Abstract
In the title compound, C16H15NOS, the thiazolidine ring, which is essentially planar [maximum deviation = 0.071 (2) Å], makes dihedral angles of 88.01 (8) and 87.21 (8)° with the terminal phenyl rings. The dihedral angle between the phenyl rings is 49.45 (5)°. In the crystal, molecules are linked by a weak intermolecular C—H⋯O hydrogen bond, forming a supramolecular chain along the b axis. Furthermore, the crystal packing is stabilized by a weak C—H⋯π interaction.
Related literature
For details and applications of thiazolidine-4-ones, see: Dutta et al. (1990 ▶); Jadhav & Ingle (1978 ▶); Gursoy & Terzioglu (2005 ▶); Rawal et al. (2007 ▶); Shrivastava et al. (2005 ▶); Look et al. (1996 ▶); Anders et al. (2001 ▶); Barreca et al. (2001 ▶); Diurno et al. (1992 ▶).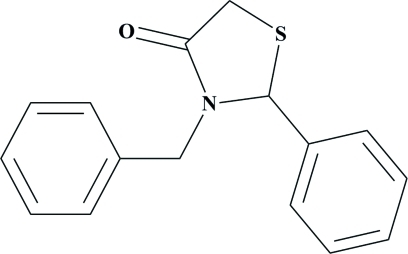
Experimental
Crystal data
C16H15NOS
M r = 269.35
Monoclinic,

a = 13.5734 (15) Å
b = 10.1402 (11) Å
c = 10.1496 (11) Å
β = 104.305 (2)°
V = 1353.6 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.23 mm−1
T = 296 K
0.41 × 0.19 × 0.06 mm
Data collection
Bruker APEXII DUO CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.913, T max = 0.985
21164 measured reflections
3990 independent reflections
2813 reflections with I > 2σ(I)
R int = 0.041
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.112
S = 1.05
3990 reflections
172 parameters
H-atom parameters constrained
Δρmax = 0.21 e Å−3
Δρmin = −0.25 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811037706/is2778sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811037706/is2778Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811037706/is2778Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 is the centroid of the C11–C16 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C14—H14A⋯O1i | 0.93 | 2.47 | 3.323 (2) | 153 |
| C2—H2A⋯Cg1ii | 0.93 | 2.99 | 3.705 (3) | 134 |
Symmetry codes: (i) 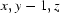 ; (ii)
; (ii)  .
.
Acknowledgments
HKF and MH thank the Malaysian Government and Universiti Sains Malaysia for the Research University grant No. 1001/PFIZIK/811160. MH thanks Universiti Sains Malaysia for a post-doctoral research fellowship.
supplementary crystallographic information
Comment
One of the main objectives of organic and medicinal chemistry is the design, synthesis and production of molecules having value as human therapeutic agents. During the past decade, combinatorial chemistry has provided access to chemical libraries based on privileged structures with heterocyclic moiety receiving special attention as they belong to a class of compounds with proven utility in medicinal chemistry. There are numerous biologically active molecules with five-membered rings, containing two hetero atoms. Among them, thiazolidin-4-ones are the most extensively investigated class of compounds, which have many interesting activity profiles namely bactericidal (Dutta et al., 1990), antifungal (Jadhav & Ingle, 1978), anticonvulsant (Gursoy & Terzioglu, 2005), anti-HIV (Rawal et al., 2007), antituberculotic (Shrivastava et al., 2005), COX-1 inhibitors (Look et al., 1996), inhibitors of the bacterial enzyme MurB (Anders et al., 2001), non-nucleoside inhibitors of HIV-RT (Barreca et al., 2001) and anti-histaminic agents (Diurno et al., 1992).
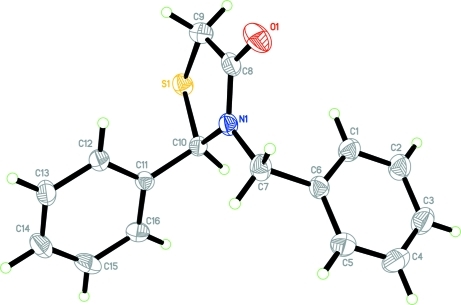
The asymmetric unit of the title compound is shown in Fig. 1. The thiazolidine (S1/N1/C8–C10) ring is essentially planar, with a maximum deviation of 0.071 (2) Å for atom C10. The central thiazolidine (S1/N1/C8–C10) ring makes dihedral angles of 88.01 (8) and 87.21 (8)° with the terminal phenyl (C1–C6) and (C11–C16) rings, respectively. The dihedral angle between the phenyl (C1–C6) and (C11–C16) rings is 49.45 (5)°.
