Abstract
An allele of the 1-aminocyclopropane-1-carboxylic acid (ACC) synthase gene (Md-ACS1), the transcript and translated product of which have been identified in ripening apples (Malus domestica), was isolated from a genomic library of the apple cultivar, Golden Delicious. The predicted coding region of this allele (ACS1-2) showed that seven nucleotide substitutions in the corresponding region of ACS1-1 resulted in just one amino acid transition. A 162-bp sequence characterized as a short interspersed repetitive element retrotransposon was inserted in the 5′-flanking region of ACS1-2 corresponding to position −781 in ACS1-1. The XhoI site located near the 3′ end of the predicted coding region of ACS1-2 was absent from the reverse transcriptase-polymerase chain reaction product, revealing that exclusive transcription from ACS1-1 occurs during ripening of cv Golden Delicious fruit. DNA gel-blot and polymerase chain reaction analyses of genomic DNAs showed clearly that apple cultivars were either heterozygous for ACS1-1 and ACS1-2 or homozygous for each type. RNA gel-blot analysis of the ACS1-2 homozygous Fuji apple, which produces little ethylene and has a long storage life, demonstrated that the level of transcription from ACS1-2 during the ripening stage was very low.
Ethylene is a gaseous plant hormone that regulates many physiological processes in plant growth and development. Its synthesis is induced not only by stress, such as wounding, but also during unique developmental stages, such as seed germination, fruit ripening, and leaf and flower abscission (Yang and Hoffman, 1984; Ky et al., 1992; Rodrigues-Pousasa et al., 1993). Because large amounts of fruit and vegetables are lost due to the effect of ethylene on plant senescence, the significance of controlling the synthesis of ethylene is obvious (Theologis, 1992). The enzyme that limits ethylene production is ACC synthase (EC 4.1.1.14), which catalyzes the formation of ACC, the immediate precursor of ethylene (Gussman et al., 1993; Gorny and Kader, 1997). In tomato ACS is encoded by at least six divergent genes (Lincoln et al., 1993), two of which, LE-ACS2 and LE-ACS4, are expressed during fruit ripening. Expression of antisense RNA derived from LE-ACS2 results in almost complete inhibition of the mRNA of both these genes and extends the longevity of the fruit (Oeller et al., 1991).
The apple is also a typical climacteric fruit, and therefore it appears to be possible to extend its storage life by inhibiting ethylene biosynthesis using antisense RNA technology (Hamilton et al., 1990). Furthermore, the molecular mechanisms responsible for the different storage properties of apple (Malus domestica) cultivars are of interest. At the commercial harvesting stage, the cv Golden Delicious already produces a considerable amount of ethylene, and the amount increases by about 5-fold or more at the climacteric stage, even during cold storage. On the other hand, some cultivars produce very little ethylene during storage and can be stored for up to 1 year at a low temperature with little or no deterioration in the physical quality of the fruit (Gussman et al., 1993). Although a clear relationship between ripening and ethylene production has been demonstrated, much less is known about the molecular mechanism responsible for good, long-term storage properties. Gussman et al. (1993) studied ethylene production and the fruit-softening rates of several apple variants as they ripened. One variant with a long storage life produced only a small amount of ethylene at the time of harvesting and throughout most of an 86-d storage period at 4°C, whereas fruit of the control, cv Golden Delicious, produced large amounts of ethylene under the same conditions. Because the variant converted ACC, but not Met, to ethylene, it is considered to lack the ability to synthesize ACC. Fuji, the most popular cultivar in Japan, has a long storage life and produces very little ethylene at harvest time and during subsequent storage at low temperature (Kondo et al., 1991). However, the molecular mechanisms responsible for this low level of ethylene production are unclear.
Dong et al. (1991) isolated a partial ACS cDNA with a predicted translation product identical to an ACS extracted from ripening apple. Lay-Yee and Knighton (1995) reported the sequence of the full-length cDNA of this ACS (Md-ACS1, M. domestica ACC synthase), and we reported its genomic sequence (Harada et al., 1997). Furthermore, we have isolated an allelic form of Md-ACS1 from a genomic library of cv Golden Delicious, and in the present communication, we describe some of its characteristics. A SINE was inserted into the promotor region of this allele, and the transcription product of the allelic gene was not observed during ripening of cv Golden Delicious fruit. Furthermore, cv Fuji, which produces little ethylene, was found to be homozygous for the allelic gene and to contain a very small amount of the transcript, suggesting that this ACS1 allelic gene may contribute to the good, long-term storage properties of cv Fuji.
MATERIALS AND METHODS
Plant Material
Young leaves of apple (Malus domestica Borkh.) cultivars and their corresponding wild species were collected from the experimental farms of Hirosaki University (Japan) and the Apple Research Center, National Institute of Fruit Tree Science (Morioka, Japan). The cvs Golden Delicious and Fuji were harvested when they were commercially mature on October 21 and November 8, respectively. Then, one-half of the apples were transported to our laboratory and stored at 20°C for 12 d to allow them to reach the climacteric stage. The IEC, flesh firmness, and titratable acidity of the apple fruits were evaluated to obtain preclimacteric and climacteric stage data. Cortical tissue from which the skin and core parts had been removed was frozen by immersion in liquid N2 and then stored in a freezer (−80°C) until isolation of the total RNA.
