Abstract
Neuroblastoma (NB) is a neural crest-derived childhood tumor characterized by a remarkable phenotypic diversity, ranging from spontaneous regression to fatal metastatic disease. Although the cancer stem cell (CSC) model provides a trail to characterize the cells responsible for tumor onset, the NB tumor-initiating cell (TIC) has not been identified. In this study, the relevance of the CSC model in NB was investigated by taking advantage of typical functional stem cell characteristics. A predictive association was established between self-renewal, as assessed by serial sphere formation, and clinical aggressiveness in primary tumors. Moreover, cell subsets gradually selected during serial sphere culture harbored increased in vivo tumorigenicity, only highlighted in an orthotopic microenvironment. A microarray time course analysis of serial spheres passages from metastatic cells allowed us to specifically “profile” the NB stem cell-like phenotype and to identify CD133, ABC transporter, and WNT and NOTCH genes as spheres markers. On the basis of combined sphere markers expression, at least two distinct tumorigenic cell subpopulations were identified, also shown to preexist in primary NB. However, sphere markers-mediated cell sorting of parental tumor failed to recapitulate the TIC phenotype in the orthotopic model, highlighting the complexity of the CSC model. Our data support the NB stem-like cells as a dynamic and heterogeneous cell population strongly dependent on microenvironmental signals and add novel candidate genes as potential therapeutic targets in the control of high-risk NB.
Introduction
Neuroblastoma (NB) is a pediatric cancer originating from neural crest-derived sympathoadrenal progenitors. This disease displays a considerable heterogeneity, reflected in the clinical outcome, ranging from spontaneous regression to extreme malignancy [1–3]. Moreover, NB exhibits a wide range of differentiated phenotypes, from undifferentiated tumors to tumors containing a neural crest-derived differentiated cell range [4].
Heterogeneity within cancer cell populations is common. However, whether it results from functional cellular hierarchy for malignancy, as proposed by the cancer stem cell (CSC) model, or from clonal evolution within the tumor is still a matter of intensive debate. In the CSC model, tumors are exclusively sustained by a minor subpopulation of tumor-initiating cells (TICs) [5–7]. This hierarchical stem cell model gained considerable attractiveness by providing a likely explanation for the multiple treatment failures and ideal cellular targets to definitively eradicate refractory tumors [5,8,9]. However, because stem cell-like cells have only been convincingly identified in a limited number of tumor types, the TIC model has been increasingly challenged [10]. The possibility that such model may not apply to all solid tumors or may reveal to be considerably more complex in some cancers is now suggested [11,12].
Because of its embryonal origin and heterogeneity, NB tumor qualifies as a pertinent model to explore functional cellular hierarchy in solid tumors. In addition, rare cells expressing neural crest stem and progenitor cells markers were identified within NB tissues and cell lines, suggesting the existence of NB CSC . However, very little is known on the existence and phenotype of NB-TICs. Moreover, none of the markers defined to prospectively identify functional TICs in several adult hematological and solid malignancies apply for putative NB-TICs identification. Investigators have therefore turned to functional stem cell-like characteristics such as self-renewal (serial sphere formation), tumorigenicity (in vivo serial tumor engraftment), and drug resistance (side population, drug survival) to isolate NB-TICs. Thus, Hansford et al. [13] and Smith et al. [14] have isolated highly tumorigenic, drug-resistant sphere-forming cell lines from NB metastatic bone marrow based on their capacities to self-renew and differentiate toward distinct lineages.
Besides, well-characterized NB cell lines are known to recapitulate the heterogeneity observed in tumors, and some of them were shown to display stem cell-like characteristics. Thus, a highly tumorigenic “side population” of cells capable to specifically efflux a Hoechst dye was isolated from NB cell lines [15–17]. However, no specific gene expression profiles have been formerly identified for NB-TICs either in primary tumors or in NB cell lines.
To characterize representative phenotypes for self-renewal and tumorigenicity, we performed a microarray time course analysis of serial sphere passages obtained from patient bone marrow metastatic NB cells. Such a dynamic and original analysis revealed a list of NB sphere-associated genes that crossed with neural crest (NCC) and neural stem cell (NSC) genes, essentially implicated in developmental and cell differentiation pathways [18–21]. Moreover, the sphere-associated gene expression profiling combined with in vivo assay indicated that NB-TICs most likely represent a heterogeneous cell population, whose phenotype is strongly dependent on the direct microenvironment.
Materials and Methods
Isolation of NB Primary Cells
The tumor material was collected from a cohort of 10 NB patients, diagnosed at the Hemato-oncology Unit of the University Hospital of Lausanne (Switzerland), after informed consent and in agreement with local institutional ethical regulations. Tumor tissues were processed (within 24 hours after resection) in a single-cell suspension by mechanical dissociation in phosphate-buffered saline containing 0.01 mg/ml collagenase II (Invitrogen, Carlsbad, CA; http://www.invitrogen.com) and 0.1 mg/ml DNaseI (Roche Diagnostics, Basel, Switzerland; http://www.roche-applied-science.com) for 30 minutes at 37°C, followed by filtration through CellTricks (50 µm; Partek, Inc, St Louis, MO; http://www.partek.com).
NB Genomic Profile Analysis
The genetic type of NB tumors was determined on biopsies with a tumor cell content superior to 60% either by array comparative genomic hybridization (CGH; Curie Institute, Paris, France) as described [22] or at the Agilent platform from the Genetic Division, University Hospital, Lausanne [23]. Alternatively, fluorescent in situ hybridization for the 1p, 2p, 7p and 7q, 11q, and 17q regions was performed [24].
In Vivo Studies
All animal experiments were carried out with athymic Swiss nude mice (BALB/C nu/nu) in accordance to the European Community guidelines (directive no. 86/609/CEE). NB cells isolated from involved bone marrow aspirations or NB patient samples xenografts were used.
In heterotopic assays, 104 cells in Dulbecco modified Eagle medium (DMEM)/F12 and BD Matrigel Basement Membrane matrix (1:1; BD Biosciences, San Diego, CA; http://www.bdbiosciences.com) were subcutaneously (s.c.) injected in mice flanks. In orthotopic assays, 104 cells in phosphate-buffered saline were implanted in the adrenal gland as previously described [25]. Tumor intake and growth were followed up using calipers every 3 days for s.c. tumors or every 2 weeks by ultrasound imaging to follow orthotopic growth (Lausanne Cardiovascular Assessment Facility). Macroscopic metastases were evaluated by gross examination.
Immunohistochemistry
Standard hematoxylin and eosin (H&E) and immunohistostaining procedures were performed on paraffin-embedded tissues, using mouse monoclonal anti-human p75 (clone ME20.4), rabbit polyclonal antihuman CD31 (Thermo Fisher Scientific, Waltham, MA; http://www.thermofisher.com), and mouse monoclonal anti-human Ki67 (clone MIB-1; Dako, Carpinteria, CA; http://www.dakousa.com).
Cell Lines and Sphere Culture Conditions
NB cell lines, including LAN-1 [26] and SK-N-BE(2)C [27], were grown in DMEM containing 10% fetal calf serum (Sigma-Aldrich, St Louis, MO; http://www.sigma-aldrich.com), 100 U/ml penicillin, and 10 µg/ml streptomycin (Invitrogen). The NB1-C cell line was established by plating the dissociated NB1 cell suspension in neural basic medium (DMEM/F12 supplemented with penicillin/streptomycin, 2% B27 [Invitrogen], human recombinant basic fibroblast growth factor [FGF; 20 ng/ml; Peprotech, Rocky Hill, NJ; http://www.peprotech.com], and EGF [20 ng/ml; Peprotech]).
NB sphere culture was performed in neural crest sphere medium, prepared as described [28,29], and cultured in poly(2-hydroxyethyl methylacrylate) (16 mg/ml in EtOH; Sigma)-coated six-well culture plates to prevent cell adhesion. NB cells (2–8 x 104 cells/ml) were plated, and spheres were passed every 7 to 10 days by dissociation in 0.05% trypsin-EDTA (Invitrogen) and filtration in CellTricks (50 µm; Partek) to obtain a single-cell suspension. This plating (104 cells/500 µl neural crest sphere medium in quadriplicates) procedure was serially repeated to assay for self-renewal.