In the crystal structure, (Fig. 2), the molecules are linked by intermolecular weak C—H···O hydrogen bonds forming supramolecular chains along the b-axis. Furthermore, the crystal packing is stabilized by weak C—H···π interactions involving the C11–C16 ring.
Experimental
To a well ground intimate mixture of triphenyl phosphine (0.43 g, 1.6 mmol) and benzaldehyde, (0.15 g, 1.5 mmol) in a microwave vial (10 ml) equipped with a magnetic stirring bar, benzylazide, (0.2 g, 1.5 mmol) was added in drop with stirring. Stirring was continued until liberation of nitrogen ceased and then mercaptoacetic acid, (0.15 g, 1.6 mmol) was added to the above mixture and the reaction vessel was sealed with a septum. It was then placed into the cavity of a focused monomode microwave reactor (CEM Discover, benchmate) and operated at 150°C (temperature monitored by a built-in IR sensor), power 80W for 10 minutes. The reaction temperature was maintained by modulating the power level of the reactor. The reaction mixture was allowed to stand at room temperature. Then the residue was purified by column chromatography on silica (petrolium ether–ethyl acetate, 94:6) to afford the 3-benzyl-2-phenylthiazolidin-4-one. Yield: 0.38g (95%); m.p. 152–155°C.
Refinement
All hydrogen atoms were positioned geometrically (C—H = 0.93–0.98 Å) and were refined using a riding model, with Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
An ORTEP view of the title compound, showing 30% probability displacement ellipsoids.
Fig. 2.
The crystal packing of the title compound, viewed along the a axis.
Crystal data
| C16H15NOS | F(000) = 568 |
| Mr = 269.35 | Dx = 1.322 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4044 reflections |
| a = 13.5734 (15) Å | θ = 2.9–24.9° |
| b = 10.1402 (11) Å | µ = 0.23 mm−1 |
| c = 10.1496 (11) Å | T = 296 K |
| β = 104.305 (2)° | Plate, colourless |
| V = 1353.6 (3) Å3 | 0.41 × 0.19 × 0.06 mm |
| Z = 4 |
Data collection
| Bruker APEXII DUO CCD area-detector diffractometer | 3990 independent reflections |
| Radiation source: fine-focus sealed tube | 2813 reflections with I > 2σ(I) |
| graphite | Rint = 0.041 |
| φ and ω scans | θmax = 30.2°, θmin = 1.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −19→19 |
| Tmin = 0.913, Tmax = 0.985 | k = −14→14 |
| 21164 measured reflections | l = −14→14 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.112 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0448P)2 + 0.2789P] where P = (Fo2 + 2Fc2)/3 |
| 3990 reflections | (Δ/σ)max = 0.001 |
| 172 parameters | Δρmax = 0.21 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.04954 (3) | 0.22379 (4) | 0.22886 (4) | 0.04505 (12) | |
| O1 | 0.16669 (11) | 0.46692 (11) | 0.51751 (13) | 0.0650 (4) | |
| N1 | 0.21308 (9) | 0.28385 (11) | 0.41853 (12) | 0.0389 (3) | |
| C1 | 0.34627 (12) | 0.44309 (17) | 0.30014 (17) | 0.0510 (4) | |