Measurement of IECs
The internal gas was withdrawn from apple cortical tissue submerged in water under a vacuum, and a 1-mL aliquot of the gas sample was used to evaluate the IEC by GC.
Genomic Library Screening and Sequence Analysis
Genomic DNA was isolated from leaves using the method of Varadarajan and Prakash (1991) and purified using the ethidium bromide-CsCl method of Sambrook et al. (1989). The genomic DNA from cv Golden Delicious was cloned as partial Sau3AI fragments into the XhoI site of FIX (Stratagene), and the library was screened with a 32P-radiolabeled ACS probe DNA (ACS1-A). The positive phage clones (AP1) were used to make a restriction map using XhoI and ApaI, and the 8.7-kb ApaI fragment (pAP1-2) containing the ACS1 gene was subcloned into the pBluescriptII KS vector (Stratagene). Nucleotide sequencing of the 5.7-kb HindIII/EcoRI fragment of pAP1-2 was carried out by subjecting the secondary subcloned fragments to dideoxynucleotide chain termination, and the sequence was determined by analyzing the overlapping clones and both strands. The accession numbers of ACS1-1 and ACS1-2 are U89156 and AB010102, respectively.
Southern-Blot Hybridization
Genomic DNAs extracted from young leaves were digested with a restriction enzyme, separated by agarose gel electrophoresis, and blotted onto a membrane (Hybond N+, Amersham). A fragment of each probe DNA was labeled with 32P-dCTP using the Prime-It II random primer labeling kit (Stratagene), and the blots were hybridized with the probe at 65°C for 15 h in a hybridization buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 6× SSC, 5× Denhardt's solution, 0.2 mg/mL salmon sperm DNA, 20 mm sodium phosphate buffer, and 1% SDS). The membrane was washed with 2× SSC/0.1% (w/v) SDS at room temperature for 30 min and 0.2× SSC/0.1% (w/v) SDS at 65°C for 15 min twice and then autoradiographed using Kodak x-ray film at −80°C and an intensifying screen.
Northern-Blot Hybridization
Total RNA was isolated from apple fruit with guanidinium thiocyanate followed by centrifugation on a CsCl gradient (Lay-Yee et al., 1990). RNA (20 μg per lane) was denatured with 50% (v/v) deionized formamide in 2.2 m formaldehyde at 65°C for 15 min, separated using 2.2 m formaldehyde and 1.2% (w/v) agarose-gel electrophoresis with 40 mm Mops, and then blotted onto membranes (Hybond N+). After the blots were fixed under UV light they were prehybridized in a solution comprising 6× SSC, 50% (v/v) formamide, 5× Denhardt's solution, 0.5% (w/v) SDS, and denatured salmon sperm DNA (Sambrook et al., 1989), and then the 32P-labeled probe DNA was hybridized with the blots at 65°C overnight in the same buffer. The membranes were washed with 2× SSC/0.1% (w/v) SDS at room temperature for 20 min, washed twice with 0.2× SSC/0.1% (w/v) SDS at 65°C for 15 min, and then autoradiographed using Kodak x-ray film at −80°C.
PCR Amplification
PCR was used to produce probe DNAs for ACS1-A, ACS1-B, and ACS3 (Rosenfield et al., 1996). The ACS1 probes were produced by amplifying the cloned fragments, and the ACS3 was derived from the genomic DNA of the apple cv McIntosh. PCR was also carried out to identify the ACS1 allelic forms in apple cultivars and their corresponding wild species. The reaction mixture (50 μL) contained about 50 ng of template genomic DNA, 0.2 μm each primer, 200 μm each deoxyribonucleotide triphosphate, 1× PCR reaction buffer, and 2.5 units of Taq DNA polymerase. It was held at 94°C for 3 min, followed by 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2.5 min with a final 10-min extension step at 72°C. RT-PCR was performed to obtain the cDNA from the 3′ region of the ACS1 transcription product as follows. The reaction mixture comprised 1 μg of poly(A+) RNA, 40 mm KCl, 1 mm DTT, 20 units of RNase inhibitor, 50 units of RT (Perkin-Elmer) and the primer ACS1-3′AR, and reverse transcription was carried out by incubating this mixture at 42°C for 1 h. The tubes were then put on ice, 80 μL of PCR reaction mixture comprising 1× buffer, the forward primer ACS1-3′AF, 2.5 units of Taq polymerase, and 2.5 mm each dNTP were added, and PCR (30 cycles of 94°C for 2 min, 45°C for 1 min, and 72°C for 3 min) was carried out. The first reaction solution and the ACS1-3′B primers were subjected to a second PCR to obtain the product. The following oligonucleotide primers (the positions being numbered according to accession no. U89156 for ACS1 and no. U73816 forACS3) were used for the above reactions: ACS1-AF, 5′-TACCATGAGGTCCACAACAC-3′ (2302–2321); ACS1-AR, 5′-GGTGAGCACTAAGTGGTTGGG-3′ (2813–2793); ACS1BF, 5′-GATGAAAGGTAGCCTGGTCTGA-3′ (4056–4077); ACS1-BR, 5′-TACACTAATCACATTGTATAGAATC-3′ (4513–4489); ACS3-F, 5′-GACAAATAGAAAGAGACCTGAGGACG-3′ (1086–1110); ACS3-R, 5′-CCATCGATTATACAAACTGATTGTG-3′ (1573–1549); ACS1-3′AF, 5′-TCCACAACACAAACGGGATTATTCA-3′ (3312–3336); ACS1-3′AR, 5′-AGGCTACCTTTCATCTACCGGGA-3′ (4070–4048); ACS1-3′BF, 5′-ATCTTCTCGAGTCATGGCTGGC-3′ (2489–2510); ACS1-3′BR, 5′-TTTCATCTACCGGGAATAGGACCGCGG-3′ (4062–4036); ACS1-5′F, 5′-AGAGAGATGCCATTTTTGTTCGTAC-3′ (861–887); and ACS1-5′R, 5′-CCTACAAACTTGCGTGGGGATTATAAGTGT-3′ (1379–1350).
