Abstract
During metazoan development, the Wnt/Wingless signal transduction pathway is activated repetitively to direct cell proliferation, fate specification, differentiation and apoptosis. Distinct outcomes are elicited by Wnt stimulation in different cellular contexts; however, mechanisms that confer context specificity to Wnt signaling responses remain largely unknown. Starting with an unbiased forward genetic screen in Drosophila, we recently uncovered a novel mechanism by which the cell-specific co-factor Earthbound 1 (Ebd1), and its human homolog jerky, promote interaction between the Wnt pathway transcriptional co-activators β-catenin/Armadillo and TCF to facilitate context-dependent Wnt signaling responses. Here, through the same genetic screen, we find an unanticipated requirement for Erect Wing (Ewg), the fly homolog of the human sequence-specific DNA-binding transcriptional activator nuclear respiratory factor 1 (NRF1), in promoting contextual regulation of Wingless signaling. Ewg and Ebd1 functionally interact with the Armadillo-TCF complex and mediate the same context-dependent Wingless signaling responses. In addition, Ewg and Ebd1 have similar cell-specific expression profiles, bind to each other directly and also associate with chromatin at shared genomic sites. Furthermore, recruitment of Ebd1 to chromatin is abolished in the absence of Ewg. Our findings provide in vivo evidence that recruitment of a cell-specific co-factor complex to specific chromatin sites, coupled with its ability to facilitate Armadillo-TCF interaction and transcriptional activity, promotes contextual regulation of Wnt/Wingless signaling responses.
Keywords: β-Catenin, Context specificity, Earthbound, Erect wing, TCF, Wnt/Wingless, Drosophila
INTRODUCTION
The evolutionarily conserved Wnt/Wingless signal transduction pathway is used repetitively during animal development to regulate cell proliferation, fate determination, differentiation and cell death; deregulation of this pathway is associated with multiple congenital disorders and types of cancer (Clevers, 2006; MacDonald et al., 2009). In response to interaction between Wnt ligands and their transmembrane receptors, the activity of a complex that includes Axin, Adenomatous Polyposis Coli (APC) and glycogen synthase kinase 3 is attenuated, thereby reducing degradation of β-catenin/Armadillo, a key Wnt pathway transcriptional co-activator. Stabilized β-catenin accumulates in cytoplasm and translocates into nucleus, where it associates with DNA-binding transcription factors of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family to activate Wnt pathway target genes (MacDonald et al., 2009).
Diverse target genes are activated upon Wnt stimulation in distinct developmental and cellular contexts, whereas universally induced Wnt target genes are relatively rare (Logan and Nusse, 2004; Clevers, 2006). As the β-catenin-TCF complex is expressed ubiquitously, how does Wnt signaling elicit distinct outcomes in different cells or in the same cell at different stages of differentiation? Extensive efforts during the past three decades have identified core Wnt pathway components that mediate signaling in all cells; yet how cell identity modulates specific Wnt signaling outcomes remains largely unknown.
To identify new factors that promote Wnt signaling, we performed unbiased forward genetic screens in Drosophila, anticipating that we might identify core Wnt pathway components as well as cell-specific factors that confer context-specificity to Wnt signaling responses. Indeed, we identified alleles of the well-characterized Wnt pathway transcriptional co-activator Legless/BCL9 (Kramps et al., 2002; Benchabane et al., 2011), as well as alleles of Apc that revealed an unexpected positive regulatory role (Takacs et al., 2008). Furthermore, we recently identified a novel cell-specific factor, Earthbound 1 (Ebd1), that functions as a context-specific regulator of Wingless signaling responses (Benchabane et al., 2011). We found that Ebd1 and its human homolog jerky, which contain centromere binding protein B (CENPB)-type DNA-binding domains, physically associate with and bridge β-catenin/Armadillo and TCF, thereby promoting stability of the β-catenin-TCF complex and recruitment of β-catenin to chromatin (Benchabane et al., 2011). These findings uncovered a novel mechanism by which cell-specific adaptors facilitate interaction between β-catenin/Armadillo and TCF to promote contextual regulation of Wnt signaling responses.
Here, through the same genetic screen, we identify an unanticipated role for the sequence-specific DNA binding transcription factor Erect Wing (Ewg), the fly homolog of human nuclear respiratory factor 1 (NRF1), in the context-specific regulation of Wingless signaling responses. Unexpectedly, Ewg mediates the same subset of Wingless-dependent developmental processes that require Ebd1. Our characterization of Ewg function sheds light on a novel mechanism in which a cell-specific co-factor complex associates with both the core Wingless pathway transcription complex and with distinct chromatin sites to facilitate contextual regulation of Wingless signaling.
MATERIALS AND METHODS
Fly stocks
ewgP1 was isolated in an ethyl methanesulfonate-based mutagenesis screen (Lewis and Bacher, 1968) for dominant suppressors of photoreceptor apoptosis in Apc1Q8 mutants. Other lines used were ewg1, ewg2, elav-ewg (DeSimone and White, 1993), Apc1Q8 (Ahmed et al., 1998), GMR-p35 (Hay et al., 1994), UAS-TCFDN (van de Wetering et al., 1997) and ebd1-Gal4 (Benchabane et al., 2011). Canton S flies were used as wild-type controls. Fly crosses were performed at 25°C unless otherwise indicated.
Mosaic clone analysis
To generate ewg2 germline clones, ewg2 FRT101 was crossed to ovoD FRT101; hs-flp flies. First and second instar larvae were heat shocked at 37°C for 2 hours and female progeny were crossed to ewg2 mutant males carrying duplication Dp(1;Y)y261l. ewg2 maternal/zygotic mutant embryos were recognized by lack of Ewg staining.
To generate ewg-null mutant eye clones, ewg2 FRT19A flies were crossed to arm-lacZ FRT19A; ey-flp; retinas from progeny were fixed at 35 to 40 hours APF and stained with Homothorax or cleaved Caspase 3 antibody. ewg mutant clones were identified by staining with Ewg antiserum or β-Gal antibody.
Plasmids
ewg cDNA was provided Kalpana White (DeSimone et al., 1996). For bacterial expression, ewg cDNA was cloned into the EcoRI sites of pGST-Parallel1 (Sheffield et al., 1999) to produce GST-Ewg. For expression in cultured cells, ewg cDNA was cloned into the EcoRI sites of pFLAG-CMV-5a (Sigma). pProEx-Ebd1 and pCMV5-Flag-Ebd1 have been described previously (Benchabane et al., 2011). cDNAs for human NRF1 (IMAGE clone 4397528) were obtained from Open Biosystems. For transgenic expression of NRF1 in flies, a V5 epitope tag (GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACG) was added to the N terminus of NRF1, and V5-tagged NRF1 was cloned into the pUAST vector (Brand and Perrimon, 1993) between XhoI and XbaI sites.
Immunohistochemistry
Embryos, larval imaginal discs, pupal retinas and pupal flight muscles were fixed and stained as described previously (Rothwell and Sullivan, 2000; Fernandes et al., 2005; Takacs et al., 2008). Primary antibodies used for immunostaining were: guinea pig anti-Ebd1, 1:4000 (Benchabane et al., 2011); rabbit anti-Ewg, 1:1000 (DeSimone and White, 1993); mouse (9F8A9) or rat (7E8A10) anti-Elav, 1:10 (DSHB) (O’Neill et al., 1994); mouse 22C10, 1:50 (DSHB) (Hummel et al., 2000), mouse anti-FasII, 1:20 (DSHB) (Grenningloh et al., 1991), mouse BP102, 1:40 (DSHB) (Seeger et al., 1993); mouse anti-Eve, 1:10 (DSHB); rabbit anti-dMef2, 1:800 (Bour et al., 1995); guinea pig anti-Homothorax, 1:500 (Abu-Shaar et al., 1999); rabbit anti-Homothorax, 1:1000 (Pai et al., 1998); and rabbit anti-cleaved Caspase 3, 1:1000 (Cell Signaling). Secondary antibodies used for immunostaining were goat or donkey Alexa Fluor 488 or 568 conjugates at 1:200 to 1:400 (Molecular Probes), and goat or donkey Cy5 conjugates at 1:200 (Jackson Immunochemicals).
Larval salivary gland polytene chromosomes were prepared as described by Christine James, Paul Badenhorst and Bryan Turner (University of Birmingham, UK) (http://www.epigenome-noe.net/WWW/researchtools/protocol.php?protid=1), with the following modifications. Briefly, salivary glands were fixed in 4% formaldehyde, 1% Triton X-100 for 30 seconds, followed by 45% acetic acid, 4% formaldehyde, 2.5% lactic acid for 2 minutes, and spread on glass slides under borosilicate cover slips using the graphite end of a pencil. Slides were blocked with 3% BSA and 0.1% Triton X-100 in PBS prior to staining. Cytological subdivisions in polytene chromosomes were mapped with reference to the DAPI staining pattern (Ashburner, 1972; Sorsa, 1988).
