Abstract
REV3 is the catalytic subunit of DNA translesion synthesis polymerase ζ. Inhibition of REV3 expression increases the sensitivity of human cells to a variety of DNA-damaging agents and reduces the formation of resistant cells. Surprisingly, we found that short hairpin RNA-mediated depletion of REV3 per se suppresses colony formation of lung (A549, Calu-3), breast (MCF-7, MDA-MB-231), mesothelioma (IL45 and ZL55), and colon (HCT116 +/-p53) tumor cell lines, whereas control cell lines (AD293, LP9-hTERT) and the normal mesothelial primary culture (SDM104) are less affected. Inhibition of REV3 expression in cancer cells leads to an accumulation of persistent DNA damage as indicated by an increase in phospho-ATM, 53BP1, and phospho-H2AX foci formation, subsequently leading to the activation of the ATM-dependent DNA damage response cascade. REV3 depletion in p53-proficient cancer cell lines results in a G1 arrest and induction of senescence as indicated by the accumulation of p21 and an increase in senescence-associated β-galactosidase activity. In contrast, inhibition of REV3 expression in p53-deficient cells results in growth inhibition and a G2/M arrest. A small fraction of the p53-deficient cancer cells can overcome the G2/M arrest, which results in mitotic slippage and aneuploidy. Our findings reveal that REV3 depletion per se suppresses growth of cancer cell lines from different origin, whereas control cell lines and a mesothelial primary culture were less affected. Thus, our findings indicate that depletion of REV3 not only can amend cisplatin-based cancer therapy but also can be applied for susceptible cancers as a potential monotherapy.
Introduction
Screening in Saccharomyces cerevisiae for mutants defective in UV-induced mutagenesis revealed the so-called reversionless phenotype (REV), which is characterized by a diminished frequency of mutations reverting a specific marker gene deficiency [1]. Two genes that confer this phenotype when absent are Rev3 and Rev7, the catalytic and the structural subunits of the DNA translesion synthesis (TLS) polymerase ζ (Pol ζ), respectively [2,3]. The mammalian REV3L gene (hereafter REV3) encodes a ∼350-kDa protein (REV3) consisting of a large C-terminal DNA polymerase subunit, which misses the characteristic proofreading activity present in other B-family DNA polymerases (reviewed in Waters et al. [4]). REV3 interacts through a specific binding domain with REV7, but no additional protein-protein interaction sites were identified. Deletion of REV3 is embryonically lethal around midgestation [5–8], whereas overexpression of REV3 leads to increased spontaneous mutation rates [9], confirming that REV3 expression has to be tightly regulated to maintain genomic integrity. Conversely, one study found that REV3 expression was downregulated in colon carcinomas compared with that in adjacent normal tissue [10], whereas another study found that REV3 expression was elevated in human glioma tissues resected before therapy compared with that in normal brain tissues [11].
Pol ζ belongs to the functional group of TLS DNA polymerases, which are characterized by a less-stringent active site and a lower processivity compared with the high-fidelity replicative DNA polymerases (reviewed in Waters et al. [4]). TLS polymerases contribute to the maintenance of the genomic integrity by allowing DNA replication to continue in the presence of DNA adducts, which otherwise could lead to DNA replication fork breakdown and subsequent gross chromosomal instability. Pol ζ is the major extender from mismatches formed when incorrect nucleotides are inserted opposite DNA ad-ducts, thereby contributing to mutation formation on the nucleotide level. Recently, it was shown that REV3 is involved not only in DNA damage tolerance but also in DNA repair mechanisms, for example, interstrand cross-link repair [12–14], homologous recombination [15], and nonhomologous end-joining as indicated by the deficiency of REV3-deleted B cells in class switching of immunoglobulin genes [16].
The unique function of REV3 is highlighted by the fact that the REV3 depletion increases sensitivity and decreases mutagenesis induced by UV light, cisplatin, and other mutagens in human and mouse fibroblasts [15,17,18]. In addition, depletion of REV3 sensitizes mouse B-cell lymphomas and lung adenocarcinomas to cisplatin [19,20]. Although disruption of mouse REV3 leads to embryonic lethality, it is possible to generate REV3-deleted mouse embryonic fibroblasts (MEFs) in a p53-deficient background [21]. Spontaneous chromosomal instability was observed in REV3-deleted MEFs and REV3-deleted cell lines [16,22,23].
DNA damage induction results in the activation of an evolutionarily conserved signal cascade known as DNA damage response (DDR) (reviewed in d'Adda di Fagagna [24]). Induction of DNA double-strand breaks (DSBs) results in recruitment and activation of ataxia-telangiectasia mutated (ATM). Activated ATM phosphorylates the histone variant H2AX at serine 139 (γH2AX) near DNA DSBs, subsequently leading to an accumulation of DDR proteins at DSBs, which can be visualized by immunofluorescence microscopy as distinct foci. Once ATM activation reaches a certain threshold, checkpoint kinase Chk2 is phosphorylated, resulting in the accumulation of p53, leading to the accumulation of the cyclin-dependent kinase inhibitor p21. Prolonged activation of p21 after DNA damage is associated with a terminal proliferation arrest, i.e., senescence.
While investigating how inhibition of REV3 expression affects cisplatin-induced mutagenesis, we observed that depletion of REV3 per se reduces cancer cell growth, whereas growth of control cells is less affected. Suppression of REV3 expression in cancer cells leads to the accumulation of persistent DNA damage independent of the p53 status. In p53-proficient cancer cells, inhibition of REV3 expression results in the activation of the ATM-dependent DDR cascade, leading to senescence induction. In p53-deficient cancer cells, depletion of REV3 results in a G2/M arrest and increases the fraction of aneuploid cells. In contrast, inhibition of REV3 expression in control cell lines and a mesothelial primary culture neither reduces colony formation nor activates the DDR cascade.
Materials and Methods
Cell Lines and Culture
All cell lines used in this study were authenticated by DNA fingerprinting (Microsynth, Balgach, Switzerland). SDM104 was maintained as described previously [25]. All other cell lines were maintained in high-glucose Dulbecco modified Eagle medium (DMEM; Sigma-Aldrich, St Louis, MO) supplemented with 2 mM l-glutamine, 1 mM sodium pyruvate, 10% fetal calf serum, and 1% (wt/vol) penicillin/streptomycin. All cells were grown at 37°C in a humidified atmosphere containing 5% CO2. Additional details can be found in Supplemental Materials and Methods.
Vector Production and Transduction
Replication-deficient lentiviral particles were produced, titrated, and used for transduction as described previously [26,27]. Additional details can be found in Supplemental Materials and Methods.
Plasmid Transfection
Cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions with pSuperior.puro containing either scrambled control short hairpin RNA (shSCR) or three distinct short hairpin RNA (shRNA) sequences targeting the REV3 messenger RNA (shREV3). Additional details can be found in Supplemental Materials and Methods.
