segments of the Ig heavy-chain were used to amplify the rearranged variable (V), diverse (D), and joining (J) segments of the Ig heavy chain. Six barcoded V(D)J libraries from independent aliquots of DNA template from each sample were amplified, pooled, and sequenced using the 454 platform with Titanium chemistry (Roche; Boyd et al., 2009). Each sample was sequenced to an mean depth of 3,300 total Ig VH sequences. Sequences with identical V, D, and J segment usage and identical V-D and D-J junctions were considered to be members of an individual B cell clone.
Design and synthesis of inferred unmutated common ancestor and phylogenetic intermediate antibodies.
For each set of clonally related antibodies, Phylip’s DNA maximum likelihood (Felsenstein, 1973, 1981) and SoDA (Volpe et al., 2006) were used in conjunction to infer RUA and unobserved phylogenetic intermediates. These inferred RUA and intermediate VH and VL genes were synthesized (GenScript) and cloned as full-length IgG1 for heavy chain and full-length κ or λ light chain genes into pcDNA3.1 plasmid (Invitrogen) using standard recombinant techniques.
Expression of VH and VL as full-length IgG1 recombinant mAb.
The isolated Ig VH and VL gene pairs were assembled by PCR into the linear full-length Ig heavy- and light-chain gene expression cassettes for production of recombinant mAbs by transfection in the human embryonic kidney cell line, 293T (American Type Culture Collection) using the methods as previously described (Liao et al., 2009). The purified PCR products of the paired Ig heavy- and light-chain gene expression cassettes were co-transfected into near confluent 293T cells grown in 6-well (2 µg of DNA for each cassette/well) tissue culture plates (Becton Dickson) using PolyFect (QIAGEN) or Effectene (QIAGEN), and the protocols recommended by the manufacturers. 6 to 8 h after transfection, the 293T cells were fed with fresh culture medium supplemented with 2% FCS and incubated at 37°C in a 5% CO2 incubator. Culture supernatants were harvested 3 d after transfection and quantified for expressed IgG levels and screened for antibody specificity. The average concentration of recombinant antibody produced in 293T cells by transient transfection and used for screening specificity of antibody was 3.2 µg (+1.7 µg; n = 2,500). These screening assays of the transfected supernatants were designed to identify higher affinity antibodies in supernatants of the transfected cell cultures when the recombinant antibodies were expressed at from 0.1 to 25 µg/ml in transient transfections. Antibodies identified as exhibiting binding activity to tested antigens in screening assays, as well as the inferred UCA and intermediate clonal antibodies, were produced on a larger scale so that screening assays could be replicated and broadened to more fully define the range of binding activity of expressed plasma cell–derived antibodies. Purified recombinant antibodies were produced in bulk cultures by transient transfection using Ig heavy-and light-chain genes cloned in pcDNA plasmids (Liao et al., 2009). The linear Ig heavy- and light-chain gene expression cassettes used for production of recombinant antibodies for initial screening were cloned into pcDNA 3.3 (Invitrogen) for production of purified recombinant mAbs using standard molecular protocol, and then co-transfected into 293T cells cultured in T175 flasks using PolyFect (QIAGEN) or polyethylenimine (Smith et al., 2009) cultured in DME supplemented with 2% FCS. Recombinant mAbs were purified from culture supernatants of the transfected 293T cells using anti–human Ig heavy chain specific antibody-agarose beads (Sigma-Aldrich) using the previously described method (Liao et al., 2009). Purified antibodies used in the study were confirmed as having the typical patterns of predominant whole IgG in SDS-PAGE and Western blots under reducing and nonreducing conditions (Liao et al., 2009).
ELISA and Luminex assays for screening and characterization of antibody specificity.