Confocal images shown in Fig. 5D-K, Fig. 6A-C, and supplementary material Fig. S3 and Fig. S7 were obtained with a Nikon A1 confocal microscope. Other fluorescent polytene chromosomes images were obtained on a Zeiss Axioskop 2 microscope. Fluorescent images of other tissues were obtained on a Leica TCS SP UV confocal microscope.
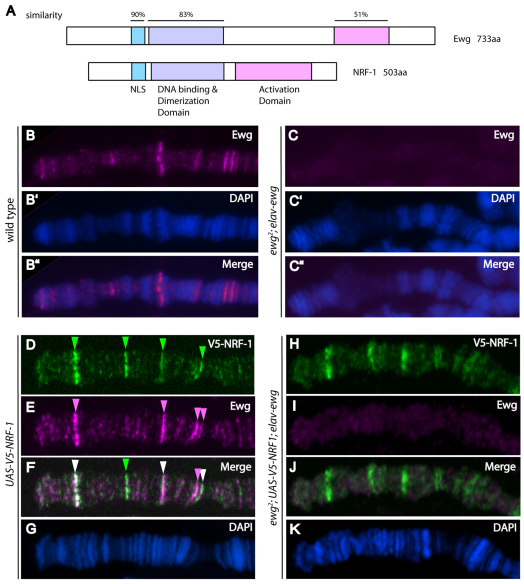
Fig. 5.
Ewg and human nuclear respiratory factor 1 share chromatin association sites in vivo. (A) Domains conserved between Ewg and mouse NRF-1 (Fazio et al., 2001). Domains in Ewg and NRF1 were aligned individually using ClustalW2. (B-C″ ) Squashes of larval third instar salivary gland polytene chromosomes, immunostained with Ewg antiserum (B,C). DNA is highlighted by DAPI staining (B’,C’). Merged images are shown in B″ and C″ . (B-B″ ) Wild type, distal tip of chromosome X. Ewg is detected on polytene chromosomes at discrete bands (B). (C-C″ ) Distal tip of chromosome X in ewg2; elav-ewg mutants. Ewg is not detected on polytenes in ewg2; elav-ewg mutant salivary glands (C). (D-K) Squashes of polytene chromosomes from third instar larvae, UAS-V5-NRF-1 (D-G) and ewg2/Y; elav-ewg/UAS-V5-NRF-1 (H-K), immunostained with V5 antibody (D,H) and Ewg antiserum (E,I). Merged images are shown in F,J. DNA is highlighted by DAPI staining (G,K). V5-NRF-1 expression results from leaky expression of the UAS-V5-NRF-1 transgene. Distal tip of chromosome 3L is shown (D-G). NRF1 retains ability to associate with chromatin (H), despite the absence of Ewg (I).
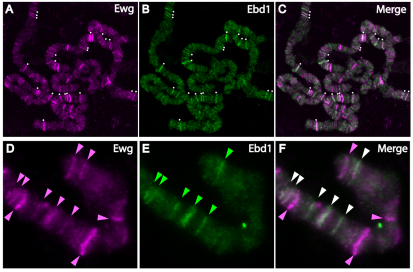
Fig. 6.
Ebd1 shares chromatin-association sites with Ewg. (A-F) Squashes of polytene chromosomes from third instar larvae, immunostained with Ewg antiserum (A,D) and Ebd1 antiserum (B,E). Merged images are shown in C and F. (A-C) Ewg and Ebd1 overlap at many sites throughout the genome. White dots indicate chromatin bands at which Ewg and Ebd1 colocalize. (D-F) A fragment from distal end of chromosome X. Ewg (pink arrowheads) and Ebd1 (green arrowheads) are both detected at discrete bands on polytene chromosomes, and colocalize at six bands (white arrowheads in F). Ewg is associated with four additional bands at which no Ebd1 is detected (pink arrowheads in F).
Histology
Adult eyes and thoraces were fixed as described previously (Cagan and Ready, 1989; Wolff, 2000; Fernandes et al., 2005), embedded in plastic resin (Durcupan, Fluka), sectioned to 1 μm and 5 μm, respectively, and stained with Toluidine Blue.
Cell lines and transfection
HEK293T cells were cultured in Dulbecco’s modified essential medium, supplemented with 10% fetal bovine serum and transiently transfected using calcium phosphate-DNA precipitation (Hayashi et al., 1997).
Immunoprecipitation and GST interaction assays
For immunoprecipitations, calcium phosphate-transfected HEK293T cells were lysed 48 hours post-transfection in lysis buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100] containing protease and phosphatase inhibitors cocktail (Thermo Scientific). Cell extracts were immunoprecipitated with anti-FLAG M2 antibody (Sigma) and collected using Protein-A/G PLUS-Agarose beads (Santa Cruz Biotechnology). Immunoprecipitates were separated by SDS-PAGE and immunoblotted with anti-V5 (Invitrogen) and anti-Flag antibodies.
For GST interaction assays, His-Ebd1 was purified from bacterial lysates using TALON metal affinity resin (Clontech). His fusion proteins were eluted off the resin using 150 mM imidazole. GST-Ewg proteins were purified using Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) and incubated with His-Ebd1. Beads were washed in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100 and associated proteins were detected by immunoblotting with anti-Ebd1 antibody.
RESULTS
Identification of Erect Wing in a forward genetic screen for factors that promote Wingless signaling
During metamorphosis, Wingless signaling induces apoptosis of photoreceptors at the Drosophila retinal periphery, thereby selectively eliminating excess neurons (Tomlinson, 2003; Lin et al., 2004). Upon loss of Adenomatous polyposis coli 1 (Apc1), constitutive Wingless pathway activation results in apoptosis of all retinal photoreceptors (Ahmed et al., 1998). This Apc1 mutant phenotype provides a sensitive assay for Wingless pathway activity, as reducing the gene dose of armadillo, TCF or the Wingless pathway transcriptional co-activator legless, by one half is sufficient to suppress the apoptosis (Ahmed et al., 1998; Benchabane et al., 2008). Therefore, to identify components that promote Wingless pathway activation, we performed genetic screens for suppressors of retinal apoptosis in the Apc1 mutant (Takacs et al., 2008; Benchabane et al., 2011).
As noted above, we recently identified Earthbound 1 (Ebd1), a novel member in a protein family containing Centromere Binding Protein B (CENPB)-type DNA binding domains (Masumoto et al., 1989) in this genetic screen (Benchabane et al., 2011). Ebd1 is expressed primarily in myocytes and neurons, and is required specifically for Wingless-dependent flight muscle development. Ebd1, and its human homolog Jerky, function as cell-specific adaptors that interact with both β-catenin/Armadillo and TCF to facilitate formation and promote activity of this core Wnt/Wingless pathway transcription complex.
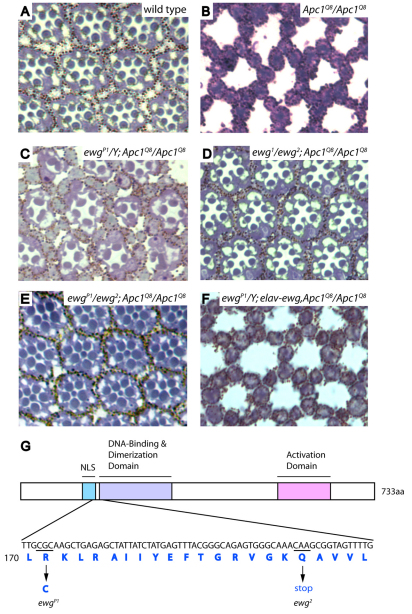
From the same genetic screen for factors that promote Wingless signaling, we identified an allele on the X chromosome that we termed P1. In Apc1 mutants, partial suppression of photoreceptor apoptosis is observed in hemizygous P1 mutant males and homozygous P1 mutant females (Fig. 1C; supplementary material Fig. S1A). Meiotic recombination revealed that P1 maps to the distal tip of the X chromosome, and is closely linked to yellow. Among genes that also map to the X chromosome tip is erect wing (ewg), the fly homolog of human nuclear respiratory factor 1 (NRF1), a sequence-specific DNA binding transcriptional activator (Fleming et al., 1982; Fazio et al., 2001; Virbasius et al., 1993). We observed that when exposed to carbon dioxide anesthesia, P1 mutant flies hold their wings in an upright or ‘erect wing’ position, and therefore we sought to determine whether P1 is an ewg mutant allele, and whether two previously isolated mutations in ewg, the null allele ewg2 and the hypomorphic allele ewg1 (Fleming et al., 1982), also suppress Apc1 mutant apoptosis. Transheterozygotes of ewg2 and ewg1 markedly suppress photoreceptor apoptosis in Apc1 mutants (Fig. 1D), as do transheterozygotes of ewg2 and P1 (Fig. 1E), suggesting that P1 is an ewg loss-of-function allele. Furthermore, expression of a transgene encoding Ewg, driven by the neuronal-specific elav promoter, restores apoptosis in P1; Apc1 double mutants (Fig. 1F), whereas expression of this ewg transgene in wild-type photoreceptors has no effect (supplementary material Fig. S1B), confirming identification of the correct gene. We sequenced the ewg coding region in ewgP1 and detected a C-to-T point mutation that results in a substitution of arginine to cysteine at amino acid 171, a residue in a region that is highly conserved in human NRF1 (Fig. 1G). Together, these data indicate that Ewg is required for Wingless pathway-dependent photoreceptor apoptosis resulting from Apc1 loss.