Colony Formation Assay
Crystal violet staining was performed after colonies were visible by eye and, the number of colonies was determined by eye, applying the same threshold for colony size to all transduced cell lines. The number of colonies obtained by mock treatment was set to 100%.
Quantitative Real-time Polymerase Chain Reaction
RNA from samples was isolated using RNeasy Mini kit (Qiagen, Germantown, MD), and reverse transcription was performed on 300 ng of RNA (QuantiTect Reverse Transcription Protocol; Qiagen). The quantitative expression of REV3 mRNA was measured by SYBR Green polymerase chain reaction (PCR) assay (PE Applied Biosystems, Foster City, CA) on a Prism 5700 detection system (SDS; PE Applied Biosystems). Additional details can be found in Supplemental Materials and Methods.
Immunofluorescence Microscopy
Immunofluorescence microscopy was essentially performed as described [28]. Details can be found in Supplemental Materials and Methods.
Flow Cytometry
Detection of bromodeoxyuridine (BrdU) incorporation in DNA-synthesizing cells was carried out using the anti-BrdU antibody (no. 555627; BD Biosciences, San Jose, CA) according to the manufacturer's instructions. Additional details can be found in Supplemental Materials and Methods.
Senescence-Associated β-Galactosidase Assay
The expression of senescence-associated (SA) β-galactosidase was determined by SA-β-galactosidase staining as described [29].
Western Analysis
Protein extracts (30 µg) were separated by 4% to 20% SDS-PAGE and transferred onto polyvinylidene fluoride membranes. Immunoblot analysis was performed as described [30]. Details can be found in Supplemental Materials and Methods.
Enzyme-Linked Immunosorbent Assay
Cells were washed three times with phosphate-buffered saline (PBS) and serum-free DMEM was added for 24 hours. Conditioned medium was filtered, and cell number was determined in every experiment by hemocytometer. Enzyme-linked immunosorbent assay (ELISA) was performed using human interleukin 6 (IL-6) Quantikine ELISA Kit (no. D6050; R&D Systems, Minneapolis, MN). Data were normalized to the cell number and reported as fold difference compared with mock-treated control.
Statistical Analysis
P values were calculated using the two-tailed Student's t test; *P < .05 and **P < .01.
Results
Depletion of REV3 Per Se Suppresses Colony Formation of Cancer Cells
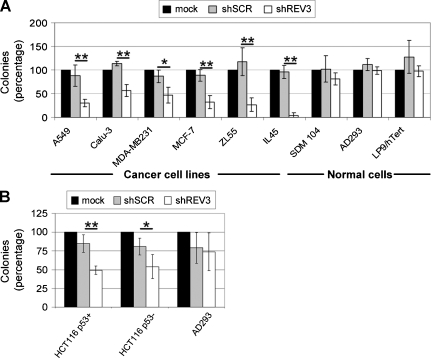
To study the effect of REV3 depletion on cisplatin-induced mutagenesis, we established a lentiviral-based system, which allowed us to significantly inhibit REV3 expression in all cell lines and the primary culture used in this study (Figure W1, A and B). Inhibition of REV3 expression did not significantly reduce colony formation of the control cell line AD293 (99% remaining colonies compared with mock-treated control), the primary mesothelial culture SDM104 (81%), and the hTERT-immortalized derivative of the mesothelial primary culture LP9 (LP9-hTERT, 98%; Figures 1A and W2A). Surprisingly, REV3 depletion per se significantly suppressed colony formation of the p53-proficient adenocarcinoma cell line A549 (30%), the p53-deficient adenocarcinoma cell line Calu-3 (57%), the p53-deficient breast cancer cell line MDA-MB-231 (47%), the p53-proficient breast cancer cell line MCF-7 (32%), the human mesothelioma cell line ZL55 (27%), and the rat mesothelioma cell line IL45 (4%) compared with the mock-treated control (Figures 1A and W2A).
Figure 1.
Inhibition of REV3 expression specifically reduces colony formation of cancer cell lines. Cells were mock treated or transduced with lentiviral-based particles containing either shSCR or shREV3. (A and B) Cells were stained by crystal violet, and total colonies were counted after 2 to 4 weeks. Colonies were counted from at least three independent experiments for all cell lines. Colony numbers of mock-treated cells were set as 100%. *P < .05. **P < .01. Shown are means ± standard deviation (SD).
In the isogenic p53-proficient and -deficient HCT116 colorectal carcinoma cell lines, there was no significant difference in the reduction of REV3 expression levels after transduction with a multiplicity of infection (MOI) of 170, as used for the cell lines described previously, or an MOI of 800 (Figure W1B). However, only the high-titer transduction significantly suppressed colony formation of p53-proficient (49%) and -deficient HCT116 (54%) compared with the mock control (Figures 1B and W2B). REV3 depletion by high-titer transduction did not significantly reduce colony formation of the control cell line AD293 (74%) compared with the mock control (Figures 1B and W2B).
Inhibition of REV3 expression by transduction with three plasmids, one encoding the same small interfering RNA (siRNA) as the lentiviral-based particles plus two plasmids encoding siRNA targeting alternative sites of the REV3 mRNA (named REV3–5 and REV3–6), significantly reduced colony formation in the mesothelioma cell line IL45, whereas the control cell line AD293 was not affected (Figure W3). Therefore, we conclude that the observed reduction in colony formation is due to the inhibition of REV3 expression and not due to an unspecific off-target effect of the REV3–4 siRNA. Thus, REV3 depletion per se significantly suppresses colony formation in cancer cell lines, whereas colony formation of control cell lines and a primary mesothelial culture is less affected.
Cancer Cells Accumulate Persistent DNA DSBs after REV3 Depletion
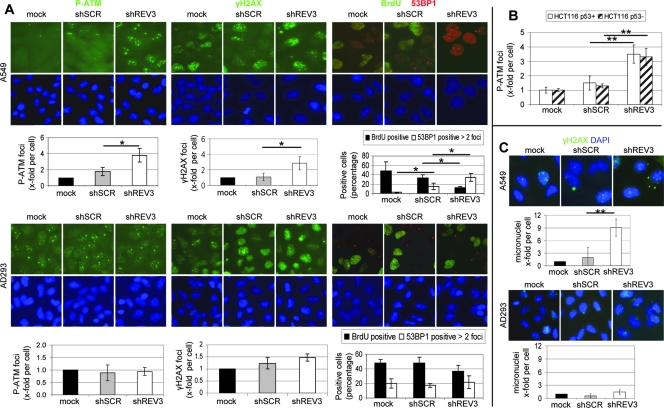
53BP1 and γH2AX foci formation is regarded as a maker for DSBs [28], and a recent study showed that their numbers were increased after persistent DNA damage induction [31]. Seven days after transduction, REV3 depletion in A549 cells increased the average number of P-ATM and γH2AX foci per cell by a factor of 3.8 and 2.3, respectively, compared with the mock control (Figure 2A). Inhibition of REV3 expression increased the fraction of A549 cells containing more than two 53BP1 foci to 34% compared with mock (2%) and scrambled (16%) control (Figure 2A). Similarly, REV3 depletion in MCF-7 breast cancer cells increased the average number of γH2AX and 53BP1 foci per cell by a factor of 3.2 and 2.5, respectively, compared with the mock control (Figure W4). P-ATM foci formation was also elevated in both p53-proficient and -deficient HCT116 cells after REV3 depletion by a factor of 2.3 and 2.5, respectively, compared with the scrambled control (Figure 2B). In contrast, inhibition of REV3 expression in the control cell line AD293 did not significantly increase P-ATM, 53BP1, or γH2AX foci formation compared with the scrambled control (Figure 2A).