Immunoblot Analysis
Protein extracts were prepared as already described [30,31] with a modified NP40 lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP40). Extracts (50 µg) were loaded on 10% SDS-PAGE and transferred on nitrocellulose membranes. Blots were labeled with rabbit polyclonal anti-c-myc antibody (N-262; Santa Cruz Biotechnology, Heidelberg, Germany) and NCMII mouse monoclonal anti-N-myc antibody [32]. Blots were revealed using secondary goat antimouse-horseradish peroxidase antibody (Jackson Immuno-Research, Suffolk, United Kingdom) or goat antirabbit immunoglobulin G-horseradish peroxidase (Nordic Immunological Laboratories, Tilburg, Netherlands). Bound antibodies were detected using the ECL Advance Kit according to the manufacturer's instructions (GE Health-care, Chalfont St Giles, United Kingdom).
Cell Viability Assay
Cells (5 x 103) were plated 24 hours before treatment in 96-well plates with 1 µg/ml doxorubicin (Sigma), 100 µg/ml etoposide (Sigma), and 16 µg/ml cisplatin (Sigma) for 48 hours. Cell viability was measured in triplicate using MTS/PMS cell proliferation kit (Promega Corp, Madison, WI; http://www.promega.com) according to the manufacturer's instructions.
Total RNA Extraction and Gene Expression Profiling
Cells were lysed with TRIzol reagent (Invitrogen), and total RNA was extracted twice with ultrapure phenol-chloroform-isoamyl alcohol (25:24:1) (Invitrogen) and purified using RNeasy MicroKit columns (Qiagen, Hilden, Germany; http://www.qiagen.com). RNA quality was verified by the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA; http://www.chem.agilent.com) before microarray hybridization.
Expression profiling experiments were performed at the DNA Array Facility Lausanne, University of Lausanne, using the Human Genome U133Plus 2.0 Affymetrix GeneChip Technology (Affymetrix, Santa Clara, CA; http://www.affymetrix.com).
The Remote Analysis Computation for gene Expression data system developed by the DNA Array Facility Lausanne was used for micro-array data analysis [33]. All chips were normalized using the robust multiarray analysis procedure [34]. Statistical analyses of differentially expressed genes were performed using the “R” statistical version 2.0.1. The statistical value for the “step” gene analysis has been calculated by the formula:
with X = “step” statistics and mean(s1:s4 - s0 = “step” change. A cutoff has been defined as the absolute value of X = 3. P values associated with the fold change (fold change ≥ 2) were corrected for multiple testing using the false discovery rate method with the cutoff set at 5%.
Complementary DNA Synthesis and Semiquantitative Real-time Polymerase Chain Reaction
Reverse transcription of total RNA was performed using random primers and SuperScript II reverse transcriptase (Invitrogen). Sphere-associated gene expression was assessed by real-time semiquantitative polymerase chain reaction (PCR, ABI PRISM 7900 HT System; Applied Biosystems, Carlsbad, CA; http://www.appliedbiosystems.com) with SYBR Green detection (Qiagen). The amount of PCR product was divided by that of human HPRT1 using the ΔΔCt method. Amplification reactions were performed with specific pairs of primers for CD133, EDNRB, GPR177, NOTCH3, MDR1, NOTCH2, ROBO1, and ABCA1 transcripts (Supplementary Materials and Methods).
Flow Cytometry
Single-cell suspensions were processed and stained as described [35] with CD133/1-APC (Clone AC133; Miltenyi Biotec, Bergisch Gladbach, Germany; http//www.miltenyibiotec.com), MDR1-PE (P-glycoprotein, clone UIC2; Beckman Coulter, Brea, CA; http://www.beckmancoulter.com) and anti-GD2 clone 7A4 [36] antibodies. For specific cell sorting, a total of 106 cells were analyzed and sorted at 4°C by FACSAria II Cell Sorter (BD Biosciences).
Statistical Analysis
Statistical analyses were carried out using GraphPad Prism 4.0 (GraphPad Software, Inc, La Jolla, CA; http://www.graphpad.com/prism). P < .05 was considered to represent significance. P < .01 and P < .001 were interpreted to be highly significant.
Results
Clinical NB Xenografts as Models to Address NB-TIC Populations
The availability of tumor material was guaranteed by in vivo serial subcutaneous xenograft NB cell transplantations. Table 1 summarizes the characteristics of the patients' tumors included in the study, comprising five stage 4 (one relapse, NB4), one stage 3 (after chemotherapy), three stage 4s samples, and one ganglioneuroma. MYCN amplification was detected in NB5 only, whereas the NB1 sample displayed a chromosomal gain in 8q identified as a c-MYC gain. Immunoblots for MYCN and c-MYC were performed with protein extracts from the NB1-NB10 tumors. Data confirmed the MYCN and c-MYC overexpression in NB5 and NB1, respectively (Figure W1). NB8 cells displayed an increased MYCN expression in the absence of gene amplification. A significant amount of c-MYC protein was also observed in the NB2 tumor without detectable amplification of the c-MYC locus. Array CGH and fluorescent in situ hybridization were performed for all samples to determine the genetic type, as proposed [37].
Table 1.
Clinical and Biologic Characteristics of NB Samples.
| Sample | Sex | Age at Diagnosis | Stage (INSS) | Specimen Type | BM Infiltration | NMYC | Genetic Type | Follow-up | Risk Group | Xeno. Tumor |
| NB1 | M | 10 mo | 4 | bm | Yes | NA | B | DOD | HR | Yes (p16) |
| NB2 | F | 44 mo | 4 | bm | Yes | NA | B | CR | HR | Yes (p10) |
| NB3 | M | 40 mo | 4 | pt | Yes | NA | B | DOD | HR | Yes (p1) |
| NB4 | M | 12 y | 4 | bm-rel | Yes | NA | B | DOD | HR | Yes (p5) |
| NB5 | M | 24 mo | 4 | bm | Yes | A | C/E | CR | HR | Yes (p2) |
| NB6 | F | 58 mo | 3 | pt-pc | No | NA | D | AD | HR | No |
| NB7 | F | 1 d | 4s | bm | Yes | NA | D | CR | LR/IR | No |
| NB8 | F | 8 mo | 4s | cut | No | NA | ND | VGPR | LR/IR | ND |
| NB9 | F | 5 d | 4s | pt | ND | NA | D | VGPR | LR/IR | No |
| NB10 | F | 22 mo | 3, GGN | pt-pc | No | NA | Flat | CR | LR | No |
NB1 to NB10 correspond to NB patients. Xeno. tumor: in vivo tumor growth capacity assessed by serial subcutaneous engraftments (px = passage number).
A indicates amplified MYCN locus; AD, alive with disease; bm, bone marrow; CR, complete remission; cut, cutaneous metastasis; DOD, dead of disease; F, female; GGN, ganglioneuroma; HR/IR/LR, high/intermediate/low-risk group; INSS, International Neuroblastoma Staging System; M, male; NA, no amplification of MYCN locus; nd, not determined; pc, postchemotherapy; pt, primary tumor; rel, relapse; VGPR, very good partial response.
To assay the tumorigenicity of patient samples, 1 million bone marrow mononuclear cells from NB1, NB2, NB4, NB5, and NB7 were s.c. injected in nude mice. Fragments of primary tumor NB3, NB6, NB9, and NB10 were s.c. grafted. Tumor growth was observed in 5 of 10 samples, all were stage 4 tumors, with a time-of-tumor detection varying from 40 to 60 days (Table 1).
Xenograft tumors were further serially propagated in nude mice (Figure 1A). The maximum passage number varied from p1 (NB3) to p16 for NB1, which showed the highest long-term in vivo serial passage capacity (Table 1). No tumor growth was obtained with stages 3 and 4s tumor samples NB6-NB10, even after 180 days of observation, demonstrating that metastatic stage 4-derived NB samples displayed the highest tumorigenicity.
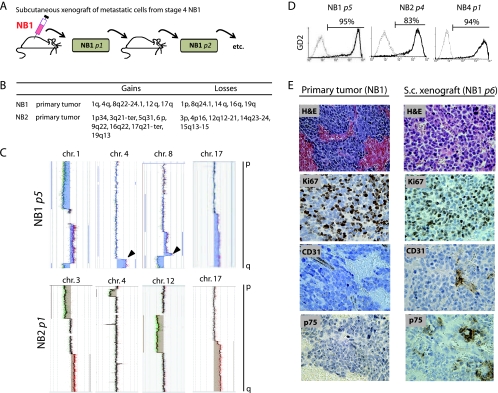
Figure 1.