Concentration of recombinant mAbs secreted in the transfected 293T cell culture in the supernatants was determined using a previously described method (Fiebig et al., 2003). The expressed recombinant mAb were assayed for antibody reactivity to HIV-1 antigens and to a panel of non–HIV-1 antigens by ELISA and Luminex assays (Liao et al., 2009; Tomaras et al., 2008). HIV-1 antigens included the Env peptides gp41 immunodominant region peptide (RVLAVERYLRDQQLLGIWGCSGKLICTTAVPWNASWSNKSLNK), gp41 MPER region peptide (QQEKNEQELLELDKWASLWN), HIV-1 MN recombinant gp41 (Immunodiagnostics), HIV-1 group M consensus gp120 (Liao et al., 2006), HIV-1 group M consensus gp140 CFI (Liao et al., 2006), p66 (Worthington Biochemical), p55 (Protein Sciences), p31 (Genway), Nef (Genway), Tat (Advanced BioScience), and AT-2–inactivated HIV-1 ADA virions (Rossio et al., 1998; a gift from J. Lifson, National Cancer Institute, Frederick, MD). In addition, all antibodies derived from subjects 684–6 and 641–7 were assayed against the corresponding autologous gp140 and gp120 Env proteins. The non–HIV-1 antigens tetanus toxoid (EMD) cardiolipin (Avanti Polar Lipids; Haynes et al., 2005) killed Cryptococcus neoformans (Perfect, 2006) and Candida albicans (Sherman, 1991; Blankenship et al., 2003) and lipid A (Avanti Polar Lipids). In addition, reactivity of recombinant antibodies to whole-cell lysates of anaerobic and aerobic bacterial extracts (Kawatsu et al., 2008) was tested. Bacterial whole-cell lysates were prepared using previously described methods (Kawatsu et al., 2008).
In brief, bacteria were inoculated from a stool specimen from 4 subjects and grown on agar plates under anaerobic or aerobic conditions at 30°C. Aerobic and anaerobic extracts were prepared separately, but the four individual samples of each were combined. Bacteria grown in confluence were harvested by adding 5 ml PBS and gently scraping the agar surface with a cell scraper (Thermo Fisher Scientific). Surfaces were then rinsed with another 5 ml PBS to remove any remaining bacteria and transferred to a 50-ml conical tube. The extract was treated with 100 µl of bacterial protease inhibitor cocktail (Sigma-Aldrich), and then sonicated with a Misonix 3000 Sonicator at maximum voltage for 15-s pulses, followed by 15-s rests on ice for 3 min. After sonication, 1.5 g of acid-washed glass beads (Sigma-Aldrich) was added to the extracts. The extracts were vortexed for 1 min at maximum setting, and then chilled for 1 min in an ice bath. This process was repeated four times for a total of five vortex-chill cycles. The resulting extracts were spun for 20 min at 3,000 g. The supernatants were harvested and measured for protein concentration by Nanodrop (Thermo Fisher Scientific). Samples were aliquoted and frozen at −80°C until they were used. In addition, all antibodies were also assayed for the reactivity to HEP-2 cells (Inverness Medical Professional Diagnostics) by indirect immunofluorescence staining (Haynes et al., 2005).
The apparent affinity of antibodies was calculated in molar concentration from EC50 values using a four parametric sigmoid curve fitting analysis. Antibodies were titrated at concentrations ranging from 50 to 0.2 µg/ml, and threefold dilutions were used in the Luminex assays. For antibodies with relatively lower affinities, EC50 values were calculated by extrapolating the binding curve to the saturation level.
SPR analysis of antibody reactivity.
SPR binding assays were performed on a BIAcore 3000 (BIAcore Inc.) maintained at 20°C. HIV-1 gp41 MN or oligomeric gp140 proteins (Con S gp140 and autologous Env gp140) were immobilized on a CM5 sensor chip by standard amine coupling, as previously described (Alam et al., 2008, 2009). In a separate CM5 sensor chip, human mAbs were captured on anti–human Fc antibody-coupled surfaces, and then human mAbs were captured to ∼200–500 response units. Specific binding responses of mAb binding were assessed by comparing nonspecific binding to either control surfaces or control antibodies (HIV-1 gp120 for Env-immobilized surfaces and control mAb IS6 [antilipid] or Synagis [anti-RSV] for mAb captured surfaces). Rate constants were measured using either the 1:1 Langmuir equation or bivalent analyte model (to account for the avidity of bivalent Ig molecules) and global curve fitting to binding curves obtained from mAb titrations. mAbs were injected at 30 µl/min for 3 min, and Glycine-HCl pH 2.0 and surfactant P20 (0.01%) were used as the regeneration buffer.