Figure 2.
REV3 depletion induces persistent DNA damage and genomic instability specifically in cancer cells. Cells were mock treated or transduced with lentiviral-based particles containing either shSCR or shREV3 and analyzed after 1 week. (A) Cells were stained for P-ATM, γH2AX, or BrdU (all green) and 53BP1 (red) and quantified by immunofluorescence microscopy. Cells containing more than two 53BP1 foci per cell were considered as 53BP1 positive. (B) Cells were stained for P-ATM, and foci per cell were quantified by immunofluorescence microscopy. (C) Cells were stained for γH2AX (green) and nuclear DNA was labeled with DAPI (blue). Micronuclei formation was identified by immunofluorescence microscopy-based analysis of DAPI staining. At least three independent experiments were analyzed. *P < .05. **P < .01. Shown are means ± SD.
DSBs, which are not repaired either due to complex DNA modifications or to deficiencies in molecular mechanisms result in the formation of persistent DSBs (reviewed in d'Adda di Fagagna [24]). P-ATM foci at persistent DSBs are significantly larger than the initial foci detectable immediately after damage initiation [32]. Microscopic analysis revealed that the DDR foci induced in REV3-depleted cells 7 days after transduction were larger compared with the background DDR foci present in the mock controls (Figure 2A).
Gross chromosomal instability indicated by an elevated number of micronuclei were observed in MEFs with REV3 deletion [21]. Similarly, the number of micronuclei increased in A549 cells by a factor of 9 after inhibition of REV3 expression compared with the mock control (Figure 2C). Micronuclei formation was not significantly elevated after inhibition of REV3 expression in AD293 cells (Figure 2C). Thus, inhibition of REV3 expression induces the formation of persistent DSBs and accumulation of gross chromosomal instability in cancer cell lines, whereas the control cell line AD293 is significantly less affected (P < .05 for γH2AX, P-ATM, and 53BP1 foci and micronuclei per cell for both A549 and MCF-7 vs AD293; numbers of P-ATM foci per cell were not determined in MCF-7 cells). In addition, our results indicate that persistent DDR foci formation after REV3 depletion is not dependent on the p53 status.
Inhibition of REV3 Expression Suppresses Proliferation of Cancer Cells
Because persistent DNA adducts block DNA replication and activate the DDR pathway, we investigated whether inhibition of REV3 expression results in reduced cellular proliferation. Labeling of newly synthesized DNA with BrdU is an established method for the assessment of cellular proliferation (reviewed in Quinn and Wright [33]).
Quantitative analysis of BrdU incorporation revealed that cellular proliferation of A549 cells was reduced by REV3 depletion to 21% compared with 37% and 38% in mock and scrambled controls, respectively (Figure 3, see also Figure 2A). Inhibition of REV3 expression reduced the proliferation of p53-proficient HCT116 cells to 25% and that of p53-deficient HCT116 cells, to a lesser extent, to 33% compared with 41% and 45% in their corresponding scrambled controls, respectively (Figure 3). Similarly, REV3 depletion also reduced the proliferation of MCF-7 breast cancer cells to 9.2% compared with 17% and 19% in mock and scrambled controls, respectively (Figure W4). In contrast, the percentage of replicating cells in the control cell line AD293 and the primary cell culture SDM104 was not diminished by the inhibition of REV3 expression (Figure 3). Thus, REV3 depletion suppresses cellular proliferation of the analyzed cancer cells, whereas proliferation of control cells is not affected.
Figure 3.
REV3 depletion changes cell cycle distribution of cancer cell lines. Cells were mock treated or transduced with lentiviral-based particles containing either shSCR or shREV3 and/or lentiviral-based particles containing shP53. After 1 week, cell cycle distribution was measured by BrdU/propidium iodide staining and subsequent FACS analysis. The averages of three independent experiments are given for A549, A549 shP53, p53-proficient HCT116, and p53-deficient HCT116 cells, whereas representative experiments are shown for SDM104 and AD293 cells. *P < .05. **P < .01. Shown are means ± SD.
REV3 Depletion Activates the DNA Damage Response Pathway in Cancer Cells
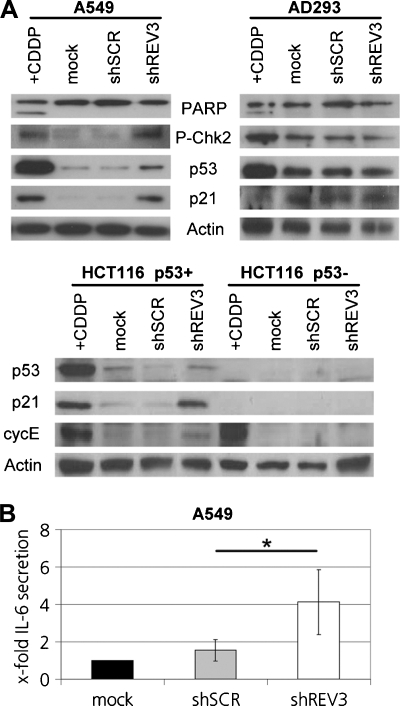
We investigated whether the observed accumulation of persistent DSBs in cancer cells results in the activation of the canonical ATM-kinase-mediated DDR pathway, which is induced by DSBs (reviewed in d'Adda di Fagagna [24]). As described here, the number of phospho-ATM foci per cell increased after the inhibition of REV3 expression compared with mock and scrambled controls in A549 and p53-proficient and -deficient HCT116 cells, whereas no significant increase occurred in AD293 control cells (Figure 2, A and B). In A549 cells, REV3 depletion resulted in increased phosphorylation of the checkpoint kinase Chk2 (P-Chk2) and the accumulation of p53 and the senescence mediator p21 (Figure 4A), which was also observed in MCF-7 breast cancer cells but not in the normal mesothelial primary culture SDM104 (Figure W5). In p53-proficient HCT116 cancer cells, inhibition of REV3 expression also resulted in an accumulation of p21, which was absent in the p53-deficient isogenic cell line (Figure 4A). Thus, in the analyzed p53-proficient cancer cells, inhibition of REV3 expression results in the activation of the canonical ATM-dependent DDR pathway.
