Abstract
In the title complex, [Zn(C7H6NO2)2(C10H8N2)], the ZnII cation is coordinated by two aminobenzoate anions and one 2,2′-bipyridine ligand in a distorted trigonal–bipyramidal geometry. The carboxylate group of one aminobenzoate anion coordinates to the ZnII cation in a monodentate manner, whereas the carboxylate group of the other aminobenzoate anion chelates the Zn cation with different Zn—O bond lengths. Intermolecular N—H⋯N and N—H⋯O hydrogen bonding is present in the crystal structure.
Related literature
For applications of Zn complexes, see: Chohan & Naseer (2007 ▶); Huang et al. (2006 ▶); Ispir et al. (2006 ▶); Lo et al. (2007 ▶); Maria et al. (1996 ▶). For a related structure, see: Wang et al. (2005 ▶).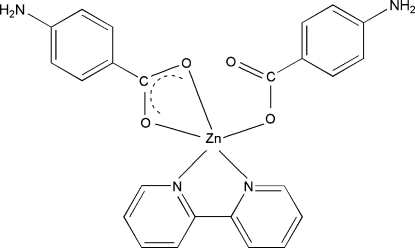
Experimental
Crystal data
[Zn(C7H6NO2)2(C10H8N2)]
M r = 493.81
Triclinic,

a = 7.9499 (14) Å
b = 10.7281 (19) Å
c = 13.905 (2) Å
α = 80.499 (2)°
β = 80.921 (2)°
γ = 70.538 (2)°
V = 1096.1 (3) Å3
Z = 2
Mo Kα radiation
μ = 1.16 mm−1
T = 295 K
0.42 × 0.23 × 0.08 mm
Data collection
Bruker SMART 1000 CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.643, T max = 0.914
8223 measured reflections
4058 independent reflections
3419 reflections with I > 2σ(I)
R int = 0.024
Refinement
R[F 2 > 2σ(F 2)] = 0.035
wR(F 2) = 0.090
S = 1.07
4058 reflections
298 parameters
H-atom parameters constrained
Δρmax = 0.31 e Å−3
Δρmin = −0.31 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811039389/xu5334sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811039389/xu5334Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Zn1—O1 | 1.9269 (18) |
| Zn1—O3 | 1.9704 (18) |
| Zn1—O4 | 2.395 (2) |
| Zn1—N3 | 2.124 (2) |
| Zn1—N4 | 2.088 (2) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1D⋯N2i | 0.88 | 2.33 | 3.202 (4) | 169 |
| N1—H1E⋯O2ii | 0.88 | 2.36 | 3.181 (4) | 155 |
| N2—H2E⋯O2iii | 0.88 | 2.28 | 3.116 (4) | 159 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
This work was supported by the Education Department Foundation of Fujian Province of China (grant No. JK2011042).
supplementary crystallographic information
Comment
In recent decades, zinc complexes have received much attention because of their interesting with biological ligands to generate stable mixed coordinated complexes, which play a key role in life process such as anti-cancer, antiseptic and anti-inflammatory (Ispir et al., 2006; Chohan & Naseer, 2007; Huang et al., 2006; Lo et al., 2007). 4-Aminobenzoic acid as an important part in the folic acid, which is a constituent of the vitamin B complex and is found in animal and plant tissues, has been shown to be a growth factor in certain microorganisms (Maria et al., 1996). In order to extend further the study of 4-aminobenzoic acid ligand coordinate to zinc ion, we synthesized the title complex and determined the crystal structure.
The asymmetric unit of 1 contains one zinc cation, two 4-aminobenzoic ion, and one 2,2'-bipyridine molecule. The Zn(II) atom is five-coordinated, forming a distorted trigonal-bipyramidal (Table 1). In the coordination polyhedron, the equatorial plane is occupied by two O(O1,O3) atoms from different 4-aba and one N(N4) atom from 2,2'-bipyridine, at the apex is situated one O(O4) atom from 4-aba and one N(N3) atom from 2,2'-bipyridine (Fig 1). The Zn(II) center is coordinated by two types of 4-aba: one behaves in an unsymmetrical chelating mode [Zn—O(3) 1.9704 (18) and Zn—O(4) 2.395 (2) Å]; the other acts as a monodentate ligand through one carboxylate oxygen atom [Zn—O(1) 1.9269 (18) and Zn—O(2) 2.841 Å], which is similar to previously reported complex {[Zn2(4,4'-bipy)2(4-aba)4](H2O)5} (Wang et al., 2005). The dihedral angle between phenyl rings of the two 4-aba is 81.83 (7)°.