Fig. 1.
Ewg is required for photoreceptor apoptosis in response to Apc1 loss. (A-F) Cross-sections through adult retinas. (A) Eight photoreceptors are present in each ommatidium with seven photoreceptors visible in this section, arrayed in a trapezoidal pattern. (B) Upon Apc1 loss, all photoreceptors undergo apoptosis. The pigment cell lattice remains intact. (C-E) Photoreceptor apoptosis is prevented in hemizygous ewgP1 (C), transheterozygous ewg1/ewg2 (D) and ewgP1/ewg2 mutants (E). (F) Exogenous ewg expression driven by the elav promoter restores apoptosis in ewgP1; Apc1Q8 double mutants. (G) Schematic representation of the Ewg protein. A C-to-T substitution is found in ewgP1, resulting in an arginine to cysteine substitution in amino acid 171. In ewg2, an early stop codon at amino acid 178 is caused by substitution of C to T; no mutation was detected in the ewg-coding region in the ewg1 allele (G. Gill, personal communication).
Ewg promotes Wingless signal transduction
To determine whether Ewg solely facilitates photoreceptor apoptosis, or also promotes other Wingless pathway-dependent processes, we analyzed additional in vivo readouts. Prior to their apoptotic death, Apc1 mutant photoreceptors display two other defects caused by constitutive activation of Wingless signaling: a reduction of photoreceptor length and a misspecification of photoreceptor fate (Ahmed et al., 1998; Benchabane et al., 2008); high level Wingless signaling is required for photoreceptor apoptosis, whereas intermediate level Wingless signaling is sufficient to induce shortened photoreceptor length and aberrant photoreceptor fate specification (Tomlinson, 2003; Benchabane et al., 2008). Reduction of the Ewg level compromises the level of Wingless signaling in Apc1 mutant retinas. A partial reduction of Ewg activity, as found in ewg1 homozygotes, reduces Wingless pathway-dependent photoreceptor apoptosis (supplementary material Fig. S2E,F); further reduction of Ewg levels, as found in transheterozygotes of the ewg2 null allele and the ewg1 hypomorphic allele, or in transheterozygotes of ewg2 and ewgP1, prevents both Wingless pathway-dependent apoptosis and shortening of photoreceptor length (Fig. S2G,H; data not shown). Together, these findings indicate that Ewg facilitates Wingless signaling.
In addition, we examined whether Ewg is also required for mis-specification of photoreceptor fate that results from constitutive Wingless pathway activation upon Apc1 loss. In wild-type flies, Wingless signaling specifies inner photoreceptors restricted to the dorsal retinal periphery, termed the dorsal rim area (DRA), to adopt a fate that permits polarized light perception, instead of color light perception (Tomlinson, 2003; Wernet et al., 2003) (Fig. 2A,D). Intermediate level Wingless signaling caused by Apc1 loss results in aberrant specification of DRA fate in all inner photoreceptors in the dorsal half of the retina, as evidenced by their enlarged rhabdomeres and expression of the transcription factor Homothorax (Hth) (Tomlinson, 2003; Wernet et al., 2003; Benchabane et al., 2008; Takacs et al., 2008) (Fig. 2B). Attenuation of Ewg activity reduces the number of inner photoreceptors adopting a DRA fate in the Apc1 mutant, as revealed by wild-type rhabdomere size and loss of ectopic Hth expression. We observe partial suppression of DRA fate upon partial reduction of Ewg levels, as found in transheterozygotes of ewgP1 and ewg2 (Fig. 2C,E-G), and nearly complete suppression upon further reduction of Ewg levels, as found in transheterozygotes of ewg1 and ewg2 (Fig. 2H). These findings provide further evidence that Ewg facilitates Wingless signaling. To determine whether Ewg is required for Wingless signaling at the retinal periphery, we generated ewg-null mutant clones in otherwise wild-type pupal retinas. We analyzed photoreceptor DRA fate specification as revealed by Homothorax expression, and photoreceptors apoptosis, as revealed by cleaved Caspase 3 (Wernet et al., 2003; Lin et al., 2004; Yu et al., 2002). We detected no defects in DRA fate determination or photoreceptors death upon the loss of Ewg (data not shown), suggesting that Ewg is dispensable for these two Wingless-dependent processes during retinal development.
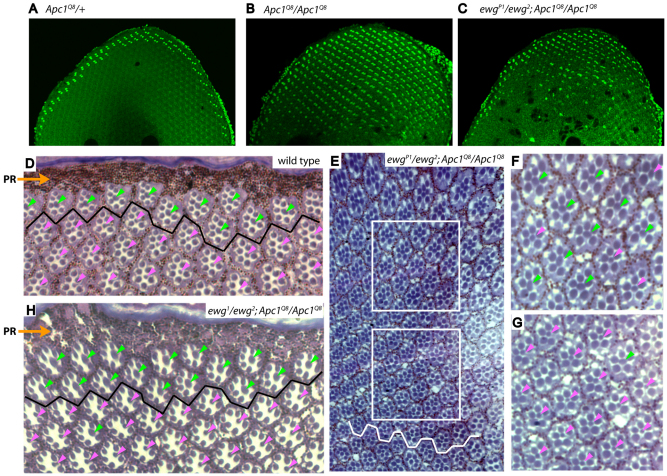
Fig. 2.
Ewg is required for aberrant photoreceptor cell fate specification in response to Apc1 loss. (A-H) Confocal images of 35-40 hour pupal retinas stained with Homothorax (Hth) antiserum (A-C) and cross-sections through adult retinas (D-H). In all panels, dorsal is towards the top. (A,D) In wild-type and Apc1 heterozygous retinas, inner photoreceptors (PRs) with DRA fate are restricted to one or two rows of ommatidia at dorsal retinal periphery, as indicated by Hth staining in pupae (A) and enlarged rhabdomeres located close to pigment rim (PR) in adults (D, ommatidia above black line, green arrowheads). (B) In Apc1Q8 mutants, constitutive activation of Wingless signaling induces all inner PRs in entire dorsal retinal half to aberrantly express Hth. (C,E-G) In ewgP1/ewg2; Apc1Q8 double mutants, partial reduction of Ewg activity leads to a reduction in the number of inner PRs expressing Hth and displaying enlarged rhabdomeres, which is most evident near dorsal-ventral equator (C,E). Higher magnifications of the upper and lower boxed areas in E are shown in F and G, respectively. Inner PRs closer to retinal dorsal edge mostly retain DRA fate, as indicated by enlarged rhabdomeres (green arrowheads in F); ommatidia closest to equator (white jagged line in E) mostly adopt a normal cell fate, as indicated by small rhabdomeres (pink arrowheads in G). (E) A montage of photographs compiled in Adobe Photoshop. (H) In ewg1/ewg2; Apc1Q8 double mutants, inner PRs adopting a DRA fate (green arrowheads) is further reduced to only two ommatidial rows adjacent to the dorsal rim (above solid black line), indicating attenuation of Ewg activity prevents ectopic Wingless signaling in Apc1 mutants.
Ewg facilitates Wingless-dependent flight muscle development
We next sought to identify Wingless-dependent developmental processes that require Ewg activity. Previous studies have revealed that Ewg is important for embryonic neuronal development and adult flight muscle formation (DeSimone and White, 1993). However, in ewg mutant embryos, we detect no defects in RP2 motor neuron specification or migration, two well-characterized processes that are dependent on Wingless signaling (Chu-LaGraff and Doe, 1993; Bhat, 2007) (supplementary material Fig. S3, Table S1). In addition, immunostaining of ewg-null mutant embryos for the neuronal markers 22C10, FasII and BP102, as well as the muscle marker Mef2, revealed no apparent defects (data not shown). Furthermore, we detect no defects in Wingless-dependent epithelial cell fate specification (Nusslein-Volhard and Wieschaus, 1980; Wieschaus and Riggleman, 1987; Baker, 1988) (data not shown).
