Abstract
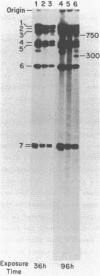
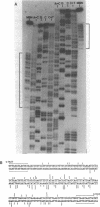
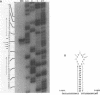
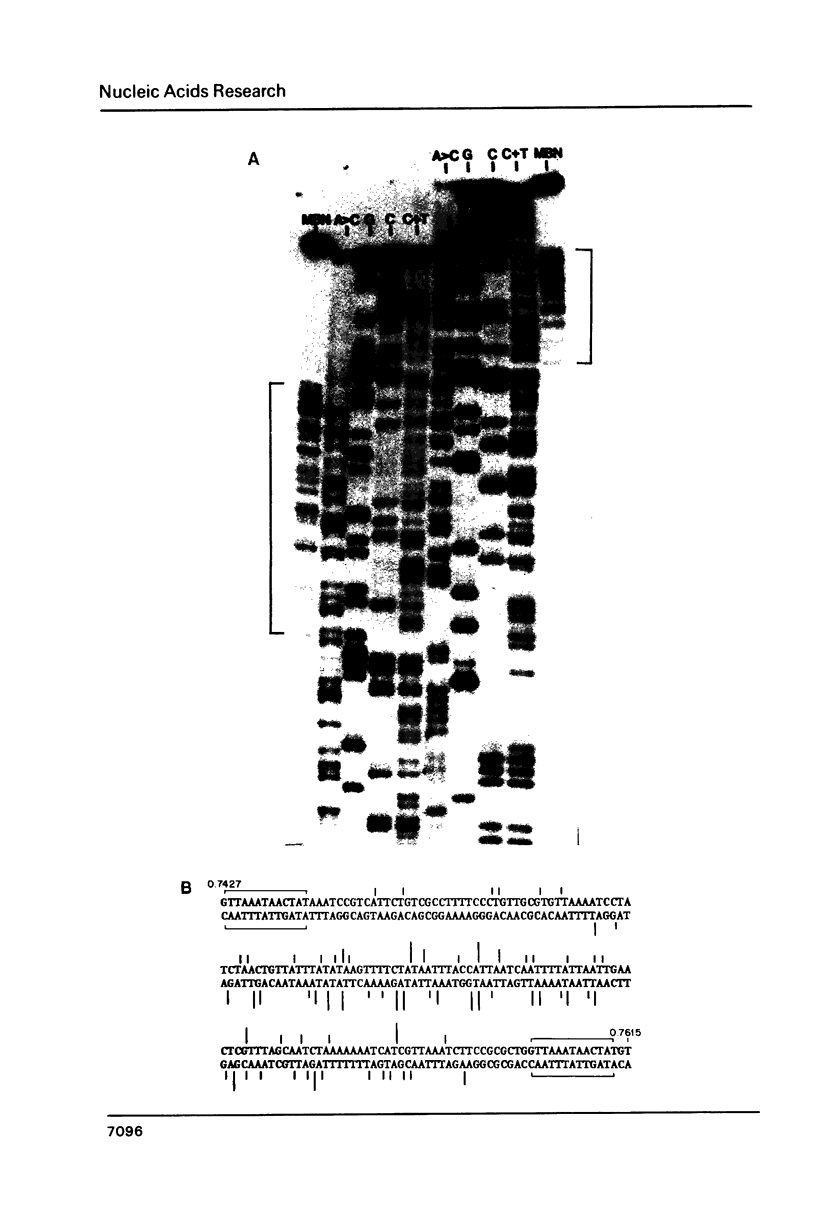
We have determined the nucleotide sequences around two alternative sites cleaved in supercoiled PM2 DNA by single-strand-specific mung bean nuclease in different ionic environments. In 10 mM Tris-HC1 (pH 7.0, 37 degrees C), the major site is a dA+dT-rich sequence which maps with a known early denaturation region at 0.75 map units. About 30 cleavages occurred in a 135 bp region. Cleavages were largely excluded at (dA)n . (dT)n (n = 3-7) sequences. Cleavage patterns of this type have not been previously observed in dA+dT-rich sequences. With the addition of 0.1 M NaC1 the major alternative site occurred in a hyphenated inverted repeat sequence 500 bp away (0.70 map units) and did not map to an early denaturation region. One major and 4 minor cleavages occurred in the region between the repeats, suggesting that a hairpin containing at most a 12 bp stem and 10 base loop is recognized. The basis for nuclease recognition of the dA+dT-rich sequence is not clear. The differences in the sequences and cleavage patterns at the alternative sites indicate that their secondary structures differ.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beard P., Morrow J. F., Berg P. Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J Virol. 1973 Dec;12(6):1303–1313. doi: 10.1128/jvi.12.6.1303-1313.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack C., Bickle T. A., Yuan R. The relation of single-stranded regions in bacteriophage PM2 supercoiled DNA to the early melting sequences. J Mol Biol. 1975 Aug 25;96(4):693–702. doi: 10.1016/0022-2836(75)90146-1. [DOI] [PubMed] [Google Scholar]
- Brack C., Eberle H., Bickle T. A., Yuan R. A map of the sites on bacteriophage PM2 DNA for the restriction endonucleases HindIII and HpaII. J Mol Biol. 1976 Jun 14;104(1):305–309. doi: 10.1016/0022-2836(76)90016-4. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Goodwin G. H. Demonstration of an S1-nuclease sensitive site near the human beta-globin gene, and its protection by HMG 1 and 2. Biochem Biophys Res Commun. 1983 Apr 29;112(2):547–554. doi: 10.1016/0006-291x(83)91499-7. [DOI] [PubMed] [Google Scholar]
- Dasgupta S., Allison D. P., Snyder C. E., Mitra S. Base-unpaired regions in supercoiled replicative form DNA of coliphage M13. J Biol Chem. 1977 Aug 25;252(16):5916–5923. [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybvig K., Clark C. D., Aliperti G., Schlesinger M. J. A chicken repetitive DNA sequence that is highly sensitive to single-strand specific endonucleases. Nucleic Acids Res. 1983 Dec 10;11(23):8495–8508. doi: 10.1093/nar/11.23.8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo R. T., Canelo E. S., Sinsheimer R. L. DNA of bacteriophage PM2: a closed circular double-stranded molecule. Proc Natl Acad Sci U S A. 1969 Aug;63(4):1164–1168. doi: 10.1073/pnas.63.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell B. E., Inman R. B. Comparison of partial denaturation maps with the known sequence of simian virus 40 and phi X174 replicative form DNA. J Mol Biol. 1979 Jun 25;131(2):331–340. doi: 10.1016/0022-2836(79)90079-2. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Vogt V. M., Hirt B. Characterization of the single-strand-specific nuclease S1 activity on double-stranded supercoiled polyoma DNA. Eur J Biochem. 1974 Apr 16;43(3):591–600. doi: 10.1111/j.1432-1033.1974.tb03446.x. [DOI] [PubMed] [Google Scholar]