Hydrogen bonds are observed between the molecules in the crystal structure (Table 2).
Experimental
An aqueous solution (5 ml) of ZnC4H6O4.2H2O (1 mmol) was added slowly to a mixed solution of 4-aminobenzoic acid (1.5 mmol) in H2O (5 ml) and 2,2'-bipyridine (1 mmol) in ethanol (95%, 5 ml). After refluxing for 3 h, the mixture was filtered off while hot. The colourless single crystals suitable for X-ray analysis were obtained by slow evaporation of the above filtrate at room temperature after a month.
Refinement
H atoms were placed geometrically and treated as riding, C—H = 0.93 and N—H = 0.88 Å, Uiso(H) = 1.2Ueq(C) and 1.5Ueq(N).
Figures
Fig. 1.
The ORTEP drawing of the title compound (I). Displacement ellipsoids are drawn at 30% probability level.
Fig. 2.
Projection showing the two-dimensional structure formed by H-bonding interaction of the compound (I).
Crystal data
| [Zn(C7H6NO2)2(C10H8N2)] | Z = 2 |
| Mr = 493.81 | F(000) = 508 |
| Triclinic, P1 | Dx = 1.496 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.9499 (14) Å | Cell parameters from 2868 reflections |
| b = 10.7281 (19) Å | θ = 2.4–24.9° |
| c = 13.905 (2) Å | µ = 1.16 mm−1 |
| α = 80.499 (2)° | T = 295 K |
| β = 80.921 (2)° | Block, colourless |
| γ = 70.538 (2)° | 0.42 × 0.23 × 0.08 mm |
| V = 1096.1 (3) Å3 |
Data collection
| Bruker SMART 1000 CCD area-detector diffractometer | 4058 independent reflections |
| Radiation source: fine-focus sealed tube | 3419 reflections with I > 2σ(I) |
| graphite | Rint = 0.024 |
| φ and ω scans | θmax = 25.5°, θmin = 2.4° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −9→9 |
| Tmin = 0.643, Tmax = 0.914 | k = −12→12 |
| 8223 measured reflections | l = −16→16 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.035 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.090 | H-atom parameters constrained |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0411P)2 + 0.2825P] where P = (Fo2 + 2Fc2)/3 |
| 4058 reflections | (Δ/σ)max < 0.001 |
| 298 parameters | Δρmax = 0.31 e Å−3 |
| 0 restraints | Δρmin = −0.31 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Zn1 | 0.14918 (4) | 0.12380 (3) | 0.19516 (2) | 0.04021 (12) | |
| O1 | −0.1044 (2) | 0.2151 (2) | 0.21855 (14) | 0.0528 (5) | |
| O2 | −0.0311 (3) | 0.3661 (2) | 0.27669 (18) | 0.0732 (7) | |
| O3 | 0.3785 (3) | 0.10054 (19) | 0.24420 (15) | 0.0558 (5) | |