| H1A | 0.2783 | 0.4676 | 0.2838 | 0.061* | |
| C2 | 0.40708 (14) | 0.49962 (19) | 0.22451 (19) | 0.0582 (4) | |
| H2A | 0.3798 | 0.5617 | 0.1580 | 0.070* | |
| C3 | 0.50706 (14) | 0.4646 (2) | 0.2471 (2) | 0.0637 (5) | |
| H3A | 0.5481 | 0.5024 | 0.1963 | 0.076* | |
| C4 | 0.54623 (15) | 0.3732 (2) | 0.3454 (3) | 0.0810 (7) | |
| H4A | 0.6143 | 0.3492 | 0.3613 | 0.097* | |
| C5 | 0.48589 (14) | 0.3161 (2) | 0.4212 (2) | 0.0650 (5) | |
| H5A | 0.5136 | 0.2538 | 0.4873 | 0.078* | |
| C6 | 0.38484 (11) | 0.35067 (14) | 0.39972 (15) | 0.0410 (3) | |
| C7 | 0.32037 (12) | 0.29270 (17) | 0.48752 (16) | 0.0479 (4) | |
| H7A | 0.3454 | 0.2052 | 0.5166 | 0.057* | |
| H7B | 0.3278 | 0.3467 | 0.5683 | 0.057* | |
| C8 | 0.14552 (12) | 0.37589 (14) | 0.43636 (15) | 0.0434 (3) | |
| C9 | 0.04128 (13) | 0.35455 (17) | 0.34511 (19) | 0.0533 (4) | |
| H9A | 0.0173 | 0.4347 | 0.2953 | 0.064* | |
| H9B | −0.0061 | 0.3309 | 0.3987 | 0.064* | |
| C10 | 0.17936 (10) | 0.17886 (13) | 0.32051 (14) | 0.0358 (3) | |
| H10A | 0.2225 | 0.1788 | 0.2560 | 0.043* | |
| C11 | 0.18426 (10) | 0.04347 (13) | 0.38516 (13) | 0.0350 (3) | |
| C12 | 0.14070 (11) | 0.02045 (14) | 0.49347 (15) | 0.0420 (3) | |
| H12A | 0.1104 | 0.0896 | 0.5291 | 0.050* | |
| C13 | 0.14219 (12) | −0.10439 (16) | 0.54857 (17) | 0.0499 (4) | |
| H13A | 0.1127 | −0.1188 | 0.6209 | 0.060* | |
| C14 | 0.18706 (13) | −0.20754 (16) | 0.49698 (19) | 0.0547 (4) | |
| H14A | 0.1879 | −0.2915 | 0.5342 | 0.066* | |
| C15 | 0.23077 (14) | −0.18557 (16) | 0.38958 (19) | 0.0561 (4) | |
| H15A | 0.2611 | −0.2550 | 0.3544 | 0.067* | |
| C16 | 0.22959 (12) | −0.06032 (15) | 0.33392 (16) | 0.0455 (4) | |
| H16A | 0.2594 | −0.0461 | 0.2619 | 0.055* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0441 (2) | 0.0416 (2) | 0.0451 (2) | −0.00081 (16) | 0.00278 (16) | 0.00479 (16) |
| O1 | 0.0919 (10) | 0.0400 (6) | 0.0633 (7) | −0.0033 (6) | 0.0195 (7) | −0.0119 (6) |
| N1 | 0.0399 (6) | 0.0325 (6) | 0.0436 (6) | −0.0060 (5) | 0.0088 (5) | −0.0002 (5) |
| C1 | 0.0404 (8) | 0.0553 (10) | 0.0561 (9) | 0.0005 (7) | 0.0099 (7) | 0.0130 (7) |
| C2 | 0.0548 (10) | 0.0599 (11) | 0.0594 (10) | −0.0048 (8) | 0.0132 (8) | 0.0154 (8) |
| C3 | 0.0508 (10) | 0.0721 (13) | 0.0721 (12) | −0.0090 (9) | 0.0225 (9) | 0.0089 (10) |
| C4 | 0.0441 (10) | 0.0872 (16) | 0.1158 (18) | 0.0105 (10) | 0.0273 (11) | 0.0296 (14) |
| C5 | 0.0480 (10) | 0.0596 (11) | 0.0852 (13) | 0.0083 (8) | 0.0119 (9) | 0.0229 (10) |
| C6 | 0.0383 (7) | 0.0363 (7) | 0.0452 (8) | −0.0057 (6) | 0.0041 (6) | −0.0011 (6) |
| C7 | 0.0456 (8) | 0.0481 (9) | 0.0450 (8) | −0.0090 (7) | 0.0020 (7) | 0.0074 (7) |
| C8 | 0.0561 (9) | 0.0309 (7) | 0.0462 (8) | −0.0036 (6) | 0.0185 (7) | 0.0023 (6) |
| C9 | 0.0496 (9) | 0.0456 (9) | 0.0662 (10) | 0.0059 (7) | 0.0173 (8) | −0.0016 (8) |
| C10 | 0.0372 (7) | 0.0346 (7) | 0.0369 (7) | −0.0029 (5) | 0.0117 (5) | −0.0001 (5) |