Figure 4.
REV3 depletion induces DDR pathway in cancer cells. Cells were cisplatin or mock treated or transduced with lentiviral-based particles containing either shSCR or shREV3. (A) After 1 week, whole-cell lysates were analyzed by Western analysis. (B) After 24 hours, IL-6 secretion in serum-free DMEM was assessed by ELISA, normalized to the cell number and reported as fold increase compared with mock-treated control. The averages of at least three independent experiments are given. Shown are means ± SD.
REV3 Depletion Induces a G1 Arrest in p53-Proficient Cancer Cells
We tested whether the activation of the DDR pathway and the reduction in BrdU incorporation due to REV3 depletion change the cell cycle distribution of cancer cells. Depletion of the S phase after REV3 depletion, as mentioned here, was accompanied by a significant increase in the fraction of A549 cells in the G1 phase of the cell cycle to 62% compared with 53% and 51% in the mock and scrambled controls, respectively (Figure 3). Similarly, the fraction of p53-proficient HCT116 cells in the G1 phase increased to 38% after inhibition of REV3 expression compared with 26% in scrambled control, respectively (Figure 3). In the control cell line AD293 and the primary cell culture SDM104, neither the fraction of cells in the S phase was decreased nor was the fraction of cells in the G1 phase increased after inhibition of REV3 expression compared with mock and scrambled controls (Figure 3). A small but significant increase in the fraction of cells in the G2 phase was observed in p53-proficient HCT116 cells after REV3 depletion (23%) compared with mock (17%) and scrambled controls (19%). In addition, protein levels of cyclin E, which accumulate during the G1 phase and are required for the transition from the G1 phase to the S phase, increased after inhibition of REV3 expression in p53-proficient but not in p53-deficient HCT116 cancer cells (Figure 4A). Thus, inhibition of REV3 expression in the investigated p53-proficient cancer cell lines induces a G1 arrest, respectively, S-phase depletion, whereas the cell cycle distribution of the investigated control cell line and the primary mesothelial culture was not affected.
Inhibition of REV3 Expression Induces Senescence in p53-Proficient Cancer Cells
Although inhibition of REV3 expression slightly increased the fraction of sub-G1 cells in p53-proficient A549 and HCT116 cells, no significant induction of apoptosis as indicated by an increased fraction of sub-G1 cells (Figure 3) or poly(ADP-ribose) polymerase (PARP) cleavage (Figure 4A) was observed in the remaining control and cancer cell lines tested in this study.
Because senescence can be induced by persistent DNA damage [31], we investigated whether cells are senescent after REV3 depletion. Induction of senescence cannot be identified by a single marker but is associated with a variety of distinct cellular and molecular changes (reviewed in Collado and Serrano [34]). Microscopic analysis after crystal violet staining revealed that the morphology of control AD293 cells was not changed 7 days after inhibition of REV3 expression compared with mock and scrambled controls (Figure W6). In the p53-proficient cancer cell lines included in this study, most colonies were smaller after REV3 depletion, and the cells of these colonies displayed morphologic changes that are associated with senescence, namely, increased cell size and flattened shape, whereas cell morphology was not affected in mock and scrambled controls (Figure W6). SA-β-galactosidase staining revealed increased SA-β-galactosidase activity in IL45, A549, and HCT116 p53-proficient cells after inhibition of REV3 expression (Figure W6 and Table 1). No increase in SA-β-galactosidase staining after inhibition of REV3 expression was detectable in the control cells AD293 or in the p53-deficient MDA-MB-231 and HCT116 cancer cell lines. As mentioned previously, G1 arrest, respectively, S-phase depletion and p21 accumulation were observed in A549 and p53-proficient HCT116 cells after REV3 depletion (Figures 3 and 4A).
Table 1.
Induction of Senescence after REV3 Inhibition Is Dependent on p53 Level.
| p53 | Cell Line | Mock | shSCR | shREV3 | t test shSCR/shREV3 |
| + | IL45 | 2.3 ± 0.7 | 2.8 ± 1.0 | 44.2 ± 4.0 | * |
| + | A549 | 5.8 ± 0.7 | 6.0 ± 1.4 | 16.1 ± 2.3 | * |
| - | A549 shP53 | 1.0 ± 0.3 | 0.9 ± 0.3 | 0.9 ± 0.3 | NS |
| - | MDA-MB-231 | 0 | 0 | 0 | N/A |
| + | AD293 (normal) | 0 | 0 | 0 | N/A |
Three independent experiments were analyzed for all cell lines. Shown are means (%) of senescent cells ± SEM.
N/A indicates not applicable; NS, not significant.
*P < .01%.
An increase in persistent DNA damage indicated by residual 53BP1/γH2AX foci is associated in human foreskin fibroblasts with a senescence-associated secretory phenotype including cytokine secretion such as IL-6 [31]. Twelve days after transduction, IL-6 secretion was increased in A549 cell after inhibition of REV3 expression compared with mock and scrambled controls (Figure 4B). In contrast, REV3 depletion in p53-deficient HCT116 cells did not result in a G1 accumulation nor did it increase p21 levels or increase SA-β-galactosidase staining (Figures 3, 4A, and W6 and Table 1). Similarly, G1 accumulation and SA-β-galactosidase staining were abolished in A549 by p53 inhibition (Figure 3 and Table 1). Thus, among the analyzed cancer cell lines, REV3 depletion per se induces senescence in p53-proficient cancer cells only.
REV3 Depletion Induces a G2/M Arrest and Aneuploidy in p53-Deficient Cancer Cells
No G1 arrest was detectable in p53-deficient HCT116 cell after inhibition of REV3 expression (Figure 3). Instead, REV3 depletion in the p53-deficient HCT116 cell line significantly increased the fraction of cells in the G2/M phase to 26% compared with 17% and 18% in the mock and scrambled controls, respectively (Figure 3). In addition, inhibition of REV3 expression also increased the fraction of aneuploid cells, which did not incorporate BrdU (AN, aneuploid nondividing) to 7% compared with 3% and 4% in the mock and scrambled controls (Figure 3). The fraction of aneuploid cells, which were still incorporating BrdU (AD, aneuploid dividing), was not increased after REV3 depletion in p53-deficient HCT116 cells compared with mock and scrambled controls (Figure 3). Thus, inhibition of REV3 expression in the investigated p53-deficient cancer cells results in the accumulation of G2/M arrested and AN cells.
In an effort to provide proof-of-principle, we inhibited p53 expression in p53-proficient A549 cancer cells (Figure W1C). Inhibition of p53 expression in A549 cells resulted in a significant increase of the cells in the G2 phase (22%) and in aneuploidy (total 14%) compared with p53-proficient A549 cells (7.5% and 1.8%, respectively; Figure 3), which is in agreement with the dominant role of p53 in the induction of the G1 arrest [35]. The dominant role of p53 in protection from aneuploidy is highlighted by the finding that additional inhibition of REV3 expression in combination with p53 inhibition did not further increase aneuploidy in A549 cancer cells.