Therefore, we also examined processes that require Ewg activity at later developmental stages. The embryonic lethality resulting from loss of Ewg can be prevented by transgenic expression of Ewg solely in neurons, driven by the elav promoter; although these rescued flies are viable, they display loss of indirect flight muscles, resulting from Ewg inactivation in myoblasts (DeSimone et al., 1996) (Fig. 3B). Similarly, hypomorphic ewg1 mutants display either complete loss of dorsal longitudinal muscles or marked reduction in flight muscle number, with enlargement of muscles that remain (Fleming et al., 1982) (Fig. 3C).
Fig. 3.
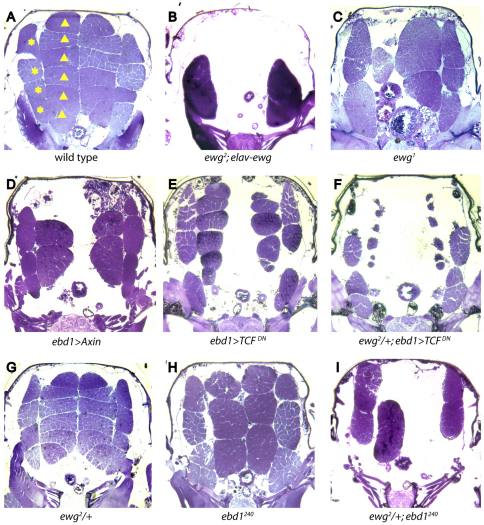
Ewg and Ebd1 are required for adult flight muscle development. Cross-sections of adult thoraces. (A) In wild type, there are six dorsal longitudinal muscles (DLMs) and seven dorsal ventral muscles (DVMs) on each side of the midline, all DLMs (triangles) and four DVMs (*) can be seen in this section. (B) DLMs and DVMs of normal number and morphology are lost, and instead are replaced by two large mis-shapen muscles in ewg2; elav-ewg mutants. (C) The majority of DLMs are lost in ewg1 homozygous mutants. (D) Disrupting Wingless signaling by overexpressing Axin results in partial loss of IFMs. (E) Disrupting Wingless signaling by overexpressing a dominant-negative form of TCF results in reduction of muscle volume. (F) Halving the gene dose of ewg enhances muscle defects caused by dominant-negative TCF. (G) Heterozygous ewg-null mutants do not have muscle defects. (H) A subset of DLMs is missing in ebd1240 homozygous mutants, and remaining muscles are abnormally enlarged. (I) Halving the gene dose of ewg markedly enhances muscle loss in ebd1240 mutants.
Wingless signaling is also required for flight muscle development (Sudarsan et al., 2001; Benchabane et al., 2011). We find that attenuation of Wingless signaling in ebd1-expressing cells, caused by overexpression of the negative regulator Axin, results in the reduction of flight muscle number and the enlargement of remaining flight muscles, a phenotype very similar to that observed in ewg mutants (Benchabane et al., 2011) (Fig. 3D). The similarity in flight muscle defects found upon attenuation of Ewg or Wingless pathway activity therefore suggested function in the same developmental process.
To determine whether the requirement of Ewg in flight muscle may reflect a role in Wingless-dependent muscle development, we examined genetic interactions between Ewg and the Wingless pathway transcriptional co-activator TCF. Attenuation of Wingless signaling, caused by expression of dominant-negative TCF in ebd1-expressing cells, results in reduction of flight muscle volume (Benchabane et al., 2011) (Fig. 3E). Halving the ewg gene dose markedly enhances severity of flight muscle defects caused by dominant-negative TCF (Fig. 3F), yet causes no muscle defects in an otherwise wild-type background (Fig. 3G), suggesting that Ewg may promote TCF activity during flight muscle development. To rule out the possibility that this phenotypic enhancement is due to an increase of Gal4 activity, we examined whether halving ewg dose affects the ebd1-Gal4 driver, but find no significant change in Gal4 mRNA levels (supplementary material Fig. S4). Thus, the genetic interaction between Ewg and TCF (Fig. 3E,F), coupled with the similar mutant phenotypes resulting from attenuation of either Ewg or Wnt pathway activity (Fig. 3C,D), support the hypothesis that Ewg facilitates Wingless-dependent flight muscle development.
Ewg acts with Ebd1 to facilitate Wingless-dependent flight muscle development
Of note, loss of Ebd1 activity also results in the reduction of flight muscle number and enlargement of remaining flight muscles, a phenotype similar to that observed in ewg mutants and upon disruption of Wingless signal transduction in ebd1-expressing cells (Benchabane et al., 2011) (Fig. 3C,D,H). Therefore, to determine whether Ewg and Ebd1 may function in the same developmental process, we used genetic interaction analyses. Halving ewg dose markedly enhances severity of muscle loss defects in ebd1 mutants, but, as noted above, causes no muscle defects in an otherwise wild-type background (Fig. 3G-I). Their strong genetic interaction with each other and with Armadillo-TCF (Fig. 3E,F) (Benchabane et al., 2011), coupled with their similar mutant muscle phenotype (Fig. 3C,H), support the model that a functional relationship between Ewg and Ebd1 promotes Wingless-dependent muscle development.
Ewg and Ebd1 are expressed in neurons and myocytes
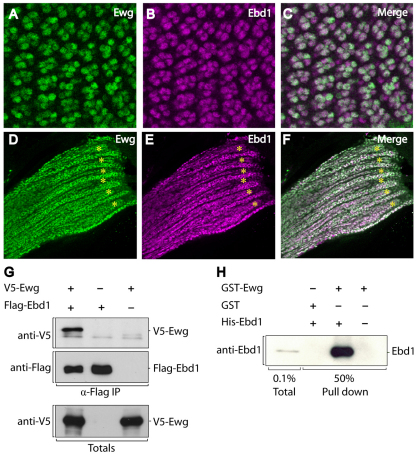
As our functional data suggest that Ewg and Ebd1 act in the same developmental process, we sought to determine whether these two proteins are also expressed in similar sets of cells. Both Ewg and Ebd1 are expressed predominantly in muscles and in the nervous system (DeSimone and White, 1993; DeSimone et al., 1996; Koushika et al., 2000; Benchabane et al., 2011). To determine whether Ebd1 and Ewg are expressed in the same subset of cells during different developmental stages, we performed immunostaining experiments using Ebd1 and Ewg antisera in embryos, larvae and pupae. Ebd1 and Ewg have identical distributions in the nervous system throughout all developmental stages examined (Fig. 4A-F; supplementary material Fig. S5A,B). During embryogenesis, Ewg and Ebd1 are expressed in both the central and peripheral nervous system (supplementary material Fig. S5A-F). In larvae and pupae, Ewg and Ebd1 are expressed in the brain, ventral ganglion, retinal photoreceptors and peripheral neurons (Fig. 4A-C, data not shown). In addition, Ewg and Ebd1 colocalize in myoblasts and developing muscle fibers during metamorphosis (Fig. 4D-F). Although not detected in previous studies, we also observe low levels of Ewg that colocalize with Ebd1 in a segmented pattern outside the nervous system in embryos (supplementary material Fig. S5B-F). Though of low level, this Ewg signal is absent in ewg-null mutant embryos, confirming specificity of the signal (supplementary material Fig. S5G-R). The pattern and morphology of these Ewg-expressing cells are consistent with embryonic myoblasts. Available antibodies to myoblast markers are all rabbit derived, as is the Ewg antiserum, precluding direct double-staining experiments. However, these Ewg-expressing cells also express Ebd1, and the vast majority of Ebd1-expressing cells outside the nervous system are myoblasts (Benchabane et al., 2011), suggesting that Ewg is also expressed in embryonic myoblast cells. Their highly similar cell-type-restricted expression patterns, coupled with their similar mutant phenotypes and genetic interactions, are consistent with the model that Ewg and Ebd1 function together in the same cells during development.
Fig. 4.
Ewg and Ebd1 display similar expression patterns and interact directly. (A-F) Confocal images of pupal retinas at 24 hours after puparium formation (A-C), pupal myofibers and associated myoblasts (D-F). Tissues are immunostained with Ewg antiserum (A,D) and Ebd1 antiserum (B,E). Merged images are shown in C,F. (A-F) During metamorphosis, Ewg and Ebd1 colocalize in neurons and myocytes, including photoreceptors (A-C), developing myofibers (asterisks) and associated myoblasts (D-F). (G) V5-tagged Ewg and Flag-tagged Ebd1 were co-transfected into HEK 293T cells. Cell lysates were subjected to immunoprecipiation with anti-Flag antibody, followed by immunoblotting with anti-V5 and anti-Flag antibody. Immunoblotting of total lysates is shown in lower panel. (H) GST interaction assay. Bacterially expressed GST-Ewg or GST control were purified and incubated with bacterially expressed and purified His-Ebd1. Protein bound to GST-Ewg or GST control was detected by immunoblotting with Ebd1 antibody.