| O4 | 0.2405 (3) | −0.0296 (2) | 0.33755 (15) | 0.0663 (6) | |
| N1 | −0.8907 (3) | 0.5767 (3) | 0.3483 (2) | 0.0678 (7) | |
| H1D | −0.9291 | 0.6259 | 0.3971 | 0.102* | |
| H1E | −0.9611 | 0.5323 | 0.3389 | 0.102* | |
| N2 | 1.0034 (4) | −0.2214 (3) | 0.5101 (2) | 0.0788 (9) | |
| H2D | 1.0548 | −0.1604 | 0.5101 | 0.118* | |
| H2E | 0.9810 | −0.2610 | 0.5693 | 0.118* | |
| N3 | 0.2165 (3) | 0.1976 (2) | 0.04781 (16) | 0.0502 (6) | |
| N4 | 0.1805 (3) | −0.0382 (2) | 0.12124 (15) | 0.0419 (5) | |
| C1 | −0.3406 (3) | 0.3914 (2) | 0.28122 (17) | 0.0362 (5) | |
| C2 | −0.4024 (4) | 0.4919 (3) | 0.3417 (2) | 0.0488 (7) | |
| H2 | −0.3201 | 0.5185 | 0.3670 | 0.059* | |
| C3 | −0.5831 (4) | 0.5529 (3) | 0.3648 (2) | 0.0537 (7) | |
| H3 | −0.6211 | 0.6196 | 0.4057 | 0.064* | |
| C4 | −0.7100 (3) | 0.5157 (3) | 0.3275 (2) | 0.0444 (6) | |
| C5 | −0.6484 (3) | 0.4166 (2) | 0.26597 (19) | 0.0417 (6) | |
| H5 | −0.7303 | 0.3914 | 0.2391 | 0.050* | |
| C6 | −0.4677 (3) | 0.3554 (2) | 0.24420 (18) | 0.0375 (5) | |
| H6 | −0.4297 | 0.2882 | 0.2036 | 0.045* | |
| C7 | −0.1451 (3) | 0.3227 (3) | 0.25832 (18) | 0.0423 (6) | |
| C8 | 0.5359 (3) | −0.0506 (2) | 0.36920 (17) | 0.0368 (5) | |
| C9 | 0.5508 (4) | −0.1572 (3) | 0.44177 (19) | 0.0479 (7) | |
| H9 | 0.4560 | −0.1919 | 0.4589 | 0.058* | |
| C10 | 0.7028 (4) | −0.2133 (3) | 0.4894 (2) | 0.0560 (8) | |
| H10 | 0.7105 | −0.2861 | 0.5372 | 0.067* | |
| C11 | 0.8444 (4) | −0.1614 (3) | 0.4663 (2) | 0.0495 (7) | |
| C12 | 0.8290 (3) | −0.0533 (3) | 0.3952 (2) | 0.0474 (6) | |
| H12 | 0.9222 | −0.0167 | 0.3796 | 0.057* | |
| C13 | 0.6791 (3) | 0.0007 (3) | 0.34731 (18) | 0.0416 (6) | |
| H13 | 0.6723 | 0.0730 | 0.2992 | 0.050* | |
| C14 | 0.3746 (3) | 0.0096 (3) | 0.3162 (2) | 0.0456 (6) | |
| C15 | 0.2298 (5) | 0.3190 (3) | 0.0162 (3) | 0.0771 (10) | |
| H15 | 0.2011 | 0.3809 | 0.0607 | 0.093* | |
| C16 | 0.2848 (7) | 0.3549 (4) | −0.0801 (3) | 0.1001 (14) | |
| H16 | 0.2920 | 0.4402 | −0.1006 | 0.120* | |
| C17 | 0.3286 (6) | 0.2642 (5) | −0.1448 (3) | 0.0988 (14) | |
| H17 | 0.3678 | 0.2865 | −0.2100 | 0.119* | |
| C18 | 0.3146 (5) | 0.1395 (4) | −0.1135 (2) | 0.0714 (9) | |
| H18 | 0.3437 | 0.0765 | −0.1571 | 0.086* | |
| C19 | 0.2563 (3) | 0.1089 (3) | −0.01574 (19) | 0.0479 (7) | |
| C20 | 0.2330 (3) | −0.0219 (3) | 0.02410 (19) | 0.0461 (6) | |