Discussion
During our study on the involvement of REV3 in chemotherapy response, we found that lentiviral-based inhibition of REV3 expression was as efficient in the analyzed cancer cell lines as in the primary mesothelial culture and the control cell lines, but surprisingly, colony formation was reduced in the cancer cell lines only. Therefore, we conclude that reduction in colony formation does not simply mirror the degree of REV3 expression inhibition relative to the scrambled control.
We found that colony formation was not significantly reduced in the control cell lines AD293 and LP9-hTERT and the primary mesothelial culture SDM104 and after inhibition of REV3 expression. This is consistent with previous studies where no deficiency in cell growth/survival was mentioned after antisense-based inhibition of REV3 expression in human nontumor cell lines [17,36]. In contrast, it was shown by different groups that REV3 knockout reduced cell growth of MEFs [21,37]. Thus, additional studies will be necessary to clarify how normal cells adapt their DDR to tolerate the loss of REV3 function. At this point, it is worth mentioning that investigations of cancer-specific pathways are usually performed using so-called normal cells as control. However, normal cells have a limited life span [38], which also applies to the primary mesothelial culture SDM104. In contrast, the control cell lines AD293 and LP9-hTERT are virally transformed or immortalized by transfection with human telomerase, respectively, to achieve unlimited proliferation in cell culture. Thus, AD293 and LP9-hTERT might not fully represent normal cells, although they have been widely used as normal controls [39,40] and their response to REV3 depletion was consistent with the reaction of the primary mesothelial cell culture SDM104.
Studies have shown controversial results on the effect of REV3 depletion on cancer cell growth. On one hand, no deficiency in cell growth/survival was mentioned after si/shRNA-based inhibition of REV3 expression in HCT116, U2OS, and HeLa cancer cells [10,14,41]. Conversely, it was shown that knockout of REV3 resulted in a pronounced growth retardation in Burkitt lymphoma cells [42]. We found that inhibition of REV3 expression per se reduced colony formation in lung, breast, mesothelioma, and colon tumor cell lines. There are two possible explanations for these apparently controversial observations on the effects of REV3 depletion in cancer cells. First, the absence of cell growth inhibition in stable cancer cell lines depleted of REV3 might be due to the genetic modifications acquired during clonal selection, namely, rewiring of cell cycle checkpoint pathways [43]. Thus, it would be interesting to identify if the clones isolated in the studies mentioned acquired genetic modifications compared with their parental cell lines. Second, when investigated, it was found that inhibition of REV3 expression per se increased DNA damage levels in cancer cells even when no effect on cell growth/survival was mentioned [10,14,42]. Thus, it is possible that the DNA damage level necessary for DDR activation is different in the tested cell lines, explaining the presence or absence of growth arrest (reviewed in Al-Ejeh et al. [44]).
The second possibility is illustrated by the fact that only inhibition of REV3 expression by high-titer transduction resulted in a reduction of colony formation in MMR-deficient HCT116 cells, although REV3 expression was not further reduced. It was shown before that activation of the DDR is impaired in MMR-deficient HCT116 cells [45]. Thus, a higher level of cellular stress in form of additional DSBs due to more viral integration events after high-titer transduction might be required in HCT116 cells for the induction of a DDR resulting in the reduced colony formation after inhibition of REV3 expression.
In addition, the p53 status influences cell fate after REV3 depletion. The p53 status did not affect the accumulation of persistent DSBs indicated by P-ATM foci after inhibition of REV3 expression in HCT116 cells. Similarly, a recent study showed that DNA damage accumulation after prolonged activation of the mitotic checkpoint is also independent of the p53 status [46]. Thus, p53 does not protect cancer cells from damage accumulation due to REV3 depletion, although the subsequent cellular outcome, as discussed below, is dependent on the p53 status.
Previously, accumulation of H2AX phosphorylation in U2OS human osteosarcoma cells was observed after REV3 depletion [10]. Microscopic analysis revealed that inhibition of REV3 expression in cancer cells resulted in the accumulation of persistent DNA damage foci, which was also observed after exposure to high-dose ionizing radiation [31], suggesting the accumulation of irreparable DSBs. Similarly, the accumulation of large 53BP1 foci was also observed after the induction of mild replication stress or the genetic ablation of the BLM helicase [47]. Interestingly, a very recent publication showed that large 53BP1 foci mark sites of replication stress, which is passed onto daughter cells [48], giving rise to the possibility that the large 53BP1 foci detected after REV3 depletion mark sites of incomplete DNA synthesis rather than DSBs due to replication fork breakdown.
Cellular senescence limits the proliferation of damaged cells that are at risk for neoplastic transformation (reviewed in Collado and Serrano [34]). Our data indicate that, at least in p53-proficient cancer cells, senescence induction after REV3 depletion might prevent further transformation of cancer cells by establishing an essentially irreversible growth arrest. It is also proposed that the senescence-associated secretory phenotype, which we observed after inhibition of REV3 expression indicated by increased IL-6 secretion, might stimulate the immune system to clear senescent cells (reviewed in Collado and Serrano [34]). However, if senescent cells are not cleared by the immune system, they remain in a “dormant” state representing a dangerous potential for tumor relapse.
A recent study showed that nocodazole (a microtubule polymerization inhibitor) treatment of p53-deficient HCT116 cells leads to prolonged mitosis and subsequent return of the mitotically arrested cells to interphase without cell division resulted in aneuploidy [46], a process known as mitotic slippage. We observed that REV3 depletion in the p53-deficient HCT116 cell line and in combination with p53 inhibition in the A549 cell line leads to an accumulation of G2/M arrested cells and an increase in the frequency of aneuploid cells, which was also described in p53-deficient REV3-null MEFs [37].
On the basis of these results, we propose a model (Figure 5) in which inhibition of REV3 expression can be tolerated in normal cells but results in the accumulation of persistent DNA damage in cancer cells harboring cancer-specific alterations. Accumulation of persistent DNA damage leads in p53-proficient cancer cells to senescence, whereas REV3 depletion in p53-deficient cells results in growth inhibition and a G2/M arrest. A small fractionofthe p53-deficient cancer cells can overcome the G2/M arrest, which results in mitotic slippage and aneuploidy.
Figure 5.
Model: REV3 depletion induces persistent DNA damage specifically in cancer cells, which subsequently results in the induction of senescence in p53-proficient cancer cells and G2/M arrest in p53-deficient cancer cells. See text for details.
The concept of “synthetic lethality,” where defects in two pathways alone can be tolerated but become lethal when combined, has been originally described in Drosophila and yeast genetic studies [49,50]. This concept has been extended by the idea of “synthetic sickness,” whereas the combined loss/mutation of function of two genes does not kill cells but significantly impairs cellular fitness [51].