RESULTS
Allele of ACS1
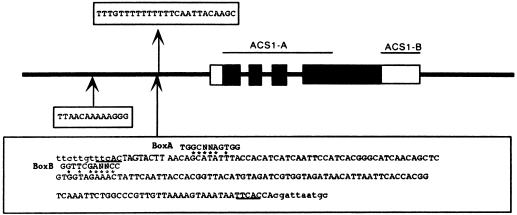
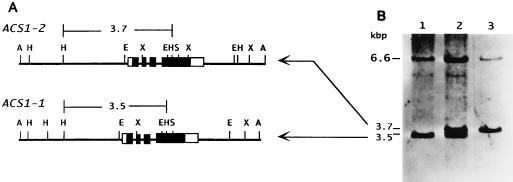
Plaque hybridization of the apple cv Golden Delicious genomic library with the ACS1-A probe (Fig. 1) yielded 13 positive clones. Restriction mapping of their subcloned fragments suggested that the sequence of one clone was very similar to that of the Md-ACS1 gene reported previously (Harada et al., 1997). Sequencing of the subcloned fragment revealed an open reading frame that was almost identical to that of the Md-ACS1 gene and its flanking regions (5676 bp, accession no. AB010102). Therefore, we designated the previously reported sequence ACS1-1 and this new sequence ACS1-2. As shown in Table I, the exons of these sequences are highly homologous and the predicted amino acid sequence of ACS1-2 differs from that of ACS1-1 by only one amino acid, because six of the seven nucleotide substitutions in the predicted ACS1-2 exon region did not affect the translation product because of their third positions in the respective codons. A guanine-to-adenine transition at position 3763 of the ACS1-1 gene changed Gly to Ser in the predicted translation product. By contrast, a significant divergence was found in the 5′-flanking region. A 13-bp sequence from 594 to 607 of ACS1-1 was tandemly repeated in ACS1-2. Furthermore, a 24-bp sequence from 1308 to 1331 of ACS1-1 was missing from ACS1-2 and a 162-bp insertion sequence was found at this site in ACS1-2 (Fig. 1). As shown in Figure 2, Southern blotting with the ACS1-A probe revealed that cv Golden Delicious had three positive fragments in its HindIII-digested genomic DNA, and two of them (3.5 and 3.7 kb) were found to correspond to the fragments containing ACS1-1 and ACS1-2, respectively. The other 6.6-kb fragment observed was thought to be derived from the homologous sequence of the gene family. However, some cultivars had either the ACS1-1 or the ACS1-2 fragment only (Fig. 2B, lanes 1 and 3). Furthermore, cv Amanishiki, which resulted from crossing the respective ACS1-1 and ACS1-2 cvs Indo and Ralus Janet, possessed both ACS1 fragments, as did cv Golden Delicious (data not shown). In light of these results, we concluded that ACS1-2 is an allele of ACS1-1 in the apple genome.
Figure 1.
Structure of Md-ACS1-2. Major differences from that of ACS1-1 are shown schematically. Solid and clear boxes indicate the coding and the untranslated regions, respectively. The lines connecting the solid boxes represent the three introns. The lines connecting the open boxes indicate the 5′- and 3′-flanking regions. The arrow from the 5′-flanking region indicates the ACS1-1 sequence, which was not observed in ACS1-2. Arrows to the 5′-flanking region show the 13-bp tandemly repeated sequence and the 162-bp insertion sequence, which were observed in ACS1-2. The flanking sequences of the insertion are also indicated in lowercase letters. Direct repeats at the flanking sequences are underlined, and two promotor motifs for RNA polymerase III are also shown. The extents of the probes used in this study are indicated as lines.
Table I.
Structural characteristics of the Md-ACS1 genes
| Region | Length
|
Identity | |
|---|---|---|---|
| ACS1-1 | ACS1-2 | ||
| bp | % | ||
| 5′ Flanking | 2112 | 2263 | 91 |
| 5′ Untranslated | 99 | 99 | 100 |
| Exon 1 | 147 | 147 | 100 |
| Intron 1 | 120 | 120 | 99 |
| Exon 2 | 132 | 132 | 100 |
| Intron 2 | 131 | 131 | 100 |
| Exon 3 | 161 | 161 | 99 |
| Intron 3 | 176 | 176 | 100 |
| Exon 4 | 982 | 982 | 99 |
| 3′ Untranslated | 453 | 454 | 98 |
| 3′ Flanking | 1013 | 1011 | 97 |
Figure 2.