Model for in vivo propagation of NB metastatic cells as xenografts. (A) The s.c. implantation of metastatic bone marrow cells from NB1 patient generated the xenograft tumor NB1 p1, further serially transplanted up to p16, in Swiss nude mice. (B) Chromosomic alterations (gains and losses) analyzed by array-CGH in NB1 and NB2 primary tumors. (C) Detailed array-CGH results for the indicated chromosomes of NB1 p5 and NB2 p1 cells. Black arrowhead indicates identical chromosomic alterations in primary and xenograft tumors. (D) FACS analysis of GD2 cell surface expression (black lines) and isotype control antibody (gray line). (E) H&E and immunohistologic staining for Ki67, CD31, and p75 markers (NB1 [primary tumor] and NB1 p6 xenografts).
The NB origin of the xenograft tumors derived from NB1 to NB5 samples was verified by array CGH analyses. NB1 p5 and NB2 p1 harbored the same NB typical alterations on the chromosomes 1, 4, 8, and 17 as the original primary tumors NB1 and NB2 (Figure 1, B and C). Moreover, the strong cell surface expression of disialoganglioside (GD2) [38] measured in NB1, NB2, and NB4 xenograft tumors confirmed their NB origin (Figure 1D). NB xenograft tumors also recapitulated the parental tumor histology and the intratumor heterogeneity as assessed by H&E staining and immunohistochemical staining for Ki67, CD31, and CD271 (p75LNGFR)(Figure 1E). NB xenograft tumors were thus considered as stable NB tissue, suitable for further phenotypic or functional studies.
High-risk Metastatic NBs Harbor Higher Sphere Formation Capacity
NB1-NB4 xenografts and clinical samples listed in Table 1 were analyzed for their ability to form serial spheres when propagated in a serum-free medium (Figure 2A). Except NB3 cells, all stages 3 and 4 samples formed serial spheres; NB1 showed the highest capacity for serial sphere formation (p5). NB7 to NB10 samples did not grow as spheres (data not shown).
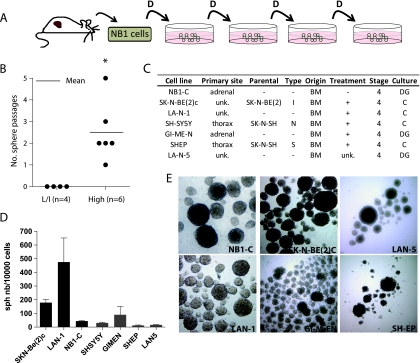
Figure 2.
The sphere-forming capacity is enriched in metastatic stage 4 NB cells. (A) Experimental model for serial spheres passage from dissociated NB xenograft tumors in a serum-free medium (e.g., NB1). D indicates dissociation/replating. (B) In vitro sphere propagation capacity of high-risk (High) and low/intermediate-risk NB groups (L/I). *P < .05, Student's t test. (C) Characteristics of NB cell lines (adapted from Thiele [77]). BM indicates bone marrow; C, direct culture; DG, density gradient; unk, unknown. (D) Self-renewal assay for indicated NB cell lines. (E) Microscopic observation (40x magnification) of secondary spheres.
The tumor samples were assigned to low/intermediate (L/I) and high-risk (HR) groups according to the current Children's Oncology Group (COG) criteria [39]. Tumors of the HR group displayed an increased ability to generate serial spheres in culture compared with tumors of the L/I group (P = .03; Figure 2B). These data supported previous reports showing that HR NB tumors display enhanced self-renewal characteristics [13].
NB cell lines are generally established from bone marrow aspirations harvested in stage 4 patients (Figure 2C) and propagated in the presence of 10% serum, which is known to induce genetic and epigenetic modifications in cancer cells, as opposed to serum-free conditions [40]. We addressed the sphere-forming capacity of six NB cell lines and NB1-C, a cell line established from the NB1 xenograft tumor in serum-free medium (Figure 2D). All cell lines were able to grow in sphere conditions up to high-passage numbers, such as NB1-C, LAN-1, and SK-N-BE(2)C cells, although variability was observed (Figure 2, D and E).
NB1 Sphere Cells (NB1-s) Are More Tumorigenic in the Adrenal Microenvironment and Harbored an In Vitro Increased Drug Resistance
Serial sphere formation has been proposed to enrich in cells with TIC property in several cancer types. Considering the previously mentioned results, we postulated that NB1 sphere cells represented a relevant model to address the existence of NB-TICs.
The criterion standard for the assessment of TIC phenotype is a serial in vivo orthotopic implantation of a putative NB-TIC population. We thus compared the tumorigenicity of NB1 spheres cells (NB1-s) with total NB1 cells orthotopically implanted. The tumor intake between NB1-s and NB1, with 79% versus 40%, respectively, was significantly different. The tumor volume (day 83) was also significantly larger in the NB1-s group (73.98 ± 36.61 mm3, n = 10) compared with the NB1 group (9.62 ± 2.16 mm3, n = 14) (P < .01; Figure 3A). In contrast, when NB1 or NB1-s cells were heterotopically injected, no significant difference in tumor intake (6/6) or detection time was measured (Figure 3B).
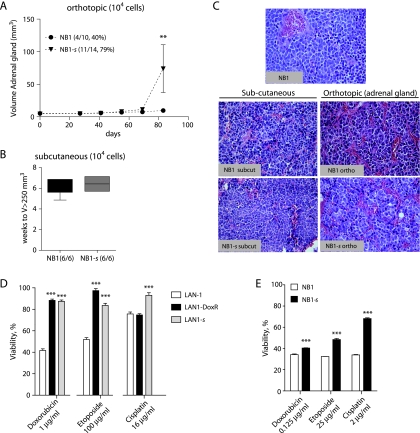
Figure 3.
NB1 sphere cells (NB1-s) harbored an increased tumorigenicity in orthotopic conditions and a resistance to chemotherapeutic drugs. (A) In vivo orthotopic tumor intake (number of mice bearing tumor/total mice) and growth (mean tumor volumes ± SEM) of NB1 and NB1-s cells. **P < .01 (two-way analysis of variance). (B) Detection time for subcutaneous tumors with a volume exceeding 250 mm3. (C) H&E staining of NB1 xenograft tumor and in vivo subcutaneous and orthotopic tumors derived from NB1 and NB1-s cells. (D) Cytotoxic assay testing the drug resistance of LAN-1, LAN-1 spheres (LAN-1-s), and LAN-1R cells. Cell viability was measured after 48 hours of indicated drug treatments. Bars are means ± SEM of two to three experiments. (E) Cytotoxic assay for drug resistance in NB1 and NB1-s cells. Dissociated NB1 tumor and NB1 spheres were treated for 48 hours with the indicated drugs. ***P < .001 (Student's t test).
To validate the influence of the environment on tumor growth, heterotopic implantations were repeated in the absence of Matrigel, similarly to the orthotopic procedure (104 NB1 and NB1-s cells). Surprisingly, no tumor intake was observed up to 133 days (data not shown), indicating that factors present in the Matrigel were necessary to support NB cell growth in the heterotopic site in contrast to that in orthotopic conditions.
Because TICs have been defined as cells able to recapitulate the parental tumor's heterogeneity, the histology of the original patient tumor NB1 and the subcutaneous (s.c.) and orthotopic NB1- and NB1-s-derived tumors was analyzed according to the pathologic criteria from the Shimada classification of NB tumors [41,42] (Figure 3C). The NB1 tumor histology revealed diffused growth patterns with lobular architecture and presented as small round cells with scanty cytoplasm. Typical for NB, a few Homer-Wright rosettes were present. The most similar histology was observed in the orthotopic or subcutaneous tumors. However, an undifferentiated histology and/or higher mitosis karyorrhexis index (MKI) was observed in the orthotopic tumors. By providing a favorable environment, orthotopic injections in the adrenal gland may thus highlight the tumorigenic properties of NB1-s cells while preserving the histology of the tumor.