| C11 | 0.0339 (7) | 0.0319 (6) | 0.0382 (7) | −0.0008 (5) | 0.0069 (5) | −0.0012 (5) |
| C12 | 0.0450 (8) | 0.0365 (7) | 0.0472 (8) | −0.0005 (6) | 0.0165 (6) | 0.0010 (6) |
| C13 | 0.0495 (9) | 0.0448 (9) | 0.0555 (9) | −0.0074 (7) | 0.0130 (7) | 0.0118 (7) |
| C14 | 0.0547 (10) | 0.0337 (8) | 0.0667 (11) | −0.0038 (7) | −0.0020 (8) | 0.0079 (7) |
| C15 | 0.0611 (11) | 0.0368 (8) | 0.0649 (11) | 0.0126 (7) | 0.0050 (9) | −0.0067 (7) |
| C16 | 0.0471 (8) | 0.0443 (8) | 0.0451 (8) | 0.0074 (7) | 0.0111 (7) | −0.0034 (6) |
Geometric parameters (Å, °)
| S1—C9 | 1.7967 (17) | C7—H7A | 0.9700 |
| S1—C10 | 1.8352 (14) | C7—H7B | 0.9700 |
| O1—C8 | 1.2231 (18) | C8—C9 | 1.503 (2) |
| N1—C8 | 1.3515 (19) | C9—H9A | 0.9700 |
| N1—C10 | 1.4518 (18) | C9—H9B | 0.9700 |
| N1—C7 | 1.4544 (19) | C10—C11 | 1.5160 (19) |
| C1—C2 | 1.383 (2) | C10—H10A | 0.9800 |
| C1—C6 | 1.383 (2) | C11—C16 | 1.3838 (19) |
| C1—H1A | 0.9300 | C11—C12 | 1.391 (2) |
| C2—C3 | 1.366 (3) | C12—C13 | 1.382 (2) |
| C2—H2A | 0.9300 | C12—H12A | 0.9300 |
| C3—C4 | 1.369 (3) | C13—C14 | 1.377 (2) |
| C3—H3A | 0.9300 | C13—H13A | 0.9300 |
| C4—C5 | 1.382 (3) | C14—C15 | 1.382 (3) |
| C4—H4A | 0.9300 | C14—H14A | 0.9300 |
| C5—C6 | 1.380 (2) | C15—C16 | 1.389 (2) |
| C5—H5A | 0.9300 | C15—H15A | 0.9300 |
| C6—C7 | 1.513 (2) | C16—H16A | 0.9300 |
| C9—S1—C10 | 93.34 (7) | C8—C9—S1 | 107.99 (11) |
| C8—N1—C10 | 119.26 (12) | C8—C9—H9A | 110.1 |
| C8—N1—C7 | 121.65 (13) | S1—C9—H9A | 110.1 |
| C10—N1—C7 | 118.93 (12) | C8—C9—H9B | 110.1 |
| C2—C1—C6 | 121.10 (15) | S1—C9—H9B | 110.1 |
| C2—C1—H1A | 119.4 | H9A—C9—H9B | 108.4 |
| C6—C1—H1A | 119.4 | N1—C10—C11 | 113.25 (11) |
| C3—C2—C1 | 120.23 (17) | N1—C10—S1 | 105.43 (9) |
| C3—C2—H2A | 119.9 | C11—C10—S1 | 112.22 (9) |
| C1—C2—H2A | 119.9 | N1—C10—H10A | 108.6 |
| C2—C3—C4 | 119.27 (17) | C11—C10—H10A | 108.6 |
| C2—C3—H3A | 120.4 | S1—C10—H10A | 108.6 |
| C4—C3—H3A | 120.4 | C16—C11—C12 | 119.00 (13) |
| C3—C4—C5 | 120.83 (18) | C16—C11—C10 | 120.14 (13) |
| C3—C4—H4A | 119.6 | C12—C11—C10 | 120.83 (12) |
| C5—C4—H4A | 119.6 | C13—C12—C11 | 120.47 (14) |
| C6—C5—C4 | 120.54 (17) | C13—C12—H12A | 119.8 |
| C6—C5—H5A | 119.7 | C11—C12—H12A | 119.8 |
| C4—C5—H5A | 119.7 | C14—C13—C12 | 120.36 (16) |
| C5—C6—C1 | 118.02 (15) | C14—C13—H13A | 119.8 |
| C5—C6—C7 | 120.33 (14) | C12—C13—H13A | 119.8 |
| C1—C6—C7 | 121.60 (14) | C13—C14—C15 | 119.60 (15) |
| N1—C7—C6 | 113.35 (12) | C13—C14—H14A | 120.2 |
| N1—C7—H7A | 108.9 | C15—C14—H14A | 120.2 |
| C6—C7—H7A | 108.9 | C14—C15—C16 | 120.29 (15) |
| N1—C7—H7B | 108.9 | C14—C15—H15A | 119.9 |
| C6—C7—H7B | 108.9 | C16—C15—H15A | 119.9 |
| H7A—C7—H7B | 107.7 | C11—C16—C15 | 120.28 (15) |
| O1—C8—N1 | 123.85 (15) | C11—C16—H16A | 119.9 |
| O1—C8—C9 | 123.53 (15) | C15—C16—H16A | 119.9 |
| N1—C8—C9 | 112.62 (13) | ||
| C6—C1—C2—C3 | 0.1 (3) | C8—N1—C10—C11 | −114.56 (14) |