A, Restriction maps of the ApaI fragments of phage clones that contain ACS1-1 or ACS1-2. The exons and the untranslated regions are represented as described in the legend of Figure 1. Restriction sites are as follows: A, ApaI; H, HindIII; E, EcoRI; X, XhoI; S, SacI. B, Genomic DNA gel-blot analyses using probe ACS-A. Lane 1, cv Indo; lane 2, cv Golden Delicious; lane 3, cv Fuji. Genomic DNAs were digested by HindIII.
Characteristics of the Insertion SINE Sequence
As shown in Figure 1, we observed that the 162-bp insertion had a flanking 5-bp direct repeat (TTCAC), which led us to assume that this insertion is probably a transposable element. We also detected an A-rich tract at the 3′ end and two conserved polymerase III motifs, boxes A and B, which were located 40 bases apart. These are characteristic features of the SINEs that have been identified in some organisms (Umeda et al., 1991; Mochizuki et al., 1992; Deragon et al., 1996). Furthermore, hybridization of the insertion sequence with digested genomic DNAs from apple, rice, and soybean confirmed that this is present only in the apple genome. When the filter was washed under low-stringency conditions after hybridization, the probe was found to hybridize with many fragments and formed no distinct bands but showed a faint, smeared background, whereas under high-stringency conditions, the probe hybridized with several fragments and distinct bands were formed (Fig. 3). These findings indicate that homologous sequences with some degree of sequence heterogeneity are interspersed in the apple genome. Therefore, we designated this element SINE-Md1. Although many families of SINEs are derived from tRNA or tRNA genes (Yoshioka et al., 1993), a search of the GenBank database using the FASTA program revealed no sequence related to SINE-Md1.
Figure 3.
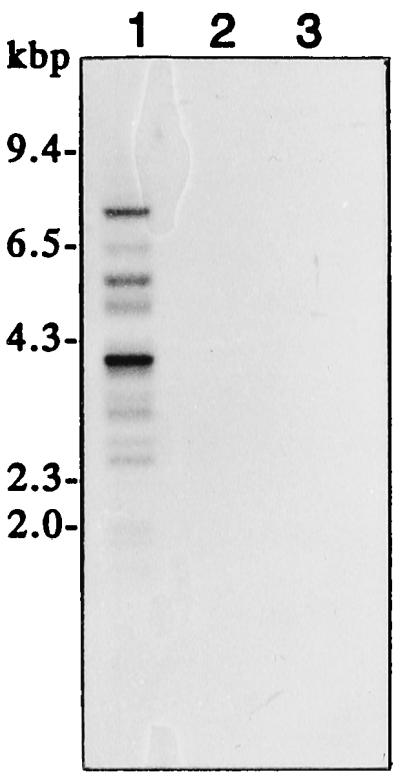
DNA gel-blot analysis of the insertion sequence SINE-Md1. Five micrograms of HindIII-digested genomic DNA was electrophoresed on a 1.2% agarose gel. Lane 1, Apple (cv Golden Delicious); lane 2, rice (cv Nipponbare); lane 3, soybean (cv Williams).
Absence of or Very Low Level of ACS1–2 Expression
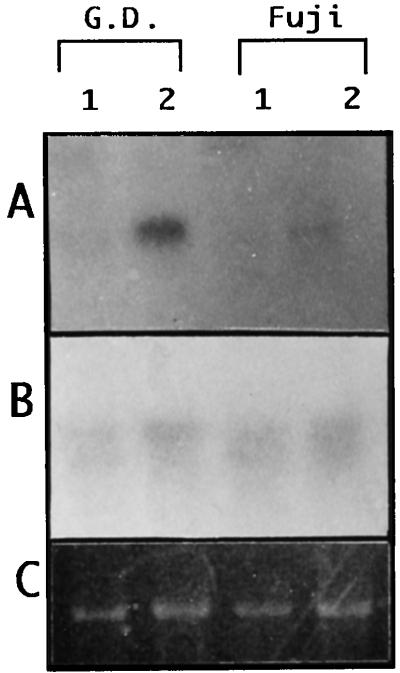
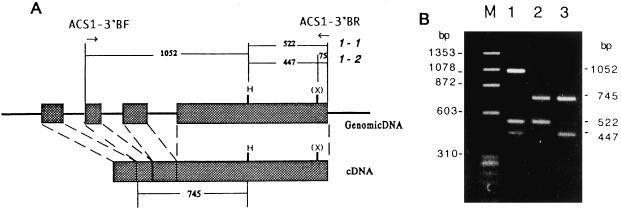
Expression of the ACS1 gene during fruit ripening was studied by carrying out RNA gel-blot analysis of the total RNAs from apple fruit. The IEC of cv Golden Delicious fruit just after harvesting was 16 μL L−1, and after storage at 20°C for 12 d it increased to 296 μL L−1, whereas the IECs of cv Fuji were 0.7 and 57 μL L−1, respectively. The total RNAs extracted from these apples were hybridized with the probe ACS1-B (Fig. 1), which DNA gel-blot analysis demonstrated was specific for the ACS1 gene (data not shown). No positive signals were detected in preclimacteric fruits of either cultivar, whereas clear 2.0-kb signals were detected in climacteric cv Golden Delicious fruit, and those in cv Fuji fruit were very faint (Fig. 4). Even after prolonged autoradiography, no signals were detected in preclimacteric fruit. Then, the same membrane was hybridized with the ACS3 probe, which, in contrast to the ACS1-B probe, yielded very similar signals in both cultivars, irrespective