Statistical analysis.
All analysis datasets were compiled, and all analysis completed in SAS v9.2 (SAS, Inc.). For continuous measures (e.g., mutation rate) there were two types of tests performed. For two group comparisons between means, Student’s t tests with Satterthwaite variance estimates were performed in PROC TTEST. Where there were more than two groups where means were compared, the least squares means were tested using one degree of freedom F-tests in PROC GLM. For comparisons of count data, χ2 tests were used. When cell counts were low, Fisher’s exact tests were performed in PROC FREQ.
Online supplemental material.
Fig. S1 shows distribution of VH somatic mutation frequencies in AHI antibodies. Fig. S2 shows reactivity of antibody members of clonal lineages 558, 572, 712, 728, and 2495 with HIV-1 gp41, gut flora, and HEp-2 epithelial cells. Fig. S3 shows reactivity of antibody members of clone 558 with HIV-1 MN gp41 and gut flora in Western blots of blue native gels and SDS-PAGE under reducing and nonreducing conditions. Fig. S4 shows reactivity of IgM antibody 1E7 isolated from bone marrow of an AHI patient. Table S1 shows Ig isotypes of HIV-1–reactive antibodies and antibodies with unknown specificity isolated from AHI. Table S2 shows comparison of VH and VL usage of HIV-1–reactive antibodies and antibodies with unknown specificity isolated from AHI. Table S3 shows CDR3 length of Ig heavy (H), k (K), and l (L) chains of antigen-reactive and nonreactive antibodies. Table S4 shows mutation frequencies of total framework and CDR regions in VH, VK, and VL gene segments of antigen-reactive and nonreactive antibodies. Table S5 shows gene family, mutation frequency, and CDR regions of anti–HIV-1 gp120 antibodies isolated from HIV-1 W61D gp120-immunized subjects. Table S6 shows HIV-1–positive clonal lineages identified from AHI patients. Tables S7–11 show VH and VL sequences of clones 558, 572, 712, 728, and 2495, respectively. Table S12 shows mutations in VH and VL CDR and framework regions of clone 558. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110363/DC1.
Acknowledgments
The authors are grateful for technical assistance from Richard Scearce, Richard Frothingham, Mark Drinker, Kenesha Luney, and Barbara Alexander for preparation of gut whole cell lysates. We thank Kelly Soderberg and Jennifer Kirscherr for program management. We thank Katherine DeRosa, Ashley Trama, and Ashlyn Dixon for isolation of Ig VH and VL genes; Andrew Foulger, Marco Harris II, Haiyan Chen, Benjiang Ma, Adam Brewer, and Radharani De for assistance with production of recombinant antibodies; Joshua Amos and Thad Gurley for blood processing and performing flow cytometry experiments and analysis; Michele J. Donathan, Robert Parks, Judith Lucas, Robert Meyerhoff, Nicole Yates, Doris Murray, Judith Lucas, and Vicki Ashley for help with design and quality control of the custom binding antibody assays using Luminex; and Joshua Eudailey for HEp-2 immunofluorescence assays. We are grateful to Myron Cohen, Joseph Eron, and Joanne Kuruc of the CHAVI clinical Core for their support of the CHAVI 012 protocol.
This work was supported by the Center for HIV/AIDS Vaccine Immunology National Institute of Allergy and Infectious Disease (NIAID) grant U19 AI067854, NIAID grants P01 AI061734 and U54 AI065359, and a Collaboration for HIV Vaccine Discovery grant from the Bill and Melinda Gates Foundation. Flow cytometry was performed in the Duke Center for AIDS Research BSL3 Flow Cytometry Core Facility supported by the National Institutes of Health grants S10RR019145, UC6 AI058607, AI64518, P30 AI051445, and AI051.