- Goding C. R., Russell W. C. S1 sensitive sites in adenovirus DNA. Nucleic Acids Res. 1983 Jan 11;11(1):21–36. doi: 10.1093/nar/11.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale P., Woodward R. S., Lebowitz J. Carbodiimide inactivation of Escherichia coli RNA polymerase promoters on supercoiled simian virus 40 and ColE1 DNAs occurs by a one-hit process at salt concentrations in the physiological range. J Biol Chem. 1983 Jun 25;258(12):7828–7839. [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Hsieh T. S., Wang J. C. Thermodynamic properties of superhelical DNAs. Biochemistry. 1975 Feb 11;14(3):527–535. doi: 10.1021/bi00674a011. [DOI] [PubMed] [Google Scholar]
- Kilpatrick M. W., Wei C. F., Gray H. B., Jr, Wells R. D. BAL 31 nuclease as a probe in concentrated salt for the B-Z DNA junction. Nucleic Acids Res. 1983 Jun 11;11(11):3811–3822. doi: 10.1093/nar/11.11.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Gelinas R., Weintraub H. Detection of an altered DNA conformation at specific sites in chromatin and supercoiled DNA. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4389–4393. doi: 10.1073/pnas.80.14.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D. A procedure for the quantitation of relaxed closed circular DNA in the presence of superhelical DNA: an improved fluorometric assay for nicking-closing enzyme. Anal Biochem. 1979 Mar;93(2):346–354. doi: 10.1016/s0003-2697(79)80161-x. [DOI] [PubMed] [Google Scholar]
- Kowalski D., Kroeker W. D., Laskowski M., Sr Mung bean nuclease I. Physical, chemical, and catalytic properties. Biochemistry. 1976 Oct 5;15(20):4457–4463. doi: 10.1021/bi00665a019. [DOI] [PubMed] [Google Scholar]
- Kowalski D., Sanford J. P. Action of mung bean nuclease on supercoiled PM2 DNA. J Biol Chem. 1982 Jul 10;257(13):7820–7825. [PubMed] [Google Scholar]
- Kroeker W. D., Kowalski D. Gene-sized pieces produced by digestion of linear duplex DNA with mung bean nuclease. Biochemistry. 1978 Aug 8;17(16):3236–3243. doi: 10.1021/bi00609a010. [DOI] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim V. I., Mazanov A. L. Tertiary structure for palindromic regions of DNA. FEBS Lett. 1978 Apr 1;88(1):118–123. doi: 10.1016/0014-5793(78)80621-8. [DOI] [PubMed] [Google Scholar]