| C21 | 0.2580 (5) | −0.1217 (3) | −0.0330 (2) | 0.0702 (9) | |
| H21 | 0.2929 | −0.1097 | −0.1000 | 0.084* | |
| C22 | 0.2307 (6) | −0.2389 (4) | 0.0103 (3) | 0.0783 (10) | |
| H22 | 0.2470 | −0.3069 | −0.0274 | 0.094* | |
| C23 | 0.1794 (5) | −0.2557 (3) | 0.1088 (2) | 0.0673 (9) | |
| H23 | 0.1607 | −0.3348 | 0.1390 | 0.081* | |
| C24 | 0.1561 (4) | −0.1535 (3) | 0.1622 (2) | 0.0534 (7) | |
| H24 | 0.1221 | −0.1648 | 0.2293 | 0.064* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Zn1 | 0.03267 (17) | 0.04633 (19) | 0.03920 (18) | −0.00652 (13) | −0.00538 (12) | −0.00961 (13) |
| O1 | 0.0335 (9) | 0.0589 (12) | 0.0619 (12) | −0.0044 (9) | −0.0064 (8) | −0.0168 (10) |
| O2 | 0.0385 (11) | 0.0841 (16) | 0.1075 (19) | −0.0255 (11) | −0.0096 (11) | −0.0263 (14) |
| O3 | 0.0480 (11) | 0.0515 (12) | 0.0688 (13) | −0.0078 (9) | −0.0261 (10) | −0.0074 (10) |
| O4 | 0.0385 (11) | 0.1067 (18) | 0.0642 (13) | −0.0291 (11) | −0.0019 (9) | −0.0299 (12) |
| N1 | 0.0407 (13) | 0.0650 (17) | 0.092 (2) | −0.0066 (12) | 0.0100 (13) | −0.0317 (15) |
| N2 | 0.0660 (18) | 0.0786 (19) | 0.0765 (19) | 0.0202 (15) | −0.0384 (15) | −0.0286 (15) |
| N3 | 0.0525 (14) | 0.0509 (14) | 0.0436 (13) | −0.0125 (11) | −0.0062 (11) | −0.0024 (11) |
| N4 | 0.0386 (11) | 0.0477 (13) | 0.0367 (12) | −0.0080 (10) | −0.0058 (9) | −0.0080 (10) |
| C1 | 0.0333 (12) | 0.0379 (13) | 0.0360 (13) | −0.0109 (10) | −0.0050 (10) | −0.0006 (11) |
| C2 | 0.0466 (16) | 0.0530 (17) | 0.0544 (17) | −0.0212 (13) | −0.0054 (13) | −0.0165 (13) |
| C3 | 0.0538 (17) | 0.0496 (17) | 0.0607 (18) | −0.0152 (14) | 0.0040 (14) | −0.0272 (14) |
| C4 | 0.0370 (13) | 0.0389 (14) | 0.0521 (16) | −0.0087 (11) | 0.0046 (12) | −0.0069 (12) |
| C5 | 0.0346 (13) | 0.0412 (14) | 0.0519 (15) | −0.0135 (11) | −0.0053 (11) | −0.0092 (12) |
| C6 | 0.0361 (13) | 0.0355 (13) | 0.0392 (13) | −0.0078 (10) | −0.0030 (10) | −0.0089 (11) |
| C7 | 0.0371 (14) | 0.0484 (16) | 0.0369 (14) | −0.0096 (12) | −0.0068 (11) | 0.0012 (12) |
| C8 | 0.0330 (12) | 0.0403 (14) | 0.0373 (13) | −0.0090 (10) | −0.0007 (10) | −0.0133 (11) |
| C9 | 0.0513 (16) | 0.0508 (16) | 0.0463 (15) | −0.0246 (13) | 0.0056 (13) | −0.0115 (13) |
| C10 | 0.074 (2) | 0.0433 (16) | 0.0413 (15) | −0.0070 (15) | −0.0095 (14) | 0.0011 (12) |
| C11 | 0.0436 (15) | 0.0522 (16) | 0.0433 (15) | 0.0072 (13) | −0.0117 (12) | −0.0200 (13) |