Evidences for the capacity for chemoresistance of CSCs have recently emerged [43–45]. Therefore, we evaluated the sphere cell's sensitivity to conventional chemotherapeutic drugs in LAN-1 cell line and in NB1 tumor cells. The doxorubicin-resistant LAN-1R counterpart of the LAN-1 cell line [46] was included as a positive control for resistance. Interestingly, LAN-1-s and NB1-s displayed a significantly increased drug resistance compared with LAN-1 and NB1 cells, respectively (Figure 3, D and E).
The Transcriptome Profile of NB1-s Reveals a Stem Cell-like Phenotype
As NB1-s cells demonstrated enhanced in vivo tumorigenicity and in vitro drug resistance, an original strategy to specifically “profile” these functional TIC characteristics was chosen.
We postulated that, with serial sphere passages from NB1 sample (NB1-s0), NB1 spheres (NB1-s1 to -s4) would progressively be enriched in cells with TIC properties. Hence, the successive NB1-s passage transcriptomes (s0–s4) were identified by a time course microarray expression analysis (Figure 4A).
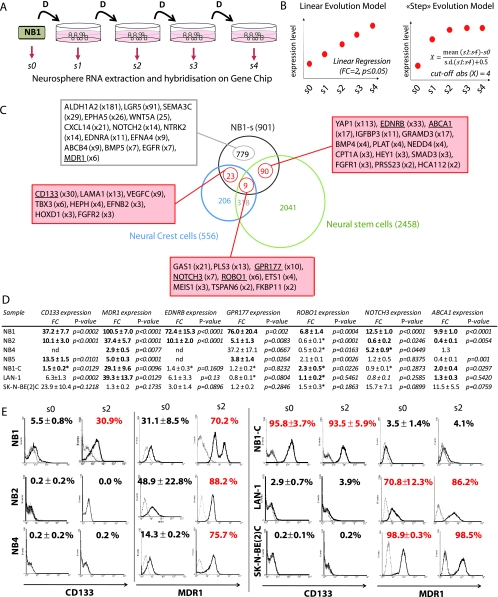
Figure 4.
Gene expression profiling of spheres derived from NB1, a high-risk tumor group. (A) Time course gene expression profiling of four sphere passages derived from NB1 (NB1-s1 to NB1-s4). (B) Graphs illustrating the two directed analyses: left indicates linear regression; right, «step-genes» analysis of the spheres' gene expression profiles. (C) Venn diagram illustrating the overlap of upregulated transcripts obtained in the NB1-s with the published genes expressed in NCCs and NSCs [18,20]. Boxes show the listed selected genes with a higher increase in spheres (fold change [FC] is indicated). The complete list of genes is available in the Supplementary Materials and Methods section. (D) Validation of the neurosphere-associated gene profile in NB samples by real-time PCR. The overexpression FC is expressed by the mean ± SEM from three independent experiments for NB1 and cell lines: two for NB2 and one for NB5. *Samples where the measured transcripts were already highly expressed in the tumor or the cell line. In three cases with “nd” (i.e., no detection), the level of the transcript extracted from the secondary spheres was too low to be detected. (E) Flow cytometry analysis of the CD133 and MDR1 cell surface expression, in NB1, NB2, and NB4 tumors (s0) and NB1-C, LAN-1 and SK-N-BE(2)C cell lines and in their derived secondary sphere (s2). Percentages of positive cells are indicated by means ± SEM of three independent measures.
The differentially expressed genes in NB1-s were monitored using two statistical approaches that reflected the two hypotheses of TIC genes enrichment in the sphere cell population. Genes with a “linear” expression evolution through s0 to s4 were selected, as well as genes showing a drastic change or “step” evolution at s1. Both scenarios allowed to in silico select genes with expression profile changes occurring with successive sphere passages (Figure 4B).
The “linear” evolution analysis performed with NB1 and NB1-s provided a list of 718 overexpressed and 540 underexpressed genes (Table W1). However, the “step” evolution analysis showed 183 over-expressed and 160 underexpressed genes. By overlapping those two results, a global list of 1601 deregulated genes in NB1-s was obtained, of which 700 were downregulated and 901 were upregulated genes (Figure 4C and Table W1).
The highest upregulated genes identified in NB1-s corresponded to the aldehyde dehydrogenase (ALDH1A2) gene and the leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5) gene, previously described as stem cell markers in human breast [47] and intestine cancers [48], respectively. Interestingly, although NB1 sample was collected before treatment, the ATP-binding cassette ABCB1 (MDR1) multidrug resistance gene showed a seven-fold overexpression in NB1-s, supporting the drug resistance of NB1-s cells shown in Figure 3E.
Finally, the NB1-s expression profile was enriched in genes involved in essential attributes of stemness, including the NOTCH, WNT, and growth hormone signaling pathways [20], as well as in neural crest development and NB tumorigenesis (Table W2).
As NB originates from sympathetic nervous system progenitors, we next postulated that NB-TICs were likely to display gene expression profiles common with sympathetic nervous system progenitors, such as NCCs and NSCs. The list of NB1-s differentially overexpressed genes was crossed with two lists of published profiles: NCC-associated genes, identified in neural crest stem cells (NCSCs, day E9.5 in the mouse embryo) and neural crest-derived progenitor cells (NCDCs, day E12), and NSC-associated genes [18,20] (Figure 4C and Table W1). The overlap generated nine genes commonly upregulated in NCC, NSC, and NB1-s, including the axon guidance receptor ROBO1 (6x); GPR177 (10x), which regulates WNT proteins sorting and secretion [49]; and NOTCH3 receptor (7x), which promotes cell self-renewal and tumor formation [50–52]. In addition, 23 genes were found commonly upregulated in NCC and NB1-s, including the CD133 (prominin-1) receptor (30x), VEGFC growth factor (9x), and FGF receptor FGFR2 (3x). Finally, a list of 90 upregulated genes was identified in both NB1-s and NSC, among which two particularly relevant genes emerged: EDNRB (33x) and ABCA1 (17x). EDNRA, receptor for endothelin 1 (EDN1), has also been found 11-fold overexpressed in NB1-s.
TIC Candidate Genes Are Overexpressed in Metastatic NB-Derived Spheres
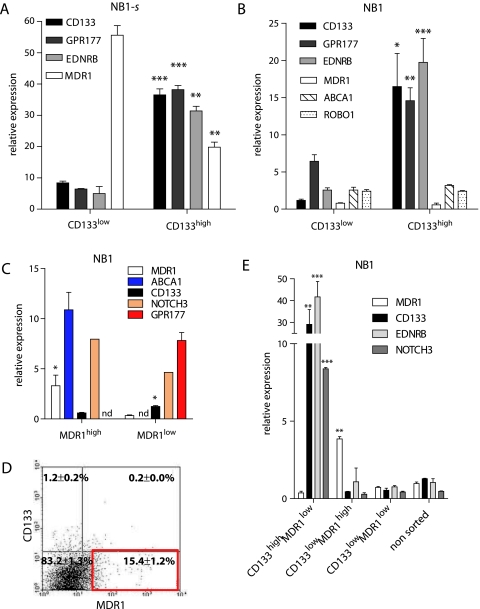
CD133, MDR1, EDNRB, GPR177, ROBO1, NOTCH3, and ABCA1 genes were selected from the overlaps between NB1 tumor sphere and NCC and NSC gene transcriptomes. Their relevance as potential TIC genes and representativeness for self-renewal was addressed. Sphere up-regulation of these genes was validated by real-time PCR and/or FACS in secondary spheres derived from NB1, NB2, NB4, and NB5 xenografts and from three NB cell lines, NB1-C, LAN-1, and SK-N-BE(2)C, demonstrating high sphere formation capacity (Figure 4D). Interestingly, CD133, MDR1, GPR177, and ROBO1 were significantly upregulated (fold change >1.5) or already highly expressed in 70% to 100% of the tested NB samples. In addition, EDNRB and NOTCH3 were highly overexpressed in NB1-s2 (72.4x and 12.5x, respectively), in NB2-s2 (EDNRB, 10.1x), and in NB4-s2 (NOTCH3, 5.2x), as well as highly expressed in NB1-C. ABCA1 overexpression was restricted to NB1-s2 (9.9x) and NB1-C spheres (2.0x). NOTCH3 and ABCA1 gene expression did not increase in NB2 and NB4 secondary spheres. Most importantly, CD133 and MDR1 were constantly increased in secondary spheres from most samples.