| C1—C2—C3—C4 | −0.1 (3) | C7—N1—C10—C11 | 69.99 (16) |
| C2—C3—C4—C5 | 0.2 (4) | C8—N1—C10—S1 | 8.50 (15) |
| C3—C4—C5—C6 | −0.3 (4) | C7—N1—C10—S1 | −166.95 (10) |
| C4—C5—C6—C1 | 0.3 (3) | C9—S1—C10—N1 | −10.43 (10) |
| C4—C5—C6—C7 | −177.10 (19) | C9—S1—C10—C11 | 113.28 (11) |
| C2—C1—C6—C5 | −0.2 (3) | N1—C10—C11—C16 | −131.16 (14) |
| C2—C1—C6—C7 | 177.17 (16) | S1—C10—C11—C16 | 109.62 (13) |
| C8—N1—C7—C6 | −98.30 (17) | N1—C10—C11—C12 | 50.93 (18) |
| C10—N1—C7—C6 | 77.04 (17) | S1—C10—C11—C12 | −68.30 (15) |
| C5—C6—C7—N1 | −151.78 (16) | C16—C11—C12—C13 | −0.4 (2) |
| C1—C6—C7—N1 | 30.9 (2) | C10—C11—C12—C13 | 177.59 (13) |
| C10—N1—C8—O1 | 179.17 (14) | C11—C12—C13—C14 | 0.1 (2) |
| C7—N1—C8—O1 | −5.5 (2) | C12—C13—C14—C15 | 0.0 (3) |
| C10—N1—C8—C9 | −1.01 (18) | C13—C14—C15—C16 | 0.0 (3) |
| C7—N1—C8—C9 | 174.31 (13) | C12—C11—C16—C15 | 0.4 (2) |
| O1—C8—C9—S1 | 172.54 (13) | C10—C11—C16—C15 | −177.53 (14) |
| N1—C8—C9—S1 | −7.28 (16) | C14—C15—C16—C11 | −0.3 (2) |
| C10—S1—C9—C8 | 10.22 (12) |
Hydrogen-bond geometry (Å, °)
| Cg1 is the centroid of the C11–C16 ring. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14A···O1i | 0.93 | 2.47 | 3.323 (2) | 153 |
| C2—H2A···Cg1ii | 0.93 | 2.99 | 3.705 (3) | 134 |
Symmetry codes: (i) x, y−1, z; (ii) x, −y−1/2, z−3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS2778).
References
- Anders, C. J., Bronson, J. J., Andrea, D. S. V., Deshpande, S. M., Falk, P. J., Grant-Young, K. A., Harte, W. E., Ho, H., Misco, P. F., Robertson, J. G., Stock, D., Sun, Y. & Walsh, A. W. (2001). Bioorg. Med. Chem. Lett. pp. 715–717. [DOI] [PubMed]
- Barreca, M. L., Chimirri, A., Luca, L. D., Monforte, A., Monforte, P., Rao, A., Zappala, M., Balzarini, J., De Clercq, E., Pannecouque, C. & Witvrouw, M. (2001). Bioorg. Med. Chem. Lett. pp. 1793–1796. [DOI] [PubMed]
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Diurno, M. V., Mazzoni, O., Calignano, P. E., Giordano, F. & Bolognese, A. (1992). J. Med. Chem. 35, 2910–2912. [DOI] [PubMed]
- Dutta, M. M., Goswami, B. N. & Kataky, J. C. (1990). J. Indian Chem. Soc. 67, 332–334.
- Gursoy, A. & Terzioglu, N. (2005). Turk. J. Chem. 29, 247–254.
- Jadhav, K. P. & Ingle, D. B. (1978). J. Indian Chem. Soc. 4, 424–426.
- Look, G. C., Schullek, J. R., Homes, C. P., Chinn, J. P., Gordon, E. M. & Gallop, M. A. (1996). Bioorg. Med. Chem. Lett. 6, 707–712.
- Rawal, R. K., Tripathi, R., Katti, S. B., Pannecouque, C. & Clercq, E. D. (2007). Bioorg. Med. Chem. 15, 1725–1731. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shrivastava, T., Gaikwad, A. K., Haq, W., Sinha, S. & Katti, S. B. (2005). Arkivoc, 2, 120–130.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811037706/is2778sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811037706/is2778Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811037706/is2778Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report