| C12 | 0.0339 (13) | 0.0607 (18) | 0.0493 (16) | −0.0137 (13) | −0.0017 (12) | −0.0166 (14) |
| C13 | 0.0380 (14) | 0.0432 (14) | 0.0421 (14) | −0.0116 (11) | −0.0056 (11) | −0.0025 (11) |
| C14 | 0.0349 (14) | 0.0535 (17) | 0.0504 (16) | −0.0071 (12) | −0.0044 (12) | −0.0270 (14) |
| C15 | 0.111 (3) | 0.059 (2) | 0.064 (2) | −0.034 (2) | −0.013 (2) | 0.0027 (17) |
| C16 | 0.150 (4) | 0.081 (3) | 0.072 (3) | −0.056 (3) | −0.009 (3) | 0.021 (2) |
| C17 | 0.128 (4) | 0.107 (3) | 0.055 (2) | −0.048 (3) | 0.008 (2) | 0.013 (2) |
| C18 | 0.083 (2) | 0.077 (2) | 0.0436 (17) | −0.0196 (19) | 0.0055 (16) | −0.0039 (16) |
| C19 | 0.0380 (14) | 0.0542 (17) | 0.0414 (15) | −0.0035 (12) | −0.0025 (11) | −0.0029 (13) |
| C20 | 0.0405 (14) | 0.0507 (16) | 0.0413 (15) | −0.0034 (12) | −0.0083 (11) | −0.0093 (12) |
| C21 | 0.095 (3) | 0.065 (2) | 0.0431 (17) | −0.0107 (19) | −0.0063 (17) | −0.0166 (15) |
| C22 | 0.114 (3) | 0.058 (2) | 0.063 (2) | −0.017 (2) | −0.016 (2) | −0.0232 (17) |
| C23 | 0.090 (2) | 0.0486 (18) | 0.066 (2) | −0.0206 (17) | −0.0171 (18) | −0.0092 (16) |
| C24 | 0.0610 (18) | 0.0525 (17) | 0.0464 (16) | −0.0170 (14) | −0.0079 (13) | −0.0054 (13) |
Geometric parameters (Å, °)
| Zn1—O1 | 1.9269 (18) | C6—H6 | 0.9300 |
| Zn1—O3 | 1.9704 (18) | C8—C9 | 1.381 (4) |
| Zn1—O4 | 2.395 (2) | C8—C13 | 1.394 (3) |
| Zn1—N3 | 2.124 (2) | C8—C14 | 1.481 (4) |
| Zn1—N4 | 2.088 (2) | C9—C10 | 1.377 (4) |
| Zn1—C14 | 2.521 (3) | C9—H9 | 0.9300 |
| O1—C7 | 1.284 (3) | C10—C11 | 1.387 (4) |
| O2—C7 | 1.223 (3) | C10—H10 | 0.9300 |
| O3—C14 | 1.282 (3) | C11—C12 | 1.380 (4) |
| O4—C14 | 1.249 (3) | C12—C13 | 1.364 (3) |
| N1—C4 | 1.372 (3) | C12—H12 | 0.9300 |
| N1—H1D | 0.8818 | C13—H13 | 0.9300 |
| N1—H1E | 0.8820 | C15—C16 | 1.375 (5) |
| N2—C11 | 1.397 (3) | C15—H15 | 0.9300 |
| N2—H2D | 0.8798 | C16—C17 | 1.358 (6) |
| N2—H2E | 0.8825 | C16—H16 | 0.9300 |
| N3—C19 | 1.332 (4) | C17—C18 | 1.372 (5) |
| N3—C15 | 1.338 (4) | C17—H17 | 0.9300 |
| N4—C24 | 1.335 (3) | C18—C19 | 1.388 (4) |
| N4—C20 | 1.350 (3) | C18—H18 | 0.9300 |
| C1—C2 | 1.388 (4) | C19—C20 | 1.481 (4) |
| C1—C6 | 1.388 (3) | C20—C21 | 1.381 (4) |
| C1—C7 | 1.490 (3) | C21—C22 | 1.371 (5) |
| C2—C3 | 1.375 (4) | C21—H21 | 0.9300 |
| C2—H2 | 0.9300 | C22—C23 | 1.367 (5) |
| C3—C4 | 1.396 (4) | C22—H22 | 0.9300 |