We therefore addressed the CD133 and MDR1 cell surface expression by FACS analysis (Figure 4E). The low or absent CD133 expression in NB2, NB4, LAN-1, and SK-N-BE(2)C cell lines as well as their derived spheres was confirmed. However, CD133 expression increased in NB1-s2 and was already very high in NB1-C and its derived spheres. MDR1 harbored a variable but important expression in NB1, NB2, and NB4, with a significant increase in the NB1-s2, NB2-s2, and NB4-s2 cells. In NB cell lines, MDR1 remained very low in NB1-C and NB1-C-s2 (3.5%–4.1%), whereas it slightly increased in LAN-1 (from 70% to 86%) and was already high in SK-N-BE(2)C (98%) and did not further increase.
These results highlighted a significant intersample heterogeneity and the influence of serum-free conditions in either induction of sphere-associated genes or selection of cells expressing these genes.
NB1-s Population Is Heterogeneous and Preexists in the Parental Tumor
NB1-s cells was identified as a population with enriched self-renewal and tumorigenic capacities and defined by a specific gene expression profile.
CD133 was characterized by a very low expression in all patient tumors analyzed and a “linear” increase evolution in the microarray analyses of NB1-s. In contrast, MDR1 showed a “step” evolution in NB1-s, suggesting that MDR1 and CD133 genes, although both increased in spheres, might not be coexpressed, revealing a potential cell heterogeneity in the sphere cell population.
To confirm this hypothesis, CD133high and CD133low- or MDR1high and MDR1low-expressing cells were selected from NB1-s by FACS. Expression of CD133, MDR1, GPR177, EDNRB, and ABCA1 was measured on sorted cells by real-time PCR (Figure 5A). CD133high population contained enriched GPR177 and EDNRB gene expression, whereas the CD133low population was clearly highly enriched in MDR1 expression. This observation confirmed the heterogeneity in the NB1-s population and revealed at least two distinct populations based on selected NB1-s markers.
Figure 5.
CD133high and MDR1high cells in the NB1 sphere cell population (NB1-s) express distinct markers. (A) Relative mRNA expression levels of CD133, GPR177, EDRNB, and MDR1 genes in CD133high- versus CD133low-sorted cells from NB1-s cells. (B) Relative mRNA expression levels of CD133, GPR177, EDRNB, MDR1, ABCA1, and ROBO1 genes in CD133high- versus CD133low-sorted cells from the dissociated NB1 xenograft tumor. (C) Relative MDR1, ABCA1, CD133, NOTCH3, and ABCA1 genes expression levels in MDR1high/low-sorted cells from the NB1 dissociated tumor. (D) MDR1 and CD133 coexpression analysis in NB1-dissociated tumor cells by flow cytometry. The percentage of CD133+MDR1-, CD133+MDR1+, and CD133-MDR1+ is indicated. (E) Relative MDR1, CD133, EDNRB, and NOTCH3 genes expression levels in the CD133highMDR1low-, CD133lowMDR1high-, and CD133lowMDR1low-sorted cells and unsorted cells from dissociated NB1 tumor. *P < .05, **P < .01, and ***P < .001, Student's t test.
To address the existence of several subpopulations in the NB1 tumor, CD133high-and CD133low-sorted cells were analyzed for CD133, MDR1, GPR177, EDNRB, ABCA1, and ROBO1 expression by real-time PCR (Figure 5B). Results show that GPR177 and EDNRB were significantly coselected within the CD133high population, whereas a very low MDR1 expression was detected in both CD133high and CD133low populations.
The reverse selection experiment was carried out in NB1 with MDR1high and MDR1low sorted populations (Figure 5C). CD133 and GPR177 were preferentially expressed in the MDR1low subset. ABCA1 was, in contrast, slightly enriched in the MDR1high subset.
When double MDR1/CD133 cell sorting of the NB1 tumor was performed, three sorted populations, CD133highMDR1low cells (1.2% ± 0.2%), CD133lowMDR1high cells (15.4% ± 1.2%), and a major population of CD133highMDR1high cells (83.2% ± 1.3%), were obtained (Figure 5D). CD133/EDNRB/NOTCH3-expressing cells were clearly coselected in the CD133high/MDR1low subset, whereas the MDR1 gene was selected in the CD133low/MDR1high population (Figure 5E). These data confirmed the anticorrelated expression of MDR1 and CD133 sphere-associated genes within the NB1 cells.
Altogether, these results supported the existence of at least two distinct cell subsets with putative TIC characteristics in NB1-s, CD133highMDR1low cells coexpressing EDNRB, GPR177, and NOTCH3 and CD133lowMDR1high cells coexpressing ABCA1, as well as their preexistence in the original NB1 sample.
CD133high and MDR1high NB Cell Populations Have Different Tumorigenic Properties
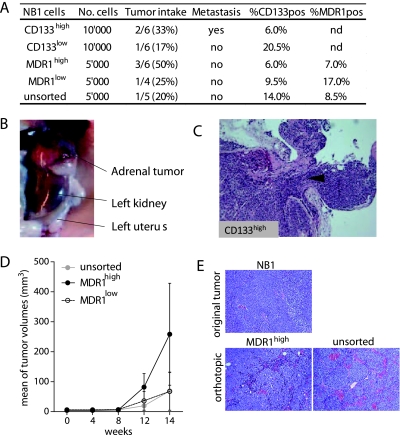
To further address the functional heterogeneity of CD133/MDR1 sorted cell populations, their in vivo tumorigenic properties were evaluated by orthotopic implantations (Figure 6).
Figure 6.
CD133high and MDR1high NB1 cells have similar tumorigenicity and showed plastic expression in orthotopic tumors. (A) In vivo tumorigenicity of CD133high/low -and MDR1high/low-sorted cells. Tumor intake is represented by the fraction and percentage of injected Swiss nude mice bearing tumors after 100 days. CD133 and MDR1 expression in dissociated mice tumors was determined by flow cytometry. nd indicates not determined. (B) Macroscopic image of a left adrenal tumor in the CD133pos group. (C) H&E staining of the NB1 CD133pos-derived orthotopic tumors showing the invasive behavior of the cells (black arrowhead and on the right) in the adrenal tissue (on the left). (D) In vivo adrenal tumor growth of unsorted MDR1high and MDR1low cells. Mean tumor volumes ± SEM are represented. (E) H&E staining of NB1 original tumor and orthotopic tumors derived from unsorted and MDR1high NB1 cells.
Tumor intake observed for CD133high and CD133low populations was not significantly different (Figure 6A). Adrenal tumors were detected in two versus one of six mice in CD133high and CD133low groups, respectively, with a contralateral ovarian metastasis in the CD133high group (Figure 6B). FACS analysis of CD133high and CD133low grown tumors revealed 6% and 21% CD133-positive cells, respectively (Figure 6A) compared with the 6% CD133-positive cells in theNB1 sample(Figure 4E), showing that CD133low tumors contained a higher number of CD133-positive cells than CD133high and NB1 tumors. These observations indicate a high plasticity of these cells in addition to their heterogeneity (Figure 6A). H&E staining of tumors grown from CD133high and CD133low populations did not reveal different histology, vascular, or differentiation patterns (Figure 6C; data not shown).
For NB1-sorted MDR1high and MDR1low populations, the follow-up revealed an increased tumor intake at 100 days after injection in the MDR1high group compared to the MDR1low and nonsorted groups, with 50% versus 25% and 20% of mice with adrenal tumors, respectively (Figure 6A). Although not significant, a faster tumor growth with MDR1high cells, with larger tumors, was observed compared with MDR1low cells and unsorted population (Figure 6D). Histologic observations have been obtained for MDR1high and unsorted cell-derived tumors (Figure 6E). The MDR1high tumors recapitulated the cellular morphology of NB1 parental cells with an undifferentiated morphology, an intermediate MKI without detectable Homer-Wright rosettes. Likewise, unsorted cells gave rise to orthotopic tumors with NB1-like undifferentiated morphology, although organized in a more lobular architecture, with an intermediate MKI. These data suggest that cells with a higher tumorigenic phenotype, compatible with TICs characteristics, are enriched within the MDR1high cell subset.