of their ripeness. The poly(A+) RNA fractions extracted from climacteric fruit and the primers ACS1-3′A (Fig. 5) were subjected to RT-PCR, and products of the 3′ region of the ACS1 mRNAs were obtained. Distinct PCR products were amplified by a second PCR using the inside primers (ACS1-3′BF and -3′BR) and first PCR product as the template DNA. Then, we checked whether the product possessed the XhoI site, which is located in the 3′ region of ACS1-2 (Fig. 5), because nucleotide transition from adenine to guanine resulted in the yield of the restriction site in ACS1-2. As shown in Figure 5B, the digestion patterns show that both alleles were present in cv Golden Delicious genomic DNA, whereas the product from the cDNA fraction of cv Golden Delicious was of the ACS1-1 type only, indicating that the mRNA had been transcribed exclusively from the ACS1-1 gene and not from ACS1-2. In contrast, the digestion pattern showed that the RT-PCR product from cv Fuji was derived from the ACS1-2 gene.
Figure 4.
Expression of Md-ACS1 (A) and Md-ACS3 (B) genes in cv Golden Delicious (G.D.) and cv Fuji fruits. Lanes 1, Preclimacteric stage; lanes 2, climacteric stage. Ethidium bromide-staining results for 18S rRNA were used as an internal control.
Figure 5.
A, Theoretical restriction fragment lengths of PCR product from the genomic DNA or cDNA. The structure of Md-ACS1 is indicated as described in the legend of Figure 1. The locations of the primers used in the PCR are shown by arrows. H, HindIII site; (X), ACS1-2-specific XhoI site. B, Fragment lengths of HindIII/XhoI double-digested PCR products. Lane M, φX174/HaeIII; lane 1, genomic DNA of cv Golden Delicious; lane 2, RT-PCR products from climacteric cv Golden Delicious fruit; lane 3, RT-PCR product from climacteric cv Fuji fruit.
ACS1-2 Homozygosity of Cultivars Producing Little Ethylene
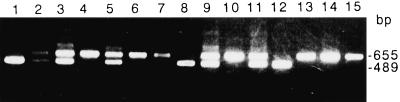
We performed a survey of the ACS1 allele in apple cultivars and their corresponding wild species. To amplify the 5′ region where the SINE-Md1 was inserted into the ACS1-2 gene, we designed a set of primers, ACS1-5′F and ACS1-5′R, for the sequences of ACS1 and carried out PCR using each genomic DNA as the template. According to the pattern of the PCR products (Fig. 6), it was possible to classify the apple species into three groups that were heterozygous or homozygous for the ACS1 alleles (Table II). All of the wild species were either ACS1-1 homozygous or ACS1-1/ACS1-2 heterozygous. It is interesting that all of the cultivars that were homozygous for ACS1-2 produced apples with good, long-term storage properties and/or that produced little ethylene, like cv Fuji.
Figure 6.
Diagnosis of the ACS1 allelic form in apple cultivars. Primers were used to amplify the SINE insert site of the ACS1 5′-flanking region and the products were electrophoresed on a 1.5% agarose gel. Lane 1, cv American Summer Peamain; lane 2, cv Golden Delicious; lane 3, cv Granny Smith; lane 4, cv Fuji; lane 5, cv Jonathan; lane 6, cv Kaori; lane 7, cv Megumi; lane 8, cv McIntosh; lane 9, cv Orin; lane 10, cv Narihoko; lane 11, cv Mutsu; lane 12, cv Cox's Orange Pippin; lane 13, cv Ralus Janet; lane 14, cv Sansa; lane 15, cv Himekami.
Table II.