| C3—H3 | 0.9300 | C23—C24 | 1.371 (4) |
| C4—C5 | 1.386 (4) | C23—H23 | 0.9300 |
| C5—C6 | 1.373 (3) | C24—H24 | 0.9300 |
| C5—H5 | 0.9300 | ||
| O1—Zn1—O3 | 141.27 (9) | C13—C8—C14 | 120.3 (2) |
| O1—Zn1—N4 | 107.51 (8) | C10—C9—C8 | 121.5 (3) |
| O3—Zn1—N4 | 108.58 (8) | C10—C9—H9 | 119.2 |
| O1—Zn1—N3 | 103.23 (9) | C8—C9—H9 | 119.2 |
| O3—Zn1—N3 | 97.43 (9) | C9—C10—C11 | 120.2 (3) |
| N4—Zn1—N3 | 78.22 (9) | C9—C10—H10 | 119.9 |
| O1—Zn1—O4 | 108.31 (8) | C11—C10—H10 | 119.9 |
| O3—Zn1—O4 | 59.37 (8) | C12—C11—C10 | 118.6 (2) |
| N4—Zn1—O4 | 88.61 (8) | C12—C11—N2 | 120.4 (3) |
| N3—Zn1—O4 | 148.23 (8) | C10—C11—N2 | 120.9 (3) |
| O1—Zn1—C14 | 129.59 (8) | C13—C12—C11 | 121.0 (3) |
| O3—Zn1—C14 | 30.11 (8) | C13—C12—H12 | 119.5 |
| N4—Zn1—C14 | 98.66 (8) | C11—C12—H12 | 119.5 |
| N3—Zn1—C14 | 124.19 (9) | C12—C13—C8 | 121.2 (2) |
| O4—Zn1—C14 | 29.29 (8) | C12—C13—H13 | 119.4 |
| C7—O1—Zn1 | 114.85 (16) | C8—C13—H13 | 119.4 |
| C14—O3—Zn1 | 99.43 (16) | O4—C14—O3 | 120.2 (3) |
| C14—O4—Zn1 | 80.92 (18) | O4—C14—C8 | 121.8 (3) |
| C4—N1—H1D | 119.3 | O3—C14—C8 | 118.0 (2) |
| C4—N1—H1E | 115.8 | O4—C14—Zn1 | 69.79 (16) |
| H1D—N1—H1E | 115.8 | O3—C14—Zn1 | 50.46 (12) |
| C11—N2—H2D | 108.4 | C8—C14—Zn1 | 167.2 (2) |
| C11—N2—H2E | 110.2 | N3—C15—C16 | 121.9 (4) |
| H2D—N2—H2E | 113.1 | N3—C15—H15 | 119.1 |
| C19—N3—C15 | 119.3 (3) | C16—C15—H15 | 119.1 |
| C19—N3—Zn1 | 114.43 (18) | C17—C16—C15 | 119.1 (4) |
| C15—N3—Zn1 | 126.2 (2) | C17—C16—H16 | 120.5 |
| C24—N4—C20 | 119.2 (2) | C15—C16—H16 | 120.5 |
| C24—N4—Zn1 | 125.67 (18) | C16—C17—C18 | 119.6 (3) |
| C20—N4—Zn1 | 115.14 (18) | C16—C17—H17 | 120.2 |
| C2—C1—C6 | 117.6 (2) | C18—C17—H17 | 120.2 |
| C2—C1—C7 | 121.3 (2) | C17—C18—C19 | 118.9 (3) |
| C6—C1—C7 | 121.1 (2) | C17—C18—H18 | 120.5 |
| C3—C2—C1 | 121.3 (2) | C19—C18—H18 | 120.5 |
| C3—C2—H2 | 119.4 | N3—C19—C18 | 121.2 (3) |
| C1—C2—H2 | 119.4 | N3—C19—C20 | 116.1 (2) |
| C2—C3—C4 | 120.8 (2) | C18—C19—C20 | 122.7 (3) |
| C2—C3—H3 | 119.6 | N4—C20—C21 | 120.7 (3) |
| C4—C3—H3 | 119.6 | N4—C20—C19 | 115.9 (2) |
| N1—C4—C5 | 120.3 (3) | C21—C20—C19 | 123.4 (3) |
| N1—C4—C3 | 121.7 (3) | C22—C21—C20 | 119.3 (3) |
| C5—C4—C3 | 118.0 (2) | C22—C21—H21 | 120.4 |
| C6—C5—C4 | 120.8 (2) | C20—C21—H21 | 120.4 |
| C6—C5—H5 | 119.6 | C23—C22—C21 | 119.9 (3) |
| C4—C5—H5 | 119.6 | C23—C22—H22 | 120.0 |