- Mace H. A., Pelham H. R., Travers A. A. Association of an S1 nuclease-sensitive structure with short direct repeats 5' of Drosophila heat shock genes. Nature. 1983 Aug 11;304(5926):555–557. doi: 10.1038/304555a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickol J. M., Felsenfeld G. DNA conformation at the 5' end of the chicken adult beta-globin gene. Cell. 1983 Dec;35(2 Pt 1):467–477. doi: 10.1016/0092-8674(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Pritchard A. E., Laskowski M., Sr Discrete fragmnets produced by limited digestion of superhelical PM2 DNA with venom phosphodiesterase. Cleavage sites and mode of generation. J Biol Chem. 1978 Sep 25;253(18):6606–6613. [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Shen C. K. Superhelicity induces hypersensitivity of a human polypyrimidine . polypurine DNA sequence in the human alpha 2-alpha 1 globin intergenic region to S1 nuclease digestion--high resolution mapping of the clustered cleavage sites. Nucleic Acids Res. 1983 Nov 25;11(22):7899–7910. doi: 10.1093/nar/11.22.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Kilpatrick M. W., Wells R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J Biol Chem. 1984 Feb 10;259(3):1963–1967. [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Strauss F., Gaillard C., Prunell A. Helical periodicity of DNA, Poly(dA) . poly(dT) and poly(dA-dT). poly(dA-dT) in solution. Eur J Biochem. 1981 Aug;118(2):215–222. doi: 10.1111/j.1432-1033.1981.tb06389.x. [DOI] [PubMed] [Google Scholar]
- Strauss H. S., Burgess R. R., Record M. T., Jr Binding of Escherichia coli ribonucleic acid polymerase holoenzyme to a bacteriophage T7 promoter-containing fragment: selectivity exists over a wide range of solution conditions. Biochemistry. 1980 Jul 22;19(15):3496–3504. doi: 10.1021/bi00556a014. [DOI] [PubMed] [Google Scholar]
- Tapper D. P., Clayton D. A. Altered mobility of polydeoxyribonucleotides in high resolution polyacrylamide gels due to removal of terminal phosphates. Nucleic Acids Res. 1981 Dec 21;9(24):6787–6794. doi: 10.1093/nar/9.24.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong B. Y., Battersby S. J. Comparison of theoretical denaturation maps of phiX174 and SV40 with their gene maps. Nucleic Acids Res. 1979 Mar;6(3):1073–1079. doi: 10.1093/nar/6.3.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VONHIPPEL P. H., FELSENFELD G. MICROCOCCAL NUCLEASE AS A PROBE OF DNA CONFORMATION. Biochemistry. 1964 Jan;3:27–39. doi: 10.1021/bi00889a006. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Larson J. E., Grant R. C., Shortle B. E., Cantor C. R. Physicochemical studies on polydeoxyribonucleotides containing defined repeating nucleotide sequences. J Mol Biol. 1970 Dec 28;54(3):465–497. doi: 10.1016/0022-2836(70)90121-x. [DOI] [PubMed] [Google Scholar]
- West R. W., Jr, Rodriguez R. L. Construction and characterization of E. coli promoter-probe plasmid vectors. II. RNA polymerase binding studies on antibiotic-resistance promoters. Gene. 1980 May;9(3-4):175–193. doi: 10.1016/0378-1119(90)90321-h. [DOI] [PubMed] [Google Scholar]
- Wingert L., Von Hippel P. H. The conformation dependent hydrolysis of DNA by micrococcal nuclease. Biochim Biophys Acta. 1968 Mar 18;157(1):114–126. doi: 10.1016/0005-2787(68)90270-0. [DOI] [PubMed] [Google Scholar]
- Woodworth-Gutai M. Recombination in SV40-infected cells: nucleotide sequences at viral-viral recombinant joints in naturally arising variants. Virology. 1981 Mar;109(2):344–352. doi: 10.1016/0042-6822(81)90505-5. [DOI] [PubMed] [Google Scholar]