A recent study showed that inhibiting specific DNA repair polymerases induces synthetic sickness/lethality specifically in MMR-deficient cells [52]. In analogy, we found that REV3 depletion induces synthetic sickness/lethality in the investigated cancer cells. It will be interesting to identify the underlying cancer-specific alteration(s), which render the investigated cancer cell lines prone to growth inhibition due to REV3 depletion. In this context it was shown that DNA repair and/or cell cycle checkpoint mechanisms are frequently abrogated in cancer cells [53], and the concentration of endogenous DNA damage is higher in human tumoral tissue compared with the corresponding adjacent normal tissue (reviewed in Croteau and Bohr [54]). Therefore, differences in repair capacity or DNA damage levels between normal and cancer cells might be the underlying cause for the observed increased sensitivity of cancer cells to REV3 depletion. Alternatively, replication stress due to the activation of oncogenes might sensitize cancer cells to the inhibition of REV3 expression. A recent study showed that overexpression of Sch9, the S. cerevisiae homolog of the mammalian proto-oncogenes Akt and S6, increases superoxide-dependent DNA damage, which subsequently leads to the REV3-dependent formation of point mutations to avoid gross chromosomal rearrangements [55]. However, we cannot exclude that the specific genetic or epigenetic alterations underlying the observed synthetic sickness/lethality after REV3 depletion might differ between the tested cancer cell lines. Indeed, a recent study showed that REV3 deletion in a S. cerevisiae strain containing a particular additional chromosome resulted in decreased colony formation [56].
Cancer cells can be addicted not only to oncogenes but also to nononcogenes (reviewed in Luo et al. [57]). “Nononcogene addiction” genes are also required for maintenance of the tumorigenic state but are in contrast to oncogenes not functionally altered or mutated. The most prominent example of a “nononcogene addiction” gene is PARP, which is essential in BRCA-deficient breast cancer cells. Thus, based on the results of our study, we propose that REV3 functions as a “nononcogene addiction” gene, whose depletion induces synthetic sickness/lethality specifically in the investigated cancer cell lines. Along those lines, we are performing a genome-wide screen to identify essential molecular pathways in cancer cells whose inhibition will further enhance cell killing in combination with REV3 inhibition.
It will be interesting to determine whether 1) DNA damage tolerance by REV3-dependent TLS, 2) REV3-dependent DNA repair, or 3) a yet-to-be-identified function of REV3 is essential for cancer cell growth. Indeed, the size of mammalian REV3 is approximately double the size of the yeast homolog, giving rise to the possibility that the nonconserved region of REV3 harbors a yet-to-be-identified functional domain, necessary only in higher organisms.
Supplemental Materials and Methods
Cell Lines
The human MPM cell line ZL55 and the primary cell culture SDM104 were generated in our laboratory [25,30]. The rat MPM cell line IL45 was generated elsewhere (Craighead et al., Am J Pathol. 1987;129:448–462). The breast cancer cell lines MDA-MB-231 and MCF-7, the adenocarcinoma Calu-3, the squamous non-small cell lung cancer cell line A549 and the HEK 293T were purchased from American Type Culture Corporation (Manassas, VA). The AD293 cell line, a HEK 293 derivative with improved cell adherence, was purchased from Stratagene (La Jolla, CA). The colorectal carcinoma cell lines HCT116 40.16 (p53+/+) and HCT116 379.2 (p53-/-) were kindly provided by Dr Bert Vogelstein (Johns Hopkins University, Baltimore, MD).
Reagents
When indicated, 20 µM cisplatin (Ebewe Pharma, Cham, Switzerland) was added for 24 hours.
To clone the short hairpin constructs into the plasmid pSuperior.puro, the following DNA oligonucleotides were ordered from Microsynth:
shREV3–4:
5′-GATCCCCCAAAGATGCTGCTACATTATTCAAGAGATAATGTAGCAGCATCTTTGTTTTTA-3′
5′-AGCTTAAAAACAAAGATGCTGCTACATTATCTCTTGAATAATGTAGCAGCATCTTTGGGG-3′
shREV3–5:
5′-GATCCCCGATATTCCATCTGTTACAATTCAAGAGATTGTAACAGATGGAATATCTTTTTA-3′
5′-AGCTTAAAAAGATATTCCATCTGTTACAATCTCTTGAATTGTAACAGATGGAATATCGGG-3′
shREV3–6:
5′-GATCCCCTAGTCAGACTTTTCAGCCTTTCAAGAGAAGGCTGAAAAGTCTGACTATTTTTA-3′
5′-AGCTTAAAAATAGTCAGACTTTTCAGCCTTCTCTTGAAAGGCTGAAAAGTCTGACTAGGG-3′
shSCR:
5′-GATCCCCATTCTAGGTGAAAGCTAATTTCAAGAGATAAGATCCACTTTCGATTATTTTTA-3′
5′-AGCTTAAAAATAATCGAAAGTGGATCTTATCTCTTGAAATTAGCTTTCACCTAGAATGGG-3′
shP53:
5′-GATCCCCGACTCCAGTGGTAATCTACTTCAAGAGAGTAGATTACCACTGGAGTCTTTTTA-3′
5′-AGCTTAAAAAGACTCCAGTGGTAATCTACTCTCTTGAAGTAGATTACCACTGGAGTCGGG-3′
To quantitatively measure the expression of REV3 mRNA by real-time PCR, the following DNA oligonucleotides were ordered from Microsynth:
REV3:
Forward 5′-TGAGTTCAAATTTGGCTGTACCT-3′
REV3:
Reverse 5′-TCTAGTCTTCAAAATTTCTTCAAGCA-3′
Histone H3:
Forward 5′-TAAAGCACCCAGGAAACAACTGGC-3′
Reverse 5′-ACCAGGCCTGTAACGATGAGGTTT-3′
P53:
Forward 5′-GCTTTGAGGTTCGTGTTTGTGCCT-3′
Reverse 5′-GCCCACGGATCTTAAGGGTGAAAT-3′
For Western analysis, the following primary antibodies were diluted at 1:1000: PARP (no. 9542; Cell Signaling, Beverly, MA), P-Chk2 (no. AF1626; R&D Systems), p53 (no. 9282; Cell Signaling), p21 (no. sc-756; Santa Cruz, Santa Cruz, CA), cyclin E (no. sc-247; Santa Cruz), and MAD2B/Rev7 (no. 612266; BD Biosciences). The primary antibody β-actin (no. 691001; MP Biomedicals, Solon, OH) was incubated 1:10,000 for 1 hour at room temperature. The secondary polyclonal antibodies coupled to horseradish peroxidase were diluted at 1:10,000.
For immunofluorescence microscopy, the following primary antibodies were used: P-ATM 1:1000 (no. sc-4526; Cell Signaling), γH2AX 1:1000 (no. 05-636; Upstate Biotechnology, Lake Placid, NY), 53BP1 1:500 (no. 4937; Cell Signaling), and BrdU 1:1000 (no. 555627; BD Biosciences).