| C5—C6—C1 | 121.6 (2) | C21—C22—H22 | 120.0 |
| C5—C6—H6 | 119.2 | C22—C23—C24 | 118.5 (3) |
| C1—C6—H6 | 119.2 | C22—C23—H23 | 120.7 |
| O2—C7—O1 | 122.4 (2) | C24—C23—H23 | 120.7 |
| O2—C7—C1 | 122.0 (3) | N4—C24—C23 | 122.4 (3) |
| O1—C7—C1 | 115.6 (2) | N4—C24—H24 | 118.8 |
| C9—C8—C13 | 117.5 (2) | C23—C24—H24 | 118.8 |
| C9—C8—C14 | 122.2 (2) | ||
| O3—Zn1—O1—C7 | 26.2 (3) | C10—C11—C12—C13 | −0.9 (4) |
| N4—Zn1—O1—C7 | −175.82 (17) | N2—C11—C12—C13 | 175.6 (2) |
| N3—Zn1—O1—C7 | −94.20 (19) | C11—C12—C13—C8 | 0.6 (4) |
| O4—Zn1—O1—C7 | 89.73 (19) | C9—C8—C13—C12 | 0.6 (4) |
| C14—Zn1—O1—C7 | 66.4 (2) | C14—C8—C13—C12 | −179.9 (2) |
| O1—Zn1—O3—C14 | 83.1 (2) | Zn1—O4—C14—O3 | 3.2 (2) |
| N4—Zn1—O3—C14 | −74.76 (17) | Zn1—O4—C14—C8 | −173.9 (2) |
| N3—Zn1—O3—C14 | −154.77 (16) | Zn1—O3—C14—O4 | −3.9 (3) |
| O4—Zn1—O3—C14 | 2.02 (14) | Zn1—O3—C14—C8 | 173.36 (18) |
| O1—Zn1—O4—C14 | −141.45 (15) | C9—C8—C14—O4 | 3.2 (4) |
| O3—Zn1—O4—C14 | −2.07 (15) | C13—C8—C14—O4 | −176.3 (2) |
| N4—Zn1—O4—C14 | 110.56 (16) | C9—C8—C14—O3 | −174.0 (2) |
| N3—Zn1—O4—C14 | 45.8 (2) | C13—C8—C14—O3 | 6.5 (3) |
| O1—Zn1—N3—C19 | −109.74 (19) | C9—C8—C14—Zn1 | −150.2 (7) |
| O3—Zn1—N3—C19 | 103.24 (19) | C13—C8—C14—Zn1 | 30.3 (9) |
| N4—Zn1—N3—C19 | −4.28 (19) | O1—Zn1—C14—O4 | 50.15 (19) |
| O4—Zn1—N3—C19 | 63.2 (3) | O3—Zn1—C14—O4 | 176.4 (3) |
| C14—Zn1—N3—C19 | 88.3 (2) | N4—Zn1—C14—O4 | −71.23 (16) |
| O1—Zn1—N3—C15 | 73.6 (3) | N3—Zn1—C14—O4 | −152.83 (15) |
| O3—Zn1—N3—C15 | −73.4 (3) | O1—Zn1—C14—O3 | −126.29 (17) |
| N4—Zn1—N3—C15 | 179.1 (3) | N4—Zn1—C14—O3 | 112.32 (16) |
| O4—Zn1—N3—C15 | −113.5 (3) | N3—Zn1—C14—O3 | 30.7 (2) |
| C14—Zn1—N3—C15 | −88.4 (3) | O4—Zn1—C14—O3 | −176.4 (3) |
| O1—Zn1—N4—C24 | −77.9 (2) | O1—Zn1—C14—C8 | −153.8 (8) |
| O3—Zn1—N4—C24 | 87.8 (2) | O3—Zn1—C14—C8 | −27.5 (8) |
| N3—Zn1—N4—C24 | −178.2 (2) | N4—Zn1—C14—C8 | 84.8 (8) |
| O4—Zn1—N4—C24 | 30.9 (2) | N3—Zn1—C14—C8 | 3.2 (9) |
| C14—Zn1—N4—C24 | 58.5 (2) | O4—Zn1—C14—C8 | 156.1 (9) |
| O1—Zn1—N4—C20 | 103.27 (18) | C19—N3—C15—C16 | −0.5 (5) |
| O3—Zn1—N4—C20 | −91.02 (18) | Zn1—N3—C15—C16 | 176.0 (3) |
| N3—Zn1—N4—C20 | 2.95 (17) | N3—C15—C16—C17 | −0.7 (7) |
| O4—Zn1—N4—C20 | −147.95 (18) | C15—C16—C17—C18 | 1.0 (7) |
| C14—Zn1—N4—C20 | −120.34 (18) | C16—C17—C18—C19 | −0.2 (6) |
| C6—C1—C2—C3 | 0.6 (4) | C15—N3—C19—C18 | 1.3 (4) |