Ewg and Ebd1 associate directly with each other
To test the hypothesis that Ewg and Ebd1 function together as a complex, we examined the physical interaction between these two proteins. We performed co-immunoprecipitation from lysates of HEK293T cells ectopically expressing Ewg and Flag-tagged Ebd1. Immunoprecipitation with Flag antibody followed by immunoblotting with Ewg antiserum revealed that Ebd1 associates with Ewg (Fig. 4G). Furthermore, bacterially expressed GST-Ewg, but not the GST control, associates with bacterially expressed Ebd1, indicating that their interaction is direct (Fig. 4H). Indeed, supporting our conclusion that Ewg and Ebd1 interact directly, Ebd1 was found to interact with Ewg in a genome-wide yeast two-hybrid screen for Drosophila protein interactions (Giot et al., 2003). Together with their similar cell-type specific expression patterns, similar loss-of-function phenotypes, and their strong genetic interactions with each other and with Armadillo-TCF, these findings support the model that direct physical interaction between Ewg and Ebd1 underlies their role in mediating context-specific responses to Wingless signal transduction.
Ewg and human NRF1 share chromatin association sites in vivo
NRF1, the human homolog of Ewg, contains a basic leucine zipper DNA-binding domain near its N terminus (Virbasius et al., 1993) (Fig. 5A). The NRF1 DNA-binding domain is highly conserved in Ewg, with more than 80% similarity (Efiok et al., 1994). Ewg has been found previously to interact with NRF1 consensus DNA-binding sequences, as revealed by gel mobility shift analysis, and to activate a transcriptional reporter containing multimerized NRF1 DNA-binding sites (Fazio et al., 2001). Together, these observations suggested that, like NRF1, Ewg might also associate with specific chromatin sites. To test this hypothesis in vivo, we took advantage of the presence of endogenous Ewg expression in third instar larval salivary glands (DeSimone and White, 1993), and analyzed the giant salivary gland polyploid chromosomes, which are produced by endoreplication and consist of ∼500 closely aligned copies of each homolog that display a highly characteristic chromatin banding pattern. To determine whether Ewg associates with chromatin and, if so, to assess the distribution of Ewg on chromatin sites throughout the genome, we performed immunostaining experiments on squashed polytene chromosomes using Ewg antiserum. As a control for specificity in these studies, we rescued the viability of ewg-null mutants with a neuronal-specific elav-ewg transgene that is not expressed in salivary glands (DeSimone and White, 1993). Ewg associates with discrete chromatin sites throughout the genome, and this signal is lost in ewg-null mutant chromosomes, confirming antibody staining specificity (Fig. 5B,C).
To determine whether Ewg and human NRF1 share similar chromatin association sites in vivo, we ectopically expressed NRF1 in larval salivary glands. We performed double-label immunostaining studies on polytene chromosomes to simultaneously detect NRF1 and endogenous Ewg. Ewg and NRF1 associate with many of the same chromatin sites throughout the genome (Fig. 5D-G). However, we wanted rule out the possibility that association of NRF1 with chromatin results solely from heterodimerization of NRF1 with Ewg, as NRF1 forms homodimers (Gugneja and Scarpulla, 1997), and the NRF1 dimerization domain is highly conserved in Ewg (Fig. 5A). Therefore, we expressed NRF1 in ewg-null salivary gland cells, and found that association of NRF1 with chromatin does not depend on the presence of Ewg (Fig. 5H-K). Thus, the similar chromatin banding patterns of Ewg and NRF1 suggest that Ewg and NRF1 may associate with specific chromatin sites through a similar mechanism.
Ebd1 associates with chromatin in vivo and shares chromatin association sites with Ewg
The physical and genetic interactions between Ewg and Ebd1 raise the possibility that these two factors are present in the same DNA-binding complex. To address this hypothesis, we sought to determine whether Ebd1 also associates with chromatin and, if so, whether Ebd1-associated chromatin sites are shared with Ewg. Immunostaining with Ebd1 antiserum also decorates discrete chromatin sites (supplementary material Fig. S6A), and this signal is lost in ebd1-null mutant chromosomes (supplementary material Fig. S6B), confirming Ebd1 antiserum specificity.
To determine whether any chromatin sites at which Ewg and Ebd1 associate are shared, we performed double labeling experiments on polytene squashes with Ebd1 and Ewg antisera. The banding pattern of Ebd1 on polytene chromosomes is highly similar to that of Ewg throughout the genome; many of the polytene bands at which Ebd1 and Ewg associate overlap with each other (Fig. 6A-C). Indeed, Ewg associates with nearly all, if not all, bands at which Ebd1 is found (supplementary material Table S2). However, Ewg associates with some polytene bands at which we detect no Ebd1 (supplementary material Table S2). For example, at the distal end of the X chromosome, Ebd1 associates with 6 of the 10 Ewg-associated bands, whereas Ebd1 is not detected on the other four Ewg-associated bands (Fig. 6F, pink arrowheads; supplementary material Table S2). Together, these findings support the model that Ebd1 and Ewg function as transcription co-factors that act as a complex at discrete promoter/enhancer sites.
Ewg is required for Ebd1 recruitment to chromatin
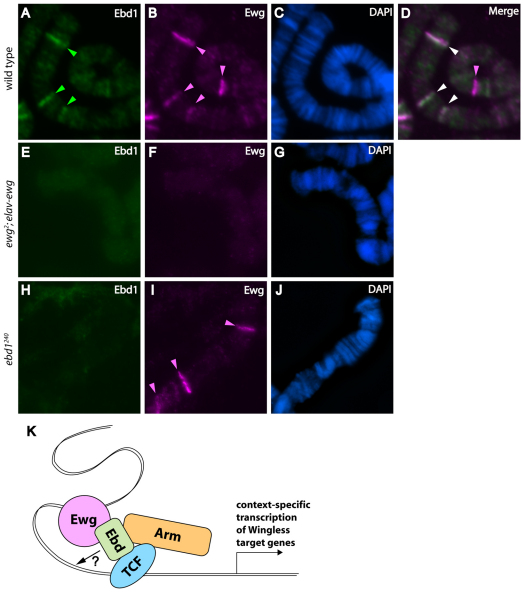
The direct physical interaction between Ebd1 and Ewg and their shared polytene banding patterns suggest that Ewg and Ebd1 associate with chromatin as a complex. To determine whether chromatin recruitment of either Ebd1 or Ewg requires the presence of the other, we analyzed the polytene banding pattern of Ebd1 in the absence of Ewg and, conversely, the banding pattern of Ewg in the absence of Ebd1. Ebd1 is readily detected on wild-type polytenes; by contrast, all association of Ebd1 with chromatin is abolished in the absence of Ewg (Fig. 7). For example, at the distal end of the left arm of chromosome 3, between cytological subdivisions 61A and 63C, Ewg and Ebd1 are both detected at the same three sites (Fig. 7A-D, arrowheads). By contrast, in ewg null mutant salivary glands, not only Ewg, but also Ebd1 is absent from these sites (Fig. 7E-G). This loss of Ebd1 chromatin association does not result from loss of Ebd1 expression, impaired Ebd1 nuclear localization, or global disruption of chromatin architecture, as Ebd1 is readily detected in nuclei of ewg-null mutant salivary gland cells, and the chromatin banding pattern, as revealed by DAPI staining, is intact in ewg-null mutants (supplementary material Fig. S7A-I; Fig. 7G). In addition, western blot analysis indicates that there is no significant reduction in Ebd1 protein level in ewg-null mutant glands (supplementary material Fig. S7J). We also sought to determine whether association of Ewg with chromatin is dependent on Ebd1. In the absence of Ebd1, immunostaining of Ewg on polytene squashes reveals distinct bands that are highly similar, if not identical to Ewg-associated chromatin bands detected in wild-type salivary glands (Fig. 7H-J). Together, these findings suggest the existence of an Ewg-Ebd1 complex that associates with distinct chromatin sites, and reveal that recruitment of Ebd1 to chromatin occurs only in the presence of Ewg (Fig. 7K).
Fig. 7.