Gerald Voss is an employee of GlaxoSmithKline Biologicals s.a. (GSK). He owns shares and options to shares in GSK. In addition, Gerald Voss is a designated inventor on a variety of patents owned by GSK. All other authors had no competing financial interests in this study.
Footnotes
Abbreviations used:
- AHI
- acute HIV-1 infection
- AT-2
- aldrithol-2
- CDR
- complementarity-determining region
- EBV
- Epstein-Barr virus
- Env
- envelope
- RUA
- reverted unmutated ancestor
- SoDA
- somatic diversification analysis
- SPR
- surface plasmon resonance
References
- Alam S.M., Scearce R.M., Parks R.J., Plonk K., Plonk S.G., Sutherland L.L., Gorny M.K., Zolla-Pazner S., Vanleeuwen S., Moody M.A., et al. 2008. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J. Virol. 82:115–125 10.1128/JVI.00927-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.M., Morelli M., Dennison S.M., Liao H.X., Zhang R., Xia S.M., Rits-Volloch S., Sun L., Harrison S.C., Haynes B.F., Chen B. 2009. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. USA. 106:20234–20239 10.1073/pnas.0908713106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar K.J., Keele B.F., Decker J., McLellan J., Salazar-Gonzales J., Salazar M., Li H., Wang S., Yang Y., Hahn B.H., Kwong P.D., Shaw G.M. 2009. P04-43. Neutralizing antibody responses against conformational envelope epitopes in early HIV-1 infection. Retrovirology. 6:P71 [Google Scholar]
- Berberian L., Goodglick L., Kipps T.J., Braun J. 1993. Immunoglobulin VH3 gene products: natural ligands for HIV gp120. Science. 261:1588–1591 10.1126/science.7690497 [DOI] [PubMed] [Google Scholar]
- Blankenship J.R., Wormley F.L., Boyce M.K., Schell W.A., Filler S.G., Perfect J.R., Heitman J. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot. Cell. 2:422–430 10.1128/EC.2.3.422-430.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S.D., Marshall E.L., Merker J.D., Maniar J.M., Zhang L.N., Sahaf B., Jones C.D., Simen B.B., Hanczaruk B., Nguyen K.D., et al. 2009. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 1:12ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley J.M., Schacker T.W., Ruff L.E., Price D.A., Taylor J.H., Beilman G.J., Nguyen P.L., Khoruts A., Larson M., Haase A.T., Douek D.C. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200:749–759 10.1084/jem.20040874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhu Z., Liao H.X., Quinnan G., Broder C.C., Haynes B.F., Dimitrov D.S. 2009. Cross-reactive human IgM monoclonal antibodies that bind HIV-1 envelope glycoproteins. Viruses. 1:1–19 10.3390/v1010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L.T., Malmborg A.C., Kristensson K., Hinkula J., Wahren B., Borrebaeck C.A. 1995. Mimicking the humoral immune response in vitro results in antigen-specific isotype switching supported by specific autologous T helper cells: generation of human HIV-1-neutralizing IgG monoclonal antibodies from naive donors. Eur. J. Immunol. 25:657–663 10.1002/eji.1830250305 [DOI] [PubMed] [Google Scholar]
- Dal Porto J.M., Haberman A.M., Kelsoe G., Shlomchik M.J. 2002. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J. Exp. Med. 195:1215–1221 10.1084/jem.20011550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- Falkoff R.J., Zhu L.P., Fauci A.S. 1983. The relationship between immunization and circulating antigen-specific plaque-forming cells. Cell. Immunol. 78:392–399 10.1016/0008-8749(83)90295-2 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1973. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am. J. Hum. Genet. 25:471–492 [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368–376 10.1007/BF01734359 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 2005. PHYLIP (phylogeny inference package) version 3.6. Department of Genome Sciences, University of Washington, Seattle [Google Scholar]