Discussion
The present report describes an original approach to identify and characterize putative TICs in a small, although well-documented panel of NB tumors. NB metastatic cells, as the most aggressive component in NB tumor progression, seemed faithful candidates to investigate self-renewing and tumor-initiating phenotypes. Thus, bone marrow-derived metastatic cells were used to generate in vivo xenografts as a continuous source of tumor tissue, further shown to recapitulate the original tumor phenotype.
Serial sphere generation, a well-established assay for self-renewal [53–55], could only be obtained from NB stage 4-derived metastatic cells, thus establishing a significant correlation between high-risk NB tumors cells and their capacity of long-term survival as spheres. However, the MYCN expression status was not related to the sphere-forming capacity of the tumor cells, whereas c-MYC seemed highly expressed in NB1 and NB2 tumor cells, which also harbored the highest passages of spheres in our panel of NB samples. The significance of this correlation should be determined on a larger panel of patients.
The sphere-forming cells (NB1-s), selected from the highly malignant NB clinical sample (NB1), survived in vitro as high passage spheres, postulating that NB1 might be highly enriched in stem-like cancer cells. NB1-s cells showed an enhanced capacity to generate orthotopic tumors in vivo with an increased tumor intake compared with NB1 total cells. This differential tumorigenicity was abolished in heterotopic conditions in the presence of Matrigel and fully lost in the absence of Matrigel, demonstrating that the adrenal gland, as a permissive microenvironment, may optimize the TIC phenotype. We therefore concluded that NB1-s population was significantly enriched in tumorigenic NB1 cells. In addition, the selective increased resistance of NB1-s cells to a panel of chemotherapeutic drugs suggests a NB-TIC role in multidrug resistance.
The self-renewing, tumorigenic, and drug-resistant characteristics of NB1-s cells were profiled by time course gene expression microarray. The analysis of gradual gene expression changes occurring through successive sphere passages identified a total of 901 upregulated genes in NB1-s. A significant part of them corresponded to essential attributes of stemness [20], neural crests development, and NB tumorigenesis.
These results corroborated the enrichment in NB1-s of cells with stem-like features. High-passage NB1-s cells and neural crest-derived stem cells indeed shared a number of upregulated genes, as shown by overlapping NB1-s-associated genes with published lists of NCSC, NSC, and NCDC genes.
Interestingly, several NB1-s genes, such as ALDH1A2, LGR5, CD133, and MDR1, were previously described as CSC markers and as key players in developmental biology processes [47,48,56]. New candidate NB-TIC markers, EDNRB and GPR177, have been identified, and their functional role in the NB-TIC “stem” features is currently investigated.
In the literature, few NB-TIC gene markers have been reported [13,16,57]. They include CSC markers, CD133, ABCG2, NES (nestin), and the neural crest marker p75 (CD271). NCSCs markers CD271 and SOX10 were expected to be expressed by NB-TICs. However, although CD271+ and SOX10+ cells were detected in undifferentiated NB tumors and NB spheres, in contrast to melanoma, another neural crest-derived tumor [58,59], CD271 surprisingly failed to select for self-renewing and tumorigenic NB cell populations in NB patient samples and cell lines (data not shown).
Expression of hematopoietic and B-cell markers CD20, CD24, and CD34, has been described on NB-TIC lines [13]. In contrast, the highly tumorigenic NB1-s cells lacked CD20, CD24, or CD34 B-cell markers (data not shown). However, the phenotype of NB-TICs in our model has been demonstrated to be clearly neuroblastic. This validation eliminates the hypothesis of a long-term culture-derived modification resulting in a bone marrow-derived TIC lines that may explain the acquisition of different markers (American Association for Cancer Research 2011 Abstract LB-366).
Seven markers (CD133, MDR1, EDNRB, GPR177, ROBO1, NOTCH3, and ABCA1) were selected from gene lists' overlaps as representative sphere-associated genes. In particular, we showed that the multidrug resistance gene MDR1, a marker associated to normal stem cells and CSCs, was uniformly increased in all spheres derived from the NB sample collection, even in the absence of any exposure to chemotherapeutic drugs. Drug resistance, an innate tumor stem cell's characteristic, ensures them a long life span [60–63]. The alternative ABC transporter ABCG2 allows selection of a so-called “side population,” a tumor cell fraction enriched in TICs [63]. However, because no SP could be identified in NB tumor or sphere samples (data not shown), increased resistance observed in NB1-s cells could likely be attributed to MDR1 gene overexpression.
Conversely, CD133 expression was also increased in NB1-s but not in every analyzed sphere derived from NB samples and cell lines. Despite unclear function, the CD133 receptor emerges as one of the first described and most frequent gene associated to TICs behavior [56,64–67]. However, the importance of CD133 as a robust stem cell marker somehow simultaneously vanished with several recent challenges on its selectivity for tumorigenic and self-renewing cells [68–72].
Intriguingly, it was not possible to select in the NB1 tumor a unique cell population coexpressing all seven markers and exclusively recapitulating the functional TIC's features. Indeed, we demonstrated an anticorrelation between the expression of CD133/EDNRB/GPR177/NOTCH3 and MDR1/ABCA1 in NB1 tumor cells and spheres, highlighting the NB1 sphere cell's heterogeneity. In essence, at least two NB1-s-selected cell populations, CD133highMDR1low and CD133lowMDR1high, could be identified in the NB1 tumor.
When assayed for tumorigenic properties in orthotopic implantations, the CD133high and CD133low NB1 subpopulations were equally tumorigenic. CD133 expression was higher in CD133low-derived tumors compared to CD133high-derived tumor, revealing its relative plasticity. Thus, CD133, although identified as a NB1 sphere marker, was not alone able to select tumorigenic NB1 cell population. This conclusion is supported by several recent data, suggesting that CD133 is not restricted to normal stem cells and CSCs and does not reliably distinguish melanoma [70] or brain tumor stem cells [71,73,74].
In contrast, the MDR1high NB1 cell subset, with a low CD133 expression, seemed to be associated to a higher in vivo tumor growth, suggesting that cells with a higher tumorigenic phenotype, compatible with TICs characteristics, are comprised within the MDR1high NB1 cell subset.
Our results suggest that NB tumorigenic cells, isolated from metastatic cell population, correspond to heterogeneous and dynamic cell subsets, whose phenotypes are strongly dependent on in vitro and in vivo conditions. Indeed, culture conditions may determine the resulting observed cell phenotype and introduce a bias in the description of stem cell characteristics, as it has been shown recently in a glioblastoma-initiating cell model [75]. The sphere-forming conditions used in our study were originally designed to sustain both stemness and plasticity (differentiation capacity) of NCCs. Hence, these conditions may temporarily favor neural crest over other stem cells' characteristics.
Moreover, the consequent in vivo plasticity of NB-TICs may be derived from the extreme cellular heterogeneity and plasticity of the NCCs from which NB originates. Heterogeneous NB cell populations are actually observed in specific and defined culture media (sphere or adherent culture media) and niches (primary tumor, subcutaneous, or orthotopic xenografts) at a given time point in the development of the original tumor. These conditions are likely to select and expand particular but not exhaustive or stable stem cell phenotypes, which may only represent a “snapshot” of the cell's phenotype and biology. It therefore addresses the question of the methodological reliability of TIC investigations in the NB model [76].
Interestingly, similar conclusions about the dynamic behavior of tumor-propagating cells have been drawn in melanoma, suggesting that embryonic-related tumors may reveal similar models of tumor development and progression [12]. Another controversial report identified at least three melanoma cell subpopulations, with heterogeneous combined CD34 and p75 expression, and harboring different self-renewal and in vivo tumorigenicity [70].
Our data suggest that the selection of NB-TIC subset on the basis of a single marker is not sufficient to isolate the NB cell population responsible for tumor initiation and sustained growth. Moreover, because of their plasticity and the influence of the environment, it is still unclear to which extent the existence of a NB-TIC “niche” is dependent on the presence of the NB-TICs themselves. Indeed, the phenotypic identity of the NB-TICs may shift as the niche changes, and vice versa, making the NB-TICs identification even more difficult.
