Abstract
We employed mtDNA and nuclear SNPs to investigate the genetic diversity of sheep breeds of three countries of the Mediterranean basin: Albania, Greece, and Italy. In total, 154 unique mtDNA haplotypes were detected by means of D-loop sequence analysis. The major nucleotide diversity was observed in Albania. We identified haplogroups, A, B, and C in Albanian and Greek samples, while Italian individuals clustered in groups A and B. In general, the data show a pattern reflecting old migrations that occurred in postneolithic and historical times. PCA analysis on SNP data differentiated breeds with good correspondence to geographical locations. This could reflect geographical isolation, selection operated by local sheep farmers, and different flock management and breed admixture that occurred in the last centuries.
Keywords: mtDNA, sheep, SNPs, Mediterranean, domestication
1. INTRODUCTION
The earliest archaeozoological evidence of domestic sheep comes from a restricted area of south-western Asia: modern Iran, Turkey, and Cyprus [1]. A pioneering genetic study examining the karyotypes of the various species of extant wild sheep [2, 3] showed that domestic sheep derive from the Asiatic mouflon (Ovis orientalis) of Anatolia, western Iran, and southwest Iran. Afterwards, a probable migration of the Neolithic farmers occurred out of the Near East and across Europe following two main routes, through the continental heartland up the Danube valley or along the Mediterranean coast [4, 5] crossing the sea to the major islands. Archaeological data and radiocarbon dates on seeds or bones provide support for an earlier arrival in Western Europe via the Mediterranean route rather than the “Danubian” route [6].
Both archaeozoological evidence and genetic evidence indicate that the domestication of