- Fiebig E.W., Wright D.J., Rawal B.D., Garrett P.E., Schumacher R.T., Peddada L., Heldebrant C., Smith R., Conrad A., Kleinman S.H., Busch M.P. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 17:1871–1879 10.1097/00002030-200309050-00005 [DOI] [PubMed] [Google Scholar]
- Goonetilleke N., Liu M.K., Salazar-Gonzalez J.F., Ferrari G., Giorgi E., Ganusov V.V., Keele B.F., Learn G.H., Turnbull E.L., Salazar M.G., et al. ; CHAVI Clinical Core B 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253–1272 10.1084/jem.20090365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Fleming J., St Clair E.W., Katinger H., Stiegler G., Kunert R., Robinson J., Scearce R.M., Plonk K., Staats H.F., et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 308:1906–1908 10.1126/science.1111781 [DOI] [PubMed] [Google Scholar]
- Kawatsu K., Kumeda Y., Taguchi M., Yamazaki-Matsune W., Kanki M., Inoue K. 2008. Development and evaluation of immunochromatographic assay for simple and rapid detection of Campylobacter jejuni and Campylobacter coli in human stool specimens. J. Clin. Microbiol. 46:1226–1231 10.1128/JCM.02170-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G., Sun C., Grayson T., Wang S., Li H., et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 105:7552–7557 10.1073/pnas.0802203105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux-Roels I., Koutsoukos M., Clement F., Steyaert S., Janssens M., Bourguignon P., Cohen K., Altfeld M., Vandepapelière P., Pedneault L., et al. 2010. Strong and persistent CD4+ T-cell response in healthy adults immunized with a candidate HIV-1 vaccine containing gp120, Nef and Tat antigens formulated in three Adjuvant Systems. Vaccine. 28:7016–7024 10.1016/j.vaccine.2010.08.035 [DOI] [PubMed] [Google Scholar]
- Levesque M.C., Moody M.A., Hwang K.K., Marshall D.J., Whitesides J.F., Amos J.D., Gurley T.C., Allgood S., Haynes B.B., Vandergrift N.A., et al. 2009. Polyclonal B cell differentiation and loss of gastrointestinal tract germinal centers in the earliest stages of HIV-1 infection. PLoS Med. 6:e1000107 10.1371/journal.pmed.1000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Sutherland L.L., Xia S.M., Brock M.E., Scearce R.M., Vanleeuwen S., Alam S.M., McAdams M., Weaver E.A., Camacho Z., et al. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 353:268–282 10.1016/j.virol.2006.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Levesque M.C., Nagel A., Dixon A., Zhang R., Walter E., Parks R., Whitesides J., Marshall D.J., Hwang K.K., et al. 2009. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods. 158:171–179 10.1016/j.jviromet.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael A.J., Borrow P., Tomaras G.D., Goonetilleke N., Haynes B.F. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat. Rev. Immunol. 10:11–23 10.1038/nri2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.L., Ranchobe N., Lambson B.E., Gray E.S., Cave E., Abrahams M.R., Bandawe G., Mlisana K., Abdool Karim S.S., Williamson C., Morris L.; CAPRISA 002 Study; NIAID Center for HIV/AIDS Vaccine Immunology (CHAVI) 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598 10.1371/journal.ppat.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H., Scheid J.F., Zoller M.J., Krogsgaard M., Ott R.G., Shukair S., Artyomov M.N., Pietzsch J., Connors M., Pereyra F., et al. 2010. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 467:591–595 10.1038/nature09385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshaw S., Kepler T.B. 2010. SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements. Bioinformatics. 26:867–872 10.1093/bioinformatics/btq056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect J.R. 2006. Cryptococcus neoformans: the yeast that likes it hot. FEM. Yeast Res. 6:463–468 10.1111/j.1567-1364.2006.00051.x [DOI] [PubMed] [Google Scholar]