| C7—C1—C2—C3 | −178.3 (2) | Zn1—N3—C19—C18 | −175.6 (2) |
| C1—C2—C3—C4 | −0.3 (4) | C15—N3—C19—C20 | −178.2 (3) |
| C2—C3—C4—N1 | −178.9 (3) | Zn1—N3—C19—C20 | 4.9 (3) |
| C2—C3—C4—C5 | −0.6 (4) | C17—C18—C19—N3 | −0.9 (5) |
| N1—C4—C5—C6 | 179.6 (2) | C17—C18—C19—C20 | 178.5 (3) |
| C3—C4—C5—C6 | 1.3 (4) | C24—N4—C20—C21 | 1.3 (4) |
| C4—C5—C6—C1 | −1.1 (4) | Zn1—N4—C20—C21 | −179.8 (2) |
| C2—C1—C6—C5 | 0.1 (4) | C24—N4—C20—C19 | 179.7 (2) |
| C7—C1—C6—C5 | 179.0 (2) | Zn1—N4—C20—C19 | −1.4 (3) |
| Zn1—O1—C7—O2 | 2.4 (3) | N3—C19—C20—N4 | −2.4 (3) |
| Zn1—O1—C7—C1 | −177.28 (15) | C18—C19—C20—N4 | 178.1 (3) |
| C2—C1—C7—O2 | −13.1 (4) | N3—C19—C20—C21 | 176.0 (3) |
| C6—C1—C7—O2 | 168.1 (3) | C18—C19—C20—C21 | −3.5 (4) |
| C2—C1—C7—O1 | 166.6 (2) | N4—C20—C21—C22 | −0.7 (5) |
| C6—C1—C7—O1 | −12.2 (3) | C19—C20—C21—C22 | −179.0 (3) |
| C13—C8—C9—C10 | −1.5 (4) | C20—C21—C22—C23 | 0.0 (6) |
| C14—C8—C9—C10 | 179.0 (2) | C21—C22—C23—C24 | 0.1 (6) |
| C8—C9—C10—C11 | 1.2 (4) | C20—N4—C24—C23 | −1.2 (4) |
| C9—C10—C11—C12 | 0.0 (4) | Zn1—N4—C24—C23 | −180.0 (2) |
| C9—C10—C11—N2 | −176.5 (2) | C22—C23—C24—N4 | 0.5 (5) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1D···N2i | 0.88 | 2.33 | 3.202 (4) | 169. |
| N1—H1E···O2ii | 0.88 | 2.36 | 3.181 (4) | 155. |
| N2—H2E···O2iii | 0.88 | 2.28 | 3.116 (4) | 159. |
Symmetry codes: (i) x−2, y+1, z; (ii) x−1, y, z; (iii) −x+1, −y, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU5334).
References
- Bruker (2001). SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chohan, Z. H. & Naseer, M. M. (2007). Appl. Organomet. Chem. 21, 728–738.
- Huang, Q.-M., Pan, Z.-Q. & Wang, P. (2006). Bioorg. Med. Chem. Lett. 16, 3030–3033. [DOI] [PubMed]
- Ispir, E., Kurtoglu, M. & Toroglu, S. (2006). Synth. React. Inorg. Met. Org. Chem. 36, 627–631.
- Lo, P.-C., Zhao, B.-Z. & Duan, W.-B. (2007). Bioorg. Med. Chem. Lett. 17, 1073–1077. [DOI] [PubMed]
- Maria, A. Z., Roberto, D. & Stefano, M. (1996). Polyhedron, pp. 277–283.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wang, R.-H., Jiang, F.-L., Zhou, Y.-F., Han, L. & Hong, M.-C. (2005). Inorg. Chim. Acta, 358, 545–554.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811039389/xu5334sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811039389/xu5334Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