Vector Production and Transduction
Short hairpin REV3–4 and scrambled (shSCR) oligos were ligated into pSuperior.puro as described by the manufacturer (OligoEngine, Seattle, WA). The shRNA and H1 promoter fragments were subsequently ligated into the constitutive expressing lentiviral vector pLVTHM (Addgene, Cambridge, MA). Replication-deficient lentiviral particles were produced and titrated as described previously [26,27].
Cells were seeded in six-well plates (colony formation, immunofluorescence, and SA-β-galactosidase assay: 500 cells/well [SDM104: 1000 cells/well]; FACS and Western analysis: 2500 cells/well [SDM104: 5000 cells/well]; real-time PCR and ELISA: 5000 cells/well) in 2 ml of medium. After 6 hours, the medium was removed, and lentivirus suspension was added for 30 minutes in 300 µl (immunofluorescence, FACS, and real-time PCR: MOI = 100; Western analysis and colony formation: MOI = 170). All transductions of HCT116 and the corresponding control AD293 cells were performed with an MOI of 800 and were incubated for 1 hour. Subsequently, medium was added to final volume of 1.5 ml. For mock treatment, 0.5-µm filtered conditioned medium from a HEK 293T culture was added. After 7 days, cells were further processed for distinct experiment as described below except for colony formation, which were incubated longer as described below.
Quantitative Real-time PCR
Real-time PCR cycle conditions were as follows: one cycle of 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds and 60°C for 1 minute, and one cycle of 95°C for 15 seconds and from 60°C slowly elevating to 95°C for several minutes for dissociation curve analysis. Histone H3 expression was used to standardize the total amount of complementary DNA, and the specificity of the PCR was confirmed by analysis of the melting curve.
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed essentially as described before [28]. In detail, for BrdU staining, cells were incubated with 10 µM BrdU for 1 hour. Fixation and permeabilization were done with 100% methanol at -20°C and acetone-methanol (50:50) at -20°C for 20 minutes at room temperature, respectively. Cells were washed 2 x 5 minutes with PBS. For BrdU staining, cells were denatured with 2 M HCl for 30 minutes at room temperature and subsequently washed 3 x 5 minutes with PBS. PBS containing 1% fetal bovine serum and 10% bovine serum albumin was used as blocking solution for 1 hour at room temperature. First, antibodies were diluted in blocking solution and incubated at 4°C overnight. The following antibodies were used: P-ATM 1:1000 (no. sc-4526; Cell Signaling), γH2AX 1:1000 (no. 05-636; Upstate), 53BP1 1:500 (no. 4937; Cell Signaling), and BrdU 1:1000 (no. 555627; BD Biosciences). Cells were washed 2 x 5 minutes in PBS. Secondary antibodies were diluted at 1:10,000 in blocking solution and incubated at 37°C for 1 hour. Cells were washed 2 x 5 minutes with PBS and mounted with ProLong Gold Antifade reagent with 4′,6-diamidino2-phenylindole (no. P36931; Invitrogen). Images were acquired with an inverse wide-field fluorescence microscope (DM IRBE; Leica, Bensheim, Germany) equipped with a black and white camera (ORKA-ER; Hamamatsu, Hamamatsu, Japan). Image processing with Photoshop (Adobe Systems) was applied to whole images only. Images used for comparison between different transductions were acquired with the same instrument settings and exposure time and were processed equally.
Flow Cytometry
Cell cycle distribution was assessed by using a FACSCalibur (FACScan, 488-nm excitation laser; BD Biosciences) and WinMDI software.
Western Analysis
Cells were lysed for 30 minutes on ice in 1 x RIPA buffer (Upstate) containing 2 x HALT protease and phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL). Cell extracts were denatured at 95°C for 5 minutes and homogenized by successive passing through a 30-gauge syringe needle, and protein concentrations were determined using bicinchoninic acid protein assay (Pierce, Rockford, IL).
Acknowledgments
The authors thank Alexandra Graf for her help in the initial experiments and Bert Vogelstein for HCT116 (p53+/+) and HCT116 (p53-/-) cell lines.
Abbreviations
- TLS
DNA translesion synthesis
- Pol ζ
DNA translesion synthesis polymerase ζ
- REV3
the mammalian REV3L gene
- MEF
mouse embryonic fibroblast
- DDR
DNA damage response
- DSBs
DNA double-strand breaks
- ATM
ataxia-telangiectasia mutated
- γH2AX
phosphorylated H2AX
- P-Chk2
phosphorylated Chk2
- AN
aneuploid nondividing
- AD
aneuploid dividing
Footnotes
This study was funded by support from the Cancer League Zurich and the Sassella Foundation to T.M.M. and from the Seroussi Foundation and the Foundation for Applied Cancer Research Zurich to R.A.S. The authors have declared that no competing interests exist.
This article refers to supplementary materials, which are designated by Figures W1 to W6 and are available online at www.neoplasia.com.