- Richman D.D., Wrin T., Little S.J., Petropoulos C.J. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA. 100:4144–4149 10.1073/pnas.0630530100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossio J.L., Esser M.T., Suryanarayana K., Schneider D.K., Bess J.W., Jr, Vasquez G.M., Wiltrout T.A., Chertova E., Grimes M.K., Sattentau Q., et al. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72:7992–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster M.Y., Topham D.J., D’Costa S., Cardin R.D., Marion T.N., Myers L.K., Doherty P.C. 2000. Analysis of the virus-specific and nonspecific B cell response to a persistent B-lymphotropic gammaherpesvirus. J. Immunol. 164:1820–1828 [DOI] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Feldhahn N., Seaman M.S., Velinzon K., Pietzsch J., Ott R.G., Anthony R.M., Zebroski H., Hurley A., et al. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 458:636–640 10.1038/nature07930 [DOI] [PubMed] [Google Scholar]
- Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 10.1016/0076-6879(91)94004-V [DOI] [PubMed] [Google Scholar]
- Shih T.A., Meffre E., Roederer M., Nussenzweig M.C. 2002. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat. Immunol. 3:570–575 10.1038/ni803 [DOI] [PubMed] [Google Scholar]
- Smith T.F., Waterman M.S. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195–197 10.1016/0022-2836(81)90087-5 [DOI] [PubMed] [Google Scholar]
- Smith K., Garman L., Wrammert J., Zheng N.Y., Capra J.D., Ahmed R., Wilson P.C. 2009. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4:372–384 10.1038/nprot.2009.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 329:112–124 10.1016/j.jim.2007.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras G.D., Yates N.L., Liu P., Qin L., Fouda G.G., Chavez L.L., Decamp A.C., Parks R.J., Ashley V.C., Lucas J.T., et al. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 10.1128/JVI.01708-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuiji M., Yurasov S., Velinzon K., Thomas S., Nussenzweig M.C., Wardemann H.A. 2006. A checkpoint for autoreactivity in human IgM+ memory B cell development. J. Exp. Med. 203:393–400 10.1084/jem.20052033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkoczy L., Moody M.A., Holl T.M., Bouton-Verville H., Scearce R.M., Hutchinson J., Alam S.M., Kelsoe G., Haynes B.F. 2009. Functional, non-clonal IgMa-restricted B cell receptor interactions with the HIV-1 envelope gp41 membrane proximal external region. PLoS ONE. 4:e7215 10.1371/journal.pone.0007215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe A., Caramaschi P., Carletto A., Pieropan S., Bambara L.M., Biasi D. 2006. Psoriasis onset during infliximab treatment: description of two cases. Rheumatol. Int. 26:1158–1160 10.1007/s00296-006-0144-1 [DOI] [PubMed] [Google Scholar]
- Wardemann H., Yurasov S., Schaefer A., Young J.W., Meffre E., Nussenzweig M.C. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377 10.1126/science.1086907 [DOI] [PubMed] [Google Scholar]
- Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S., et al. 2003. Antibody neutralization and escape by HIV-1. Nature. 422:307–312 10.1038/nature01470 [DOI] [PubMed] [Google Scholar]
- Weitkamp J.H., Kallewaard N., Kusuhara K., Bures E., Williams J.V., LaFleur B., Greenberg H.B., Crowe J.E., Jr 2003. Infant and adult human B cell responses to rotavirus share common immunodominant variable gene repertoires. J. Immunol. 171:4680–4688 [DOI] [PubMed] [Google Scholar]
- Wrammert J., Smith K., Miller J., Langley W.A., Kokko K., Larsen C., Zheng N.Y., Mays I., Garman L., Helms C., et al. 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 453:667–671 10.1038/nature06890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J., Koutsonanos D., Li G.M., Edupuganti S., Sui J., Morrissey M., McCausland M., Skountzou I., Hornig M., Lipkin W.I., et al. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 208:181–193 10.1084/jem.20101352 [DOI] [PMC free article] [PubMed] [Google Scholar]