Ewg is required to recruit Ebd1 to chromatin. (A-J) Squashes of polytene chromosomes from third instar larvae, immunostained with Ebd1 (A,E,H) and Ewg antiserum (B,F,I). DNA is highlighted by DAPI staining (C,G,J). (A-D) Ewg and Ebd1 overlap at the majority of their binding sites in wild-type polytenes. (E-G) Ebd1 is not detectable on chromatin in the absence of Ewg. (H-J) Ewg association with chromatin is maintained in absence of Ebd1. (K) A model depicting function of Ewg and Ebd1 in context-specific Wingless signaling. Ewg associates with specific chromatin sites and recruits Ebd1. Ebd1 bridges Arm and TCF, stabilizing this complex, facilitating its recruitment to chromatin, and thereby promoting context-specific activation of a subset of Wingless target genes.
DISCUSSION
An unbiased forward genetic screen in Drosophila uncovers a novel mechanism that mediates context-specific Wnt/Wingless signal transduction
Genome-wide expression profiling has revealed that the vast majority of Wnt target genes are context specific, and, by contrast, globally activated target genes are quite rare (Clevers, 2006). Factors that confer context-dependent target gene activation in response to activation of this widely used signal transduction pathway are of fundamental clinical importance, as they provide putative targets for therapeutic intervention; however, the identity of these factors, and their mechanism of action remain poorly understood, and have posed a daunting challenge for the Wnt signaling field (Gordon and Nusse, 2006).
We have taken an unbiased forward genetic approach in Drosophila to identify genes that promote Wingless signal transduction. The design of our genetic screen allowed identification of any gene that impacts Wingless signaling downstream, or at the level of the Apc/Axin destruction complex, which in principle could encompass a wide range of functions. Surprisingly, however, among the relatively small number of genetic complementation groups revealed in our screen, we have identified two cell-specific chromatin-associated co-factors that had not been implicated in Wingless signaling previously, but directly associate with each other and probably mediate the same context-specific Wingless-dependent developmental process. The combined activities of the two factors in this complex not only result in its recruitment to discrete chromatin sites, but also promote activity of the Armadillo-TCF transcription complex. Together, these unanticipated findings uncover a novel mechanism that facilitates context-specific regulation of Wingless signaling.
The Ewg-Ebd1 complex associates with discrete chromatin sites to facilitate context-specific Wingless signaling responses
We recently discovered that the novel Drosophila CENPB domain protein Ebd1 and its human homolog Jerky contribute to Wnt/Wingless target gene activation by enhancing β-catenin-TCF complex formation and β-catenin recruitment to chromatin (Benchabane et al., 2011). As Ebd1 is expressed in a cell-restricted pattern, these findings provided evidence that context-dependent responses to Wnt/Wingless stimulation are facilitated by cell-specific co-factors that interact with both β-catenin/Armadillo and TCF to enhance complex formation and activity. However, whether Ebd1 conferred context specificity solely by functioning as a cell-specific Armadillo-TCF adaptor or also by associating with distinct chromatin sites was not known.
Here, we provide multiple lines of evidence consistent with the model that the putative sequence-specific DNA-binding transcriptional activator Ewg is a physical and functional partner of Ebd1 in regulating context-specific Wingless signaling, and is required for chromatin recruitment of the Ewg-Ebd1 complex. Together, our studies suggest that Ebd1 and Ewg act together in the same context-specific Wingless-dependent developmental process, interact functionally with Armadillo-TCF, are expressed in similar cell-restricted patterns, associate physically with each other and are recruited to similar chromatin sites throughout the genome. Furthermore, all recruitment of Ebd1 to chromatin is abolished in the absence of Ewg. Based on these findings, we propose a model in which context specificity in Wingless signaling is achieved in part through association of Ewg with distinct enhancer sites, and recruitment of Ebd1 to these enhancers (Fig. 7K). In addition, our previous work suggested that by binding both Armadillo and TCF, Ebd1 stabilizes the Armadillo-TCF complex (Benchabane et al., 2011). Together, our findings support the model that Ebd1 promotes recruitment of the Armadillo-TCF complex to enhancers containing Ewg-binding sites, thereby facilitating context specificity in Wingless signal transduction. Alternatively, Ebd1 might also function in an Ewg-independent manner by solely providing an Armadillo-TCF adaptor function that stabilizes the Armadillo-TCF complex and thereby facilitates transcriptional activation of a subset of Wingless target genes requiring relatively high Armadillo-TCF levels.
Ewg, a putative sequence-specific DNA-binding protein, promotes recruitment of Ebd1, a cell-specific Armadillo-TCF adaptor, to discrete chromatin sites
Our ability to visualize endogenous Ebd1 and Ewg on larval salivary gland polytene chromosomes reveals that Ewg is essential for association of Ebd1 with chromatin, and that nearly all, if not all, chromatin sites with which Ebd1 associates are also bound by Ewg. By contrast, the association of Ewg with chromatin is not dependent on Ebd1; furthermore, not all Ewg-associated chromatin sites are bound by Ebd1, which probably reflects the broader role for Ewg in muscle and neuronal development. Together, these findings suggest that Ewg is necessary but not sufficient for recruitment of Ebd1 to chromatin, and either additional factors or specific DNA sequences are important for association of Ebd1 with chromatin. In particular, the presence of CENPB-type DNA-binding domains in Ebd1 indicates that Ebd1 may bind DNA directly, and therefore the association of Ebd1 with chromatin may require not only Ewg, but also sequence-specific Ebd1 DNA-binding sites. If so, Ewg might bind and remodel specific chromatin sites, thereby allowing Ebd1 to access nearby cognate binding sites or, alternatively, physical association with Ewg may result in a conformational change in Ebd1 that enables DNA binding. Similarly, association of Ebd1 with the Armadillo-TCF complex may facilitate association of Ebd1 and/or Armadillo-TCF with distinct chromatin sites; currently, we are unable to test this hypothesis in vivo, as our fixation and immunostaining conditions do not detect the indirect association of endogenous Armadillo with polytene chromosomes. Nonetheless, the extensive physical and genetic interactions between Ewg, Ebd1 and Wingless pathway transcriptional components, coupled with association of Ewg-Ebd1 with distinct chromatin sites support the model that context-specific activation of Wingless target genes is facilitated by interaction of Armadillo-TCF with cell-specific co-activator complexes at specific sites in chromatin.
The Ebd1-Ewg complex promotes context-specific Wingless-dependent adult muscle development
Our findings also suggest that Ebd1-Ewg complex function is required for only a context-specific subset of the Wingless-dependent processes that direct IFM development (Sudarsan et al., 2001; Benchabane et al., 2011). For example, Wingless secreted from larval wing imaginal disc epithelia specifies the fate of myoblasts associated with the wing disc that are important for IFM development (Sudarsan et al., 2001). In addition, Wingless signaling regulates the specification of tendon cells and the position of flight muscle attachment sites (Ghazi et al., 2003). Further evidence for Wingless signaling in IFM development was revealed by hypomorphic wingless mutants, some of which display a morphologically normal external appearance, but are nonetheless flightless (Sharma and Chopra, 1976; Deák, 1978). Indeed, our analysis of these hypomorphic wingless mutants reveals defects in late steps of muscle differentiation that result in partial loss of IFMs (Benchabane et al., 2011). In addition, we find strong genetic interactions between Ewg and the Armadillo-TCF complex (Fig. 3E,F). Together with the genetic and physical interactions between Ewg and Ebd1, as well as our previous findings that Ebd1 facilitates Wingless signaling in adult muscle development (Benchabane et al., 2011), the genetic interaction between Ewg and TCF provides further evidence supporting the model that Ewg and Ebd1 facilitate Wingless-dependent adult flight muscle development. However, as documented previously (DeSimone et al., 1996), we do not detect Ewg expression in IFM precursors until pupation, suggesting that Ewg does not participate in early steps of Wingless-dependent specification of IFM myoblasts or tendon cells, but instead promotes later steps in IFM growth and differentiation. Previous studies revealed that context specificity in Wnt signaling responses is facilitated by either crosstalk between signal transduction pathways or replacement of TCF with tissue-specific DNA-binding transcription co-factors that associate with β-catenin (Halfon et al., 2000; Guss et al., 2001; Kioussi et al., 2002; Notani et al; 2010; Olson et al., 2006) (see also MacDonald et al., 2009; McNeill and Woodgett, 2010). Thus, our findings suggest a novel mechanism in which a sequence-specific DNA-binding transcription factor recruits a cell-specific Armadillo-TCF adaptor to modulate context-specific Wingless signaling responses.