References
- 1.Lemontt JF. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence CW, Das G, Christensen RB. REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet. 1985;200:80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- 4.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bemark M, Khamlichi AA, Davies SL, Neuberger MS. Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr Biol. 2000;10:1213–1216. doi: 10.1016/s0960-9822(00)00724-7. [DOI] [PubMed] [Google Scholar]
- 6.Esposito G, Godindagger I, Klein U, Yaspo ML, Cumano A, Rajewsky K. Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr Biol. 2000;10:1221–1224. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- 7.Wittschieben J, Shivji MK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, Wood RD. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr Biol. 2000;10:1217–1220. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- 8.O-Wang J, Kajiwara K, Kawamura K, Kimura M, Miyagishima H, Koseki H, Tagawa M. An essential role for REV3 in mammalian cell survival: absence of REV3 induces p53-independent embryonic death. Biochem Biophys Res Commun. 2002;293:1132–1137. doi: 10.1016/S0006-291X(02)00341-8. [DOI] [PubMed] [Google Scholar]
- 9.Rajpal DK, Wu X, Wang Z. Alteration of ultraviolet-induced mutagenesis in yeast through molecular modulation of the REV3 and REV7 gene expression. Mutat Res. 2000;461:133–143. doi: 10.1016/s0921-8777(00)00047-1. [DOI] [PubMed] [Google Scholar]
- 10.Brondello JM, Pillaire MJ, Rodriguez C, Gourraud PA, Selves J, Cazaux C, Piette J. Novel evidences for a tumor suppressor role of Rev3, the catalytic subunit of Pol ζ. Oncogene. 2008;27:6093–6101. doi: 10.1038/onc.2008.212. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Zhang SY, Wang S, Lu J, Wu W, Weng L, Chen D, Zhang Y, Lu Z, Yang J, et al. REV3L confers chemoresistance to cisplatin in human gliomas: the potential of its RNAi for synergistic therapy. Neuro Oncol. 2009;11:790–802. doi: 10.1215/15228517-2009-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki M, et al. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- 13.Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Gueranger Q, Glover TW, Canman CE. Differential roles for DNA polymerases ε, ζ, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol. 2010;30:1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu F, Lin X, Okuda T, Howell SB. DNA polymerase ζ regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res. 2004;64:8029–8035. doi: 10.1158/0008-5472.CAN-03-3942. [DOI] [PubMed] [Google Scholar]
- 16.Schenten D, Kracker S, Esposito G, Franco S, Klein U, Murphy M, Alt FW, Rajewsky K. Pol ζ ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J Exp Med. 2009;206:477–490. doi: 10.1084/jem.20080669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbs PE, McGregor WG, Maher VM, Nisson P, Lawrence CW. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc Natl Acad Sci USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diaz M, Watson NB, Turkington G, Verkoczy LK, Klinman NR, McGregor WG. Decreased frequency and highly aberrant spectrum of ultraviolet-induced mutations in the hprt gene of mouse fibroblasts expressing antisense RNA to DNA polymerase ζ. Mol Cancer Res. 2003;1:836–847. [PubMed] [Google Scholar]
- 19.Xie K, Doles J, Hemann MT, Walker GC. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci USA. 2010;107:20792–20797. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, Hemann MT. Suppression of Rev3, the catalytic subunit of Pol{ζ}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci USA. 2010;107:20786–20791. doi: 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res. 2006;66:134–142. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- 22.Van Sloun PP, Varlet I, Sonneveld E, Boei JJ, Romeijn RJ, Eeken JC, De Wind N. Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol Cell Biol. 2002;22:2159–2169. doi: 10.1128/MCB.22.7.2159-2169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamaizumi M, Yagi T, Verkaik NS, van Gent DC, Takata M, et al. Multiple roles of Rev3, the catalytic subunit of pol ζ in maintaining genome stability in vertebrates. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 25.Thurneysen C, Opitz I, Kurtz S, Weder W, Stahel RA, Felley-Bosco E. Functional inactivation of NF2/merlin in human mesothelioma. Lung Cancer. 2009;64:140–147. doi: 10.1016/j.lungcan.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Reed SE, Staley EM, Mayginnes JP, Pintel DJ, Tullis GE. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J Virol Methods. 2006;138:85–98. doi: 10.1016/j.jviromet.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Hum Genet. 2007 doi: 10.1002/0471142905.hg1210s54. Chapter 12, Unit 12 10. [DOI] [PubMed] [Google Scholar]
- 28.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylationwithinthe G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopkins-Donaldson S, Ziegler A, Kurtz S, Bigosch C, Kandioler D, Ludwig C, Zangemeister-Wittke U, Stahel R. Silencing of death receptor and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ. 2003;10:356–364. doi: 10.1038/sj.cdd.4401157. [DOI] [PubMed] [Google Scholar]
- 31.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamauchi M, Oka Y, Yamamoto M, Niimura K, Uchida M, Kodama S, Watanabe M, Sekine I, Yamashita S, Suzuki K. Growth of persistent foci of DNA damage checkpoint factors is essential for amplification of G1 checkpoint signaling. DNA Repair. 2008;7:405–417. doi: 10.1016/j.dnarep.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Quinn CM, Wright NA. The clinical-assessment of proliferation and growth in human tumors—evaluation of methods and applications as prognostic variables. J Pathol. 1990;160:93–102. doi: 10.1002/path.1711600202. [DOI] [PubMed] [Google Scholar]
- 34.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Zhang H, McManus TP, McCormick JJ, Lawrence CW, Maher VM. hREV3 is essential for error-prone translesion synthesis past UV or benzo[a]pyrene diol epoxide-induced DNA lesions in human fibroblasts. Mutat Res. 2002;510:71–80. doi: 10.1016/s0027-5107(02)00253-1. [DOI] [PubMed] [Google Scholar]
- 37.Zander L, Bemark M. Immortalized mouse cell lines that lack a functional Rev3 gene are hypersensitive to UV irradiation and cisplatin treatment. DNA Repair. 2004;3:743–752. doi: 10.1016/j.dnarep.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 38.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 39.Tu Y, Kim JS. Selective gene transfer to hepatocellular carcinoma using homing peptide-grafted cationic liposomes. J Microbiol Biotechnol. 2010;20:821–827. [PubMed] [Google Scholar]
- 40.Hillegass JM, Shukla A, MacPherson MB, Bond JP, Steele C, Mossman BT. Utilization of gene profiling and proteomics to determine mineral pathogenicity in a human mesothelial cell line (LP9/TERT-1) J Toxicol Environ Health A. 2010;73:423–436. doi: 10.1080/15287390903486568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X, Howell SB. DNA mismatch repair and p53 function are major determinants of the rate of development of cisplatin resistance. Mol Cancer Ther. 2006;5:1239–1247. doi: 10.1158/1535-7163.MCT-05-0491. [DOI] [PubMed] [Google Scholar]
- 42.Gueranger Q, Stary A, Aoufouchi S, Faili A, Sarasin A, Reynaud CA, Weill JC. Role of DNA polymerases ε, ı and ζ in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair. 2008;7:1551–1562. doi: 10.1016/j.dnarep.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM-and ATR-mediated checkpoint signaling through the p38MAPK/ MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Ejeh F, Kumar R, Wiegmans A, Lakhani SR, Brown MP, Khanna KK. Harnessing the complexity of DNA-damage response pathways to improve cancer treatment outcomes. Oncogene. 2010;29:6085–6098. doi: 10.1038/onc.2010.407. [DOI] [PubMed] [Google Scholar]
- 45.Brown KD, Rathi A, Kamath R, Beardsley DI, Zhan Q, Mannino JL, Baskaran R. The mismatch repair system is required for S-phase checkpoint activation. Nat Genet. 2003;33:80–84. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- 46.Dalton WB, Yu B, Yang VW. p53 suppresses structural chromosome instability after mitotic arrest in human cells. Oncogene. 2010;29:1929–1940. doi: 10.1038/onc.2009.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 48.Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, Fraser P, Jackson SP. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol. 2011;193:97–108. doi: 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobzhansky T. Genetics of natural populations. XIII. Recombination and variability in populations of Drosophila pseudoobscura. Genetics. 1946;31:269–290. doi: 10.1093/genetics/31.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartman JLT, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 51.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 52.Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell. 2010;17:235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 54.Croteau DL, Bohr VA. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J Biol Chem. 1997;272:25409–25412. doi: 10.1074/jbc.272.41.25409. [DOI] [PubMed] [Google Scholar]
- 55.Madia F, Wei M, Yuan V, Hu J, Gattazzo C, Pham P, Goodman MF, Longo VD. Oncogene homologue Sch9 promotes age-dependent mutations by a superoxide and Rev1/Pol ζ-dependent mechanism. J Cell Biol. 2009;186:509–523. doi: 10.1083/jcb.200906011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.