Types of ACS1 alleles of apple cultivars and Malus species
| ACS1-1/ACS1-1 | ACS1-1/ACS1-2 | ACS1-2/ACS1-2 |
|---|---|---|
| M. baccata | M. prunifolia | Anbishas |
| M. florentina | M. toringoides | Fuji |
| M. floribunda | M. yunnanesis | Himekami |
| M. hupehensis | Amanishiki | Iwakami |
| M. sargentii | Antonovka | Kaori |
| M. sieboldii | Bramley's Seedling | Megumi |
| M. spectabilis | Delicious | Narihoko |
| M. pumila | Golden Delicious | Ralus Janet |
| Amassia | Golden Melon | Sansa |
| American Summer Pearmain | Granny Smith Jonathan | |
| Cox's Orange | Mutsu | |
| Pippin | Orei | |
| Hatsuaki | Orin | |
| Indo | Red Gold | |
| Kitakami | Sturmer Pippin | |
| Reinette du | Toko | |
| Canada | Tsugaru | |
| Rome Beauty | White Winter Permain | |
| Northern Spy | Yellow Newtown | |
| McIntosh | York Imperial |
DISCUSSION
An ACS1 allele characterized by the presence of a SINE in its 5′-flanking region was found in apples. SINEs are retroposons, copies of which are made by the retroposition of RNA catalyzed by RNA polymerase III (Weiner et al., 1986). Because it has been suggested that random nicking in A+T-rich regions produces staggered breaks that serve as retroposon insertion sites (Rogers, 1985), the deletion of a 24-bp A+T-rich sequence (19/24) from the SINE insertion site in the ACS1-2 gene is considered to be associated with the insertion mechanism. The putative A- and B-box sequences, internal cis elements for RNA polymerase III, showed low homology with the corresponding elements in the consensus sequence (Galli et al., 1981). Furthermore, our survey for the presence of the SINE in the ACS1-flanking region revealed that some wild apple species also possess ACS1-2, indicating that the insertion event occurred before the apple cultivars were established. DNA gel-blot analysis showed that the SINE-Md1 homologs were distributed in the genomes of Pyrus as well as Malus species (data not shown). Although domesticated apples are thought to be complex hybrids of several Malus species (Rehder, 1940), very little is known about their phylogenetic relationships. Characterization of the retropositional events at other orthologous loci where SINE-Md1 has integrated will help to resolve such phylogenetic relationships, for which molecular evidence has already been demonstrated in animals (Shimamura et al., 1997).
RT-PCR analysis revealed no expression of the ACS1-2 allele in the ripening fruit of cv Golden Delicious, a result that agrees with the cDNA sequence data reported by two groups (Dong et al., 1991; Lay-Yee and Knighton, 1995); both cDNAs derived from the same cultivar were identical to ACS1-1, but not to ACS1-2. SINE-Md1 integration into ACS1-2 occurred at a position corresponding to −751 ACS1-1 of the wild-type counterpart, and the flanking A+T-rich 24-bp sequence was deleted concomitantly. However, six nucleotide substitutions between the SINE and the transcription start point were detected. Although clear sequence motifs have not been observed in the 5′-flanking regions of the ACS genes investigated so far, the possibility that the sequence acting positively as a cis element is changed by the substitution cannot be ruled out. We consider that the promoter of the ACS1-2 allele still functions, since a very low level of transcription was detected in climacteric-stage cv Fuji fruit. Blume and Grierson (1997) carried out experiments on the expression of ACC oxidase promoter-GUS fusion products in transgenic plants and demonstrated that the region containing nucleotides −124 to +97 of the gene was sufficient to increase in the GUS activity markedly during fruit ripening, although the activity levels were very low. However, fusion of 1825 bp of the gene upstream sequence resulted in strong and specific induction of GUS expression. Nicholass et al. (1995) reported that the proximal 150 bp of the tomato polygalacturonase promoter showed no detectable transcriptional activity, although the distal 3.4 kb of the 5′ promoter was shown to be necessary for high levels of ripening-specific gene expression. Furthermore, examination of the Arabidopsis ACS1 promoter-GUS fusion system revealed that 1.4 kb of the 5′ region upstream from the ATG start codon was enough to produce high expression levels in a transgenic system (Rodrigues-Pousada et al., 1993). Analysis of the 5′-flanking region of Md-ACS1 using transgenic or transient assay systems should reveal the causes of low ACS1-2 expression levels in ripening fruit.
ACS gene analysis of higher plants indicated that the ACS enzyme is encoded by a highly divergent multigene family, the members of which are subject to differential regulation by more than one inducer (Felix et al., 1991; Olson et al., 1991; Rottmann et al., 1991; Kim et al., 1992; Liang et al., 1992; Yip et al., 1992; Abel et al., 1995). However, in tomato at least two of the six members of the ACS gene family are responsible for the burst of ethylene production during fruit ripening (Lincoln et al., 1993), whereas expression of three of them can be strongly induced by an elicitor (Oetiker et al., 1997). So far, three cDNAs of the ACS genes, including ACS1, have been isolated from ripening apples (Rosenfield et al., 1996). Our RNA gel-blot experiments showed that the Md-ACS1 expression was induced strongly during ripening, as described previously (Dong et al., 1991), whereas Md-ACS3 was expressed constitutively, irrespective of the ripening stage. The expression of Md-ACS2 was not analyzed because our attempt to obtain the probe was unsuccessful. In any event, the levels of the ACS1 transcript alone do not seem to account for the different IECs of the cultivars. Analysis of Md-ACS2 expression is also necessary to clarify the relative contributions of the ACS gene family members to ethylene production. On the other hand, the sum of the abundant transcripts of all the ACS genes expressed in the tomato did not account for increased ethylene production (Oetiker et al., 1997). Some workers have described mechanisms by which ACS activity is regulated posttranslation, such as proteolytic processing (Li et al., 1996) and protein phosphorylation/dephosphorylation (Spanu et al., 1994). Therefore, the ACS activity in ripening apples may be regulated by coordinated mechanisms at the transcriptional level and after transcription/translation.