Thus, the correlation between the gene expression profile of in vitro selected sphere-forming cells and the in vivo tumorigenic function represents a pertinent approach that addresses a malignant proliferative cell function rather than an unstable cell entity. Such strategy may reveal a more efficient procedure to target the highest malignant NB subsets for the development of targeted therapies and definitive eradication of the tumor.
Supplementary Materials and Methods
Forward and reverse primers for human sequence amplification by real-time PCR:
HPRT1: 5′-TGACACTGGCAAAACAATGCA-3′ and 5′-GGTCCTTTTCACCAGCAAGCT-3′
CD133: 5′-CATGGCAACAGCGATCAAG-3′ and 5′-AGCACAGAGGGTCATTGAG-3′
MDR1: 5′-TTCTGGGAAGAT-3′ and 5′-TATGGTACCTGCAAACTCTG-3′
EDNRB: 5′-CGAAACGGTCCCAATATC-3′ and 5′-CCAGCTTACACATCTCAG-3′
NOTCH3: 5′-AGTGGCGACCTCACTTACGACT GTGCCTGTC-3′ and 5′-GGGCACTGGCAGTTATAG-3′
GPR177: 5′-CTGGATGCTGCTGTTTGG-3′ and 5′-TACCCTGCGATGTGGTTC-3′
ROBO1: 5′-TTGCTTTGGGACGGACTG-3′ and 5′-ATCGGCTGGATGACTGTG-3′
ABCA1: 5′-GACATCCTGAAGCCAATC-3′ and 5′-AGAGTCCCAAGACTATGC-3′
Acknowledgments
The authors thank Maja Beck-Popovic, Nicolas von der Weid (Paediatric Hemato-oncology Unit, Lausanne), Hulya Ozsahin (Paediatric Hemato-oncology Unit, Geneva), and Roland Amman (Kindespital, Bern) for their collaboration in patient sample collection. The authors also thank Véronique Noguet for her valuable help in immunohistologic analyses of NB tumors. The authors thank Ivan Stamenkovic for his helpful discussions and scientific advice and acknowledge Daniel Sengstag and Sylvain Pradervand for their help in analyzing the microarray experiments.
Abbreviations
- CGH
comparative genomic hybridization
- CSC
cancer stem cell
- NB
neuroblastoma
- NCC
neural crest cell
- NCDC
neural crest-derived progenitor cell
- NCSC
neural crest stem cell
- NSC
neural stem cell
- TIC
tumor-initiating cell
Footnotes
This work was supported by grants from the Swiss National Scientific Foundation, from FORCE Foundation for Children Cancer Research (grant “Loterie Romande” to N.G.), and from the Swiss Cancer League (to L.S.). The authors indicate no potential conflicts of interest.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figure W1 and are available online at www.neoplasia.com.
References
- 1.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 2.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 3.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edsjo A, Holmquist L, Pahlman S. Neuroblastoma as an experimental model for neuronal differentiation and hypoxia-induced tumor cell dedifferentiation. Semin Cancer Biol. 2007;17:248–256. doi: 10.1016/j.semcancer.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Al-Hajj M, Wicha MS, Ito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 8.Dirks P. Cancer stem cells: invitation to a second round. Nature. 2010;466:40–41. doi: 10.1038/466040a. [DOI] [PubMed] [Google Scholar]
- 9.Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 10.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quintana E, Shackleton M, Foster HR, Fullen DR, Sabel MS, Johnson TM, Morrison SJ. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18:510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansford LM, McKee AE, Zhang L, George RE, Gerstle JT, Thorner PS, Smith KM, Look AT, Yeger H, Miller FD, et al. Neuroblastoma cells isolated from bone marrow metastases contain a naturally enriched tumor-initiating cell. Cancer Res. 2007;67:11234–11243. doi: 10.1158/0008-5472.CAN-07-0718. [DOI] [PubMed] [Google Scholar]
- 14.Smith KM, Datti A, Fujitani M, Grinshtein N, Zhang L, Morozova O, Blakely KM, Rotenberg SA, Hansford LM, Miller FD, et al. Selective targeting of neuroblastoma tumour-initiating cells by compounds identified in stem cell-based small molecule screens. EMBO Mol Med. 2010;2:371–384. doi: 10.1002/emmm.201000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das B, Tsuchida R, Malkin D, Koren G, Baruchel S, Yeger H. Hypoxia enhances tumor stemness by increasing the invasive and tumorigenic side population fraction. Stem Cells. 2008;26:1818–1830. doi: 10.1634/stemcells.2007-0724. [DOI] [PubMed] [Google Scholar]
- 16.Mahller YY, Williams JP, Baird WH, Mitton B, Grossheim J, Saeki Y, Cancelas JA, Ratner N, Cripe TP. Neuroblastoma cell lines contain pluripotent tumor initiating cells that are susceptible to a targeted oncolytic virus. PLoS One. 2009;4:e4235. doi: 10.1371/journal.pone.0004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross RA, Spengler BA, Domenech C, Porubcin M, Rettig WJ, Biedler JL. Human neuroblastoma I-type cells are malignant neural crest stem cells. Cell Growth Differ. 1995;6:449–456. [PubMed] [Google Scholar]
- 18.Buchstaller J, Sommer L, Bodmer M, Hoffmann R, Suter U, Mantei N. Efficient isolation and gene expression profiling of small numbers of neural crest stem cells and developing Schwann cells. J Neurosci. 2004;24:2357–2365. doi: 10.1523/JNEUROSCI.4083-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morozova O, Vojvodic M, Grinshtein N, Hansford LM, Blakely KM, Maslova A, Hirst M, Cezard T, Morin RD, Moore R, et al. System-level analysis of neuroblastoma tumor-initiating cells implicates AURKB as a novel drug target for neuroblastoma. Clin Cancer Res. 2010;16:4572–4582. doi: 10.1158/1078-0432.CCR-10-0627. [DOI] [PubMed] [Google Scholar]
- 20.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 22.Schleiermacher G, Janoueix-Lerosey I, Ribeiro A, Klijanienko J, Couturier J, Pierron G, Mosseri V, Valent A, Auger N, Plantaz D, et al. Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol. 2010;28:3122–3130. doi: 10.1200/JCO.2009.26.7955. [DOI] [PubMed] [Google Scholar]
- 23.Valsesia A, Rimoldi D, Martinet D, Ibberson M, Benaglio P, Quadroni M, Waridel P, Gaillard M, Pidoux M, Rapin B, et al. Network-guided analysis of genes with altered somatic copy number and gene expression reveals pathways commonly perturbed in metastatic melanoma. PLoS One. 2011;6:e18369. doi: 10.1371/journal.pone.0018369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flahaut M, Meier R, Coulon A, Nardou KA, Niggli FK, Martinet D, Beckmann JS, Joseph JM, Muhlethaler-Mottet A, Gross N. The Wnt receptor FZD1 mediates chemoresistance in neuroblastoma through activation of the Wnt/β-catenin pathway. Oncogene. 2009;28:2245–2256. doi: 10.1038/onc.2009.80. [DOI] [PubMed] [Google Scholar]
- 25.Joseph JM, Gross N, Lassau N, Rouffiac V, Opolon P, Laudani L, Auderset K, Geay JF, Muhlethaler-Mottet A, Vassal G. In vivo echographic evidence of tumoral vascularization and microenvironment interactions in metastatic orthotopic human neuroblastoma xenografts. Int J Cancer. 2005;113:881–890. doi: 10.1002/ijc.20681. [DOI] [PubMed] [Google Scholar]
- 26.Seeger RC, Rayner SA, Banerjee A, Chung H, Laug WE, Neustein HB, Benedict WF. Morphology, growth, chromosomal pattern and fibrinolytic activity of two new human neuroblastoma cell lines. Cancer Res. 1977;37:1364–1371. [PubMed] [Google Scholar]
- 27.Ciccarone V, Spengler BA, Meyers MB, Biedler JL, Ross RA. Phenotypic diversification in human neuroblastoma cells: expression of distinct neural crest lineages. Cancer Res. 1989;49:219–225. [PubMed] [Google Scholar]
- 28.Fuchs S, Herzog D, Sumara G, Buchmann-Moller S, Civenni G, Wu X, Chrostek-Grashoff A, Suter U, Ricci R, Relvas JB, et al. Stage-specific control of neural crest stem cell proliferation by the small rho GTPases Cdc42 and Rac1. Cell Stem Cell. 2009;4:236–247. doi: 10.1016/j.stem.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 29.Stemple DL, Anderson DJ. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell. 1992;71:973–985. doi: 10.1016/0092-8674(92)90393-q. [DOI] [PubMed] [Google Scholar]