Wnt signaling and NRF-1 in vertebrate brain and retinal development
Ewg activity in neuronal differentiation can be functionally replaced by its human homolog nuclear respiratory factor 1 (NRF1) (Haussmann et al., 2008), a sequence-specific DNA-binding transcription factor important for activation of genes required in mitochondrial biogenesis and respiration, and for regulation of histone gene expression (Calzone et al., 1991; Virbasius et al., 1993; Gomez-Cuadrado et al., 1995; Dhar et al., 2008). Functional analysis of vertebrate NRF1 during development has been hampered by early lethality, as mouse Nrf1-null embryos die near the time of uterine implantation (Huo and Scarpulla, 2001). Fortuitously, however, inactivation of the zebrafish NRF1 homolog, not really finished (nrf), has improved our understanding of NRF1 function during vertebrate development; fish nrf-null mutants survive up to 14 days of development with the primary developmental defect restricted to markedly smaller brains and retinas (Becker et al., 1998). The tissue-restricted nature of the fish nrf-null mutant phenotype was unexpected, given the broader expression of the fish nrf gene (Becker et al., 1998) and the global roles for NRF1 in mitochondrial and histone function revealed by in vitro studies. However, these in vivo analyses revealed that fish Nrf, like fly Ewg, acts in specific tissue-restricted developmental processes. Of note, Wnt signaling is required for patterning of the mouse central nervous system (Thomas and Capecchi, 1990; Moon et al., 2004; Ciani and Salinas, 2005). Similarly, Wnt signaling is also crucial for brain and eye development in zebrafish (Lekven et al., 2001; Lekven et al., 2003; Dorsky et al., 2003; Nyholm et al., 2007), thus raising the possibility that, like Ewg, vertebrate Nrf may also facilitate a context-specific subset of Wnt-dependent developmental processes.
Supplementary Material
Acknowledgments
We thank E. Wieschaus for generous help with the mutagenesis screen, C. Pikielny, G. Deshpande and S. Ogden for thoughtful comments on the manuscript; K. White for providing Ewg antiserum and ewg cDNA clone; K. White, M. Soller, K. VijayRaghavan and the Bloomington Drosophila Stock Center for flies; G. Gill for sharing sequencing results of ewg alleles; J. Fernandes for muscle histology advice; and M. de la Roche, L. Pile and C. Grimaud for polytene staining advice. Elav, Eve, 22C10, FasII and BP102 monoclonal antibodies were obtained from Developmental Studies Hybridoma Bank. FlyBase provided important information used in this work.
Footnotes
Funding
This work was supported by Emerald Foundation, Norris Cotton Cancer Center, General Motors Cancer Research Foundation, American Cancer Society [IRG-82-003-21 to Y.A.], National Cancer Institute [KO8CA078532 and RO1CA105038 to Y.A.], Howard Hughes Medical Institute (through an award to Dartmouth Medical School [76200-560801]) and National Science Foundation ([DBI-1039423] for purchase of a Nikon A1RSi confocal). Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.068890/-/DC1
References
- Abu-Shaar M., Ryoo H. D., Mann R. S. (1999). Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 13, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Y., Hayashi S., Levine A., Wieschaus E. (1998). Regulation of Armadillo by a Drosophila APC inhibits neuronal apoptosis during retinal development. Cell 93, 1171–1182 [DOI] [PubMed] [Google Scholar]
- Ashburner M. (1972). Puffing patterns in Drosophila melanogaster and related species. In Developmental Studies on Giant Chromosomes. Results and Problems in Cell Differentiation (ed. Beermann W.), pp. 101–151 Springer-Verlag: Heidelberg & Berlin, Germany; [DOI] [PubMed] [Google Scholar]
- Baker N. E. (1988). Embryonic and imaginal requirements for wingless, a segment-polarity gene in Drosophila. Dev. Biol. 125, 96–108 [DOI] [PubMed] [Google Scholar]
- Becker T. S., Burgess S. M., Amsterdam A. H., Allende M. L., Hopkins N. (1998). not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human Nuclear Respiratory Factor-1 and avian Initiation Binding Repressor. Development 125, 4369–4378 [DOI] [PubMed] [Google Scholar]
- Benchabane H., Hughes E. G., Takacs C. M., Baird J. R., Ahmed Y. (2008). Adenomatous polyposis coli is present near the minimal level required for accurate graded responses to the Wingless morphogen. Development 135, 963–971 [DOI] [PubMed] [Google Scholar]
- Benchabane H., Xin N., Tian A., Hafler B. P., Nguyen K., Ahmed A., Ahmed Y. (2011). Jerky/Earthbound facilitates cell-specific Wnt/Wingless signalling by modulating beta-catenin-TCF activity. EMBO J. 30, 1444–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K. M. (2007). Wingless activity in the precursor cells specifies neuronal migratory behavior in the Drosophila nerve cord. Dev. Biol. 311, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour B. A., O’Brien M. A., Lockwood W. L., Goldstein E. S., Bodmer R., Taghert P. H., Abmayr S. M., Nguyen H. T. (1995). Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 9, 730–741 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- Cagan R. L., Ready D. F. (1989). Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 3, 1099–1112 [DOI] [PubMed] [Google Scholar]
- Calzone F. J., Hoog C., Teplow D. B., Cutting A. E., Zeller R. W., Britten R. J., Davidson E. H. (1991). Gene regulatory factors of the sea urchin embryo. I. Purification by affinity chromatography and cloning of P3A2, a novel DNA-binding protein. Development 112, 335–350 [DOI] [PubMed] [Google Scholar]
- Chang W.-T., Chen H.-i., Chiou R.-J., Chen C.-Y., Huang A. M. (2005). A novel function of transcription factor α-Pal/NRF-1: increasing neurite outgrowth. Biochem. Biophys. Res. Commun. 334, 199–206 [DOI] [PubMed] [Google Scholar]
- Chu-LaGraff Q., Doe C. (1993). Neuroblast specification and formation regulated by wingless in the Drosophila CNS. Science 261, 1594–1597 [DOI] [PubMed] [Google Scholar]
- Ciani L., Salinas P. C. (2005). WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat. Rev. Neurosci. 6, 351–362 [DOI] [PubMed] [Google Scholar]
- Clem R. J., Fechheimer M., Miller L. K. (1991). Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254, 1388–1390 [DOI] [PubMed] [Google Scholar]
- Clevers H. (2006). Wnt/beta-Catenin signaling in development and disease. Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- Deák I. I. (1978). Thoracic duplications in the mutant wingless of Drosophila and their effect on muscles and nerves. Dev. Biol. 66, 422–441 [DOI] [PubMed] [Google Scholar]
- DeSimone S. M., White K. (1993). The Drosophila erect wing gene, which is important for both neuronal and muscle development, encodes a protein which is similar to the sea urchin P3A2 DNA binding protein. Mol. Cell. Biol. 13, 3641–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSimone S., Coelho C., Roy S., VijayRaghavan K., White K. (1996). ERECT WING, the Drosophila member of a family of DNA binding proteins is required in imaginal myoblasts for flight muscle development. Development 121, 31–39 [DOI] [PubMed] [Google Scholar]
- Dhar S. S., Ongwijitwat S., Wong-Riley M. T. T. (2008). Nuclear Respiratory Factor 1 regulates all ten nuclear-encoded subunits of Cytochrome c oxidase in neurons. J. Biol. Chem. 283, 3120–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky R. I., Itoh M., Moon R. T., Chitnis A. (2003). Two tcf3 genes cooperate to pattern the zebrafish brain. Development 130, 1937–1947 [DOI] [PubMed] [Google Scholar]
- Efiok B. J., Chiorini J. A., Safer B. (1994). A key transcription factor for eukaryotic initiation factor-2 alpha is strongly homologous to developmental transcription factors and may link metabolic genes to cellular growth and development. J. Biol. Chem. 269, 18921–18930 [PubMed] [Google Scholar]
- Fazio I. K., Bolger T. A., Gill G. (2001). Conserved regions of the Drosophila Erect Wing protein contribute both positively and negatively to transcriptional activity. J. Biol. Chem. 276, 18710–18716 [DOI] [PubMed] [Google Scholar]
- Fernandes J. J., Atreya K. B., Desai K. M., Hall R. E., Patel M. D., Desai A. A., Benham A. E., Mable J. L., Straessle J. L. (2005). A dominant negative form of Rac1 affects myogenesis of adult thoracic muscles in Drosophila. Dev. Biol. 285, 11–27 [DOI] [PubMed] [Google Scholar]
- Fleming R. J., Zusman S. B., White K. (1982). Developmental genetic analysis of lethal alleles at the ewg locus and their effects on muscle development in Drosophila melanogaster. Dev. Genet. 3, 347–363 [Google Scholar]