Nevertheless, the allelic gene of ACS1, which shows dramatically reduced transcriptional activity in comparison with the parent gene, is considered to contribute to the low level of ethylene production by some cultivars at the ripening stage. In preliminary studies we have found that apple cultivars homozygous for ACS1-2 have lower IECs than other ACS1 allelic cultivars at the climacteric stage (T. Harada, T. Sunaka, J. Soejima, T. Sato, and M. Niizeki, unpublished data). To elucidate the relationship between the IECs and ACS1 allelic forms of apples, studies of many cultivars with different commercial harvesting seasons are now underway in our laboratory.
ACKNOWLEDGMENTS
We are grateful to J. Soejima and T. Masuda of the National Institute of Fruit Tree Science (Morioka, Japan) and T. Sato and T. Kon of the Aomori Prefectural Apple Experiment Station (Kuroishi, Japan) for providing the plant materials used in this investigation. We also thank S. Jin and T. Sakuma for their technical assistance. This work was carried out at the Gene Research Center of Hirosaki University (Japan).
Abbreviations:
- ACS
ACC synthase
- IEC
internal ethylene concentration
- RT
reverse transcriptase
- SINE
short interspersed DNA element
Footnotes
This work was supported in part by a Grant-in-Aid from the Ministry of Education, Science and Culture, Japan.
LITERATURE CITED
- Abel S, Nguyen MD, Chow W, Theologis A. J Biol Chem. 1995;270:19093–19099. doi: 10.1074/jbc.270.32.19093. [DOI] [PubMed] [Google Scholar]
- Blume B, Grierson D. Expression of ACC oxidase promoter-GUS fusions in tomato and Nicotiana plumbaginifoliaregulated by developmental and environmental stimuli. Plant J. 1997;12:731–746. doi: 10.1046/j.1365-313x.1997.12040731.x. [DOI] [PubMed] [Google Scholar]
- Deragon JM, Gilbert N, Rouquet L, Lenoir A, Amaud P, Picard G. A transcriptional analysis of the S1Bn (Brassica napus) family of SINE retroposons. Plant Mol Biol. 1996;32:869–878. doi: 10.1007/BF00020484. [DOI] [PubMed] [Google Scholar]
- Dong JG, Kim WT, Yip WK, Thompson GA, Li L, Bennett AB, Yang SF. Cloning of a cDNA encoding 1-aminocyclopropane-1-carboxylate synthase and expression of its mRNA in ripening apple fruit. Planta. 1991;185:38–45. doi: 10.1007/BF00194512. [DOI] [PubMed] [Google Scholar]
- Felix G, Grosskopf DG, Regenass M, Basse CW, Boller T. Elicitor-induced ethylene biosynthesis in tomato cells. Plant Physiol. 1991;97:19–25. doi: 10.1104/pp.97.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G, Hofstetter H, Birnstiel ML. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981;294:626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Gorny JR, Kader AA. Low oxygen and elevated carbon dioxide atmospheres inhibit ethylene biosynthesis in preclimacteric and climacteric apple fruit. J Am Soc Hortic Sci. 1997;122:542–546. [Google Scholar]
- Gussman CD, Goffredz JC, Gianfagna TJ. Ethylene production and fruit-softening rates in several apple fruit ripening variants. HortScience. 1993;28:135–137. [Google Scholar]
- Hamilton AJ, Lycett GW, Grieson D. Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plant. Nature. 1990;346:284–287. [Google Scholar]
- Harada T, SunakoT, Sakuraba W, Goto S, Senda M, Akada S, Niizeki M. Genomic nucleotide sequence of a ripening-related 1-aminocyclopropane-1-carboxylate synthase gene (MdACS-1) in apple (accession no. U89156) (PGR 97-066) Plant Physiol. 1997;113:1465. [Google Scholar]
- Kim WT, Silverstone A, Yip WK, Dong JG, Yang SF. Induction of 1-aminocyclopropane-1-carboxylate synthase mRNA by auxin in mung bean hypocotyls and cultured apple shoots. Plant Physiol. 1992;98:465–471. doi: 10.1104/pp.98.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Uthaibutra J, Gemma H. Comparison of 1-aminocyclopropane-1-carboxylic acid, abscisic acid and anthocyanin content of some apple cultivars during fruit growth and maturation. J Jpn Soc Hortic Sci. 1991;60:505–511. [Google Scholar]
- Ky YP, Drory A, Woodsson W. Molecular cloning of a 1-aminocyclopropane-1-carboxylate synthase from senescing carnation flower petals. Plant Mol Biol. 1992;18:377–386. doi: 10.1007/BF00034964. [DOI] [PubMed] [Google Scholar]
- Lay-Yee M, DellaPenna D, Ross GS. Changes in mRNA and protein during ripening in apple fruit (Malus domesticaBorkh. cv Golden Delicious) Plant Physiol. 1990;94:850–853. doi: 10.1104/pp.94.2.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay-Yee M, Knighton ML. A full-length cDNA encoding 1-aminocyclopropane-1-carboxylate synthase from apple (accession no. L31347) (PID:g606759) Plant Physiol. 1995;107:1017–1018. doi: 10.1104/pp.107.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Huxtable S, Yang SF, Kung SD. Effects of N-terminal deletions on 1-aminocyclopropane-1-carboxylate synthase activity. FEBS Lett. 1996;378:286–290. doi: 10.1016/0014-5793(95)01464-0. [DOI] [PubMed] [Google Scholar]