- 30.Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N. Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2000;60:4315–4319. [PubMed] [Google Scholar]
- 31.Ikegaki N, Kennett RH. Glutaraldehyde fixation of the primary antibody-antigen complex on nitrocellulose paper increases the overall sensitivity of immunoblot assay. J Immunol Methods. 1989;124:205–210. doi: 10.1016/0022-1759(89)90354-2. [DOI] [PubMed] [Google Scholar]
- 32.Ikegaki N, Kennett RH. Molecular genetic characterization of epitopespecific monoclonal antibodies against the myc family proteins. Oncogene. 1990;5:397–403. [PubMed] [Google Scholar]
- 33.Psarros M, Heber S, Sick M, Thoppae G, Harshman K, Sick B. RACE: Remote Analysis Computation for gene Expression data. Nucleic Acids Res. 2005;33:W638–W643. doi: 10.1093/nar/gki490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier R, Muhlethaler-Mottet A, Flahaut M, Coulon A, Fusco C, Louache F, Auderset K, Bourloud KB, Daudigeos E, Ruegg C, et al. The chemokine receptor CXCR4 strongly promotes neuroblastoma primary tumour and metastatic growth, but not invasion. PLoS One. 2007;2:e1016. doi: 10.1371/journal.pone.0001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gross N, Beck D, Portoukalian J, Favre S, Carrel S. New anti-GD2 monoclonal antibodies produced from gamma-interferon-treated neuroblastoma cells. Int J Cancer. 1989;43:665–671. doi: 10.1002/ijc.2910430421. [DOI] [PubMed] [Google Scholar]
- 37.Janoueix-Lerosey I, Schleiermacher G, Michels E, Mosseri V, Ribeiro A, Lequin D, Vermeulen J, Couturier J, Peuchmaur M, Valent A, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–1033. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 38.Gross N, Beck D, Portoukalian J, Carrel S, Favre S. Monoclonal antibodies to gamma-interferon treated LAN-1 cells detect modulation of ganglioside GD2 exposure on human neuroblastoma cells. Anticancer Res. 1989;9:1519–1524. [PubMed] [Google Scholar]
- 39.Weinstein JL, Katzenstein HM, Cohn SL. Advances in the diagnosis and treatment of neuroblastoma. Oncologist. 2003;8:278–292. doi: 10.1634/theoncologist.8-3-278. [DOI] [PubMed] [Google Scholar]
- 40.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 41.Joshi VV. Peripheral neuroblastic tumors: pathologic classification based on recommendations of International Neuroblastoma Pathology Committee (modification of Shimada classification) Pediatr Dev Pathol. 2000;3:184–199. doi: 10.1007/s100240050024. [DOI] [PubMed] [Google Scholar]
- 42.Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B, Stram DO, Gerbing RB, Lukens JN, Matthay KK, et al. The International Neuroblastoma Pathology Classification (the Shimada system) Cancer. 1999;86:364–372. [PubMed] [Google Scholar]
- 43.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 45.Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flahaut M, Muhlethaler-Mottet A, Martinet D, Fattet S, Bourloud KB, Auderset K, Meier R, Schmutz NB, Delattre O, Joseph JM, et al. Molecular cytogenetic characterization of doxorubicin-resistant neuroblastoma cell lines: evidence that acquired multidrug resistance results from a unique large amplification of the 7q21 region. Genes Chromosomes Cancer. 2006;45:495–508. doi: 10.1002/gcc.20312. [DOI] [PubMed] [Google Scholar]
- 47.Alison MR, Guppy NJ, Lim SM, Nicholson LJ. Finding cancer stem cells: are aldehyde dehydrogenases fit for purpose? J Pathol. 2010;222:335–344. doi: 10.1002/path.2772. [DOI] [PubMed] [Google Scholar]
- 48.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 49.Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125:509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 50.Dang L, Yoon K, Wang M, Gaiano N. Notch3 signaling promotes radial glial/progenitor character in the mammalian telencephalon. Dev Neurosci. 2006;28:58–69. doi: 10.1159/000090753. [DOI] [PubMed] [Google Scholar]
- 51.Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, Santini D, Ceccarelli C, Chieco P, Bonafe M. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, Shimizu K, Honma R, Yanagisawa Y, Nishikawa A, et al. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res. 2008;68:1881–1888. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- 53.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 54.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizrak D, Brittan M, Alison MR. CD133: molecule of the moment. J Pathol. 2008;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 57.Walton JD, Kattan DR, Thomas SK, Spengler BA, Guo HF, Biedler JL, Cheung NK, Ross RA. Characteristics of stem cells from human neuroblastoma cell lines and in tumors. Neoplasia. 2004;6:838–845. doi: 10.1593/neo.04310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boiko AD, Razorenova OV, van de Rijn M, Swetter SM, Johnson DL, Ly DP, Butler PD, Yang GP, Joshua B, Kaplan MJ, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Civenni G, Walter A, Kobert N, Mihic-Probst D, Zipser M, Belloni B, Seifert B, Moch H, Dummer R, van den Broek M, et al. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Cancer Res. 2011;71:3098–3109. doi: 10.1158/0008-5472.CAN-10-3997. [DOI] [PubMed] [Google Scholar]
- 60.Glavinas H, Krajcsi P, Cserepes J, Sarkadi B. The role of ABC transporters in drug resistance, metabolism and toxicity. Curr Drug Deliv. 2004;1:27–42. doi: 10.2174/1567201043480036. [DOI] [PubMed] [Google Scholar]
- 61.Donnenberg VS, Donnenberg AD. Multiple drug resistance in cancer revisited: the cancer stem cell hypothesis. J Clin Pharmacol. 2005;45:872–877. doi: 10.1177/0091270005276905. [DOI] [PubMed] [Google Scholar]
- 62.Kim M, Turnquist H, Jackson J, Sgagias M, Yan Y, Gong M, Dean M, Sharp JG, Cowan K. The multidrug resistance transporter ABCG2 (breast cancer resistance protein 1) effluxes Hoechst 33342 and is overexpressed in hematopoietic stem cells. Clin Cancer Res. 2002;8:22–28. [PubMed] [Google Scholar]
- 63.Scharenberg CW, Harkey MA, Torok-Storb B. The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood. 2002;99:507–512. doi: 10.1182/blood.v99.2.507. [DOI] [PubMed] [Google Scholar]
- 64.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 66.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De MR. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 67.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 68.Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 69.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 70.Held MA, Curley DP, Dankort D, McMahon M, Muthusamy V, Bosenberg MW. Characterization of melanoma cells capable of propagating tumors from a single cell. Cancer Res. 2010;70:388–397. doi: 10.1158/0008-5472.CAN-09-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ogden AT, Waziri AE, Lochhead RA, Fusco D, Lopez K, Ellis JA, Kang J, Assanah M, McKhann GM, Sisti MB, et al. Identification of A2B5+C. Neurosurgery. 2008;62:505–514. doi: 10.1227/01.neu.0000316019.28421.95. [DOI] [PubMed] [Google Scholar]
- 72.Wu Y, Wu PY. CD133 as a marker for cancer stem cells: progresses and concerns. Stem Cells Dev. 2009;18:1127–1134. doi: 10.1089/scd.2008.0338. [DOI] [PubMed] [Google Scholar]
- 73.Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 74.Joo KM, Kim SY, Jin X, Song SY, Kong DS, Lee JI, Jeon JW, Kim MH, Kang BG, Jung Y, et al. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 2008;88:808–815. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- 75.Jiang Y, Boije M, Westermark B, Uhrbom L. PDGF-B can sustain self-renewal and tumorigenicity of experimental glioma-derived cancer-initiating cells by preventing oligodendrocyte differentiation. Neoplasia. 2011;13:492–503. doi: 10.1593/neo.11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 77.Thiele CJ. Masters JRW, Palsson B. Human Cell Culture. Kluwer Academic Publishers; 1999. Neuroblastoma; pp. 21–53. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.