- Ghazi A., Paul L., VijayRaghavan K. (2003). Prepattern genes and signaling molecules regulate stripe expression to specify Drosophila flight muscle attachment sites. Mech. Dev. 120, 519–528 [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., et al. (2003). A Protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- Gomez-Cuadrado A., Martin M., Noel M., Ruiz-Carrillo A. (1995). Initiation binding repressor, a factor that binds to the transcription initiation site of the histone h5 gene, is a glycosylated member of a family of cell growth regulators. Mol. Cell. Biol. 15, 6670–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M. D., Nusse R. (2006). Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J. Biol. Chem. 281, 22429–22433 [DOI] [PubMed] [Google Scholar]
- Grenningloh G., Rehm E. J., Goodman C. S. (1991). Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell 67, 45–57 [DOI] [PubMed] [Google Scholar]
- Gugneja S., Scarpulla R. C. (1997). Serine phosphorylation within a concise amino-terminal domain in Nuclear Respiratory Factor 1 enhances DNA binding. J. Biol. Chem. 272, 18732–18739 [DOI] [PubMed] [Google Scholar]
- Guss K. A., Nelson C. E., Hudson A., Kraus M. E., Carroll S. B. (2001). Control of a genetic regulatory network by a selector gene. Science 292, 1164–1167 [DOI] [PubMed] [Google Scholar]
- Halfon M. S., Carmena A., Gisselbrecht S., Sackerson C. M., Jimenez F., Baylies M. K., Michelson A. M. (2000). Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103, 63–74 [DOI] [PubMed] [Google Scholar]
- Haussmann I., White K., Soller M. (2008). Erect wing regulates synaptic growth in Drosophila by integration of multiple signaling pathways. Genome Biol. 9, R73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B. A., Wolff T., Rubin G. M. (1994). Expression of baculovirus P35 prevents cell death in Drosophila. Development 120, 2121–2129 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Abdollah S., Qiu Y., Cai J., Xu Y. Y., Grinnell B. W., Richardson M. A., Topper J. N., Gimbrone M. A., Jr, Wrana J. L., et al. (1997). The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell 89, 1165–1173 [DOI] [PubMed] [Google Scholar]
- Hummel T., Krukkert K., Roos J., Davis G., Klambt C. (2000). Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron 26, 357–370 [DOI] [PubMed] [Google Scholar]
- Huo L., Scarpulla R. C. (2001). Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of Nuclear Respiratory Factor 1 in mice. Mol. Cell. Biol. 21, 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussi C., Briata P., Baek S. H., Rose D. W., Hamblet N. S., Herman T., Ohgi K. A., Lin C., Gleiberman A., Wang J., et al. (2002). Identification of a Wnt/Dvl/β-Catenin-Pitx2 pathway mediating cell-type-specific proliferation during development. Cell 111, 673–685 [DOI] [PubMed] [Google Scholar]
- Koushika S. P., Soller M., White K. (2000). The neuron-enriched splicing pattern of Drosophila erect wing is dependent on the presence of ELAV protein. Mol. Cell. Biol. 20, 1836–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramps T., Peter O., Brunner E., Nellen D., Froesch B., Chatterjee S., Murone M., Züllig S., Basler K. (2002). Wnt/Wingless signaling requires BCL9/Legless-mediated recruitment of Pygopus to the nuclear β-Catenin-TCF complex. Cell 109, 47–60 [DOI] [PubMed] [Google Scholar]
- Lekven A. C., Thorpe C. J., Waxman J. S., Moon R. T. (2001). Zebrafish wnt8 encodes two Wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev. Cell 1, 103–114 [DOI] [PubMed] [Google Scholar]
- Lekven A. C., Buckles G. R., Kostakis N., Moon R. T. (2003). Wnt1 and wnt10b function redundantly at the zebrafish midbrain-hindbrain boundary. Dev. Biol. 254, 172–187 [DOI] [PubMed] [Google Scholar]
- Lewis E. B., Bacher F. (1968). Method for feeding ethyl-methane sulfonate (EMS) to Drosophila males. Dros. Inf. Serv. 43, 193 [Google Scholar]
- Lin H. V., Rogulja A., Cadigan K. M. (2004). Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development 131, 2409–2418 [DOI] [PubMed] [Google Scholar]
- Logan C. Y., Nusse R. (2004). The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- MacDonald B. T., Tamai K., He X. (2009). Wnt/β-Catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H., Masukata H., Muro Y., Nozaki N., Okazaki T. (1989). A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H., Woodgett J. R. (2010). When pathways collide: collaboration and connivance among signalling proteins in development. Nat. Rev. Mol. Cell Biol. 11, 404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004). WNT and beta-catenin signalling: diseases and therapies. Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- Notani D., Gottimukkala K. P., Jayani R. S., Limaye A. S., Damle M. V., Mehta S., Purbey P. K., Joseph J., Galande S. (2010). Global regulator SATB1 recruits beta-catenin and regulates T(H)2 differentiation in Wnt-dependent manner. PLoS Biol. 8, e1000296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Wieschaus E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 [DOI] [PubMed] [Google Scholar]
- Nyholm M. K., Wu S.-F., Dorsky R. I., Grinblat Y. (2007). The zebrafish zic2a-zic5 gene pair acts downstream of canonical Wnt signaling to control cell proliferation in the developing tectum. Development 134, 735–746 [DOI] [PubMed] [Google Scholar]
- Olson L. E., Tollkuhn J., Scafoglio C., Krones A., Zhang J., Ohgi K. A., Wu W., Taketo M. M., Kemler R., Grosschedl R., et al. (2006). Homeodomain-mediated beta-catenin-dependent switching events dictate cell-lineage determination. Cell 125, 593–605 [DOI] [PubMed] [Google Scholar]
- O’Neill E. M., Rebay I., Tjian R., Rubin G. M. (1994). The activities of two Ets-related transcription factors required for drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78, 137–147 [DOI] [PubMed] [Google Scholar]
- Pai C. Y., Kuo T. S., Jaw T. J., Kurant E., Chen C. T., Bessarab D. A., Salzberg A., Sun Y. H. (1998). The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 12, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell W. F., Sullivan W. (2000). Fluorescent analysis of Drosophila embryos. In Drosophila Protocols (ed. Sullivan W., Ashburner M., Hawley R. S.), pp. 141–157 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Sharma R. P., Chopra V. L. (1976). Effect of the wingless (wg1) mutation on wing and haltere development in Drosophila melanogaster. Dev. Biol. 48, 461–465 [DOI] [PubMed] [Google Scholar]
- Seeger M., Tear G., Ferres-Marco D., Goodman C. S. (1993). Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron 10, 409–426 [DOI] [PubMed] [Google Scholar]
- Sheffield P., Garrard S., Derewenda Z. (1999). Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Prot. Exp. Purification 15, 34–39 [DOI] [PubMed] [Google Scholar]
- Sorsa V. (1988). Chromosome Maps of Drosophila. Boca Raton, FL: CRC Press; [Google Scholar]
- Sudarsan V., Anant S., Guptan P., VijayRaghavan K., Skaer H. (2001). Myoblast diversification and ectodermal signaling in Drosophila. Dev. Cell 1, 829–839 [DOI] [PubMed] [Google Scholar]
- Takacs C. M., Baird J. R., Hughes E. G., Kent S. S., Benchabane H., Paik R., Ahmed Y. (2008). Dual positive and negative regulation of Wingless signaling by Adenomatous Polyposis Coli. Science 319, 333–336 [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. (1990). Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature 346, 847–850 [DOI] [PubMed] [Google Scholar]
- Tomlinson A. (2003). Patterning the peripheral retina of the fly: decoding a gradient. Dev. Cell 5, 799–809 [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Cavallo R., Dooijes D., van Beest M., van, Es J., Loureiro J., Ypma A., Hursh D., Jones T., Bejsovec A., et al. (1997). Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88, 789–799 [DOI] [PubMed] [Google Scholar]
- Virbasius C. A., Virbasius J. V., Scarpulla R. C. (1993). NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 7, 2431–2445 [DOI] [PubMed] [Google Scholar]
- Wernet M. F., Labhart T., Baumann F., Mazzoni E. O., Pichaud F., Desplan C. (2003). Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell 115, 267–279 [DOI] [PubMed] [Google Scholar]
- Wieschaus E., Riggleman R. (1987). Autonomous requirements for the segment polarity gene armadillo during Drosophila embryogenesis. Cell 49, 177–184 [DOI] [PubMed] [Google Scholar]
- Wolff T. (2000). Histological techniques for the Drosophila eye. In The Development of Drosophila Melanogaster (ed. Sullivan W., Ashburner M., Hawley R. S.), pp. 1277–1325 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Yu S. Y., Yoo S. J., Yang L., Zapata C., Srinivasan A., Hay B. A., Baker N. E. (2002). A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development 129, 3269–3278 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.