- Liang X, Abel S, Keller JA, Shen NF, Theologis A. The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1992;89:11046–11050. doi: 10.1073/pnas.89.22.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln JE, Campbell AD, Oetiker J, Rottmann WH, Oeller PW, Shen NF, Theologis A. LE-ACS4, a fruit ripening and wound-induced 1-aminocyclopropane-1-carboxylate synthase gene of tomato (Lycopersicon esculentum) J Biol Chem. 1993;268:19422–19430. [PubMed] [Google Scholar]
- Mochizuki K, Umeda M, Ohtsubo H, Ohtsubo E. Characterization of a plant SINE, p-SINE1, in rice genomes. Jpn J Genet. 1992;57:155–166. doi: 10.1266/jjg.67.155. [DOI] [PubMed] [Google Scholar]
- Nicholass FJ, Smith CJS, Schuch W, Bird CR, Gruersin D. High levels of ripening-specific reporter gene expression directed by tomato fruit polygalacturonase gene-flanking regions. Plant Mol Biol. 1995;28:423–435. doi: 10.1007/BF00020391. [DOI] [PubMed] [Google Scholar]
- Oeller PW, Lu MW, Taylor LP, Pike AD, Theologis A. Reversible inhibition of tomato fruit senescence by antisense RNA. Science. 1991;254:437–439. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]
- Oetiker JH, Olson DC, Shiu OY, Yang SF. Differential induction of seven 1-aminocyclopropane-1-carboxylate synthase genes by elicitor in suspension cultures of tomato (Lycopersicon esculentum) Plant Mol Biol. 1997;34:275–286. doi: 10.1023/a:1005800511372. [DOI] [PubMed] [Google Scholar]
- Olson DC, White JA, Edelman L, Harkins RN, Kende H. Differential expression of two genes for 1-aminocyclopropane-1-carboxylate synthase in tomato fruit. Proc Natl Acad Sci USA. 1991;88:5340–5344. doi: 10.1073/pnas.88.12.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehder A (1947) Manual of Cultivated Trees and Shrubs, Ed 2. Macmillan, New York
- Rodrigues-Pousada RAR, Rycke RD, Dedonder A, Caeneghem WV, Engler G, Montagu MV, Straeten VD. The Arabidopsis1-aminocyclopropane-1-carboxylate synthase gene 1 is expressed during early development. Plant Cell. 1993;5:897–911. doi: 10.1105/tpc.5.8.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JH. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Rosenfield CL, Kiss E, Hrazdina G. MdACS-2 (accession no. U73815) and MdACS-3 (accession no. U73816). Two new 1-aminocyclopropane-1-carboxylate synthases in ripening apple fruit (PGR 96-122) Plant Physiol. 1996;112:1735. [Google Scholar]
- Rottmann WH, Peter GF, Oeller PW, Keller JA, Shen NF, Nagy BP, Taylor LP, Campbell AD, Theologis A. 1-Aminocyclopropane-1-carboxylate synthase in tomato is encoded by a multigene family whose transcription is induced during fruit and floral senescence. J Mol Biol. 1991;222:937–961. doi: 10.1016/0022-2836(91)90587-v. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shimamura M, Yasue H, Ohshima K, Abe H, Kato H, Kishiro T, Goto M, Munechika I, Okada N. Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature. 1997;388:666–670. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- Spanu P, Grosskopf DG, Felix G, Boller T. The apparent turnover of 1-aminocyclopropane-1-carboxylate synthase in tomato cells is regulated by protein phosphorylation and dephosphorylation. Plant Physiol. 1994;106:529–535. doi: 10.1104/pp.106.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A. One rotten apple spoils the whole bushel: the role of ethylene in fruit ripening. Cell. 1992;70:181–184. doi: 10.1016/0092-8674(92)90093-r. [DOI] [PubMed] [Google Scholar]
- Umeda M, Ohtusubo H, Ohtsubo E. Diversification of the rice waxy gene by insertion of mobile DNA elements into introns. Jpn J Genet. 1991;66:569–589. doi: 10.1266/jjg.66.569. [DOI] [PubMed] [Google Scholar]
- Varadarajan GS, Prakash CS. A rapid and efficient method for the extraction of total DNA from the potato and its related species. Plant Mol Biol Rep. 1991;9:6–12. [Google Scholar]
- Weiner AM, Derininger PL, Efstratuadus A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- Yip WK, Moore T, Yang SF. Differential accumulation of transcripts for four tomato 1-aminocyclopropane-1-carboxylate synthase homologs under various conditions. Proc Natl Acad Sci USA. 1992;89:2475–2479. doi: 10.1073/pnas.89.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka Y, Matsumoto S, Kojima S, Oshima K, Okada N, Machida Y. Molecular characterization of a short interspersed element from tobacco that exhibits sequence homology to specific tRNAs. Proc Natl Acad Sci USA. 1993;90:6562–6566. doi: 10.1073/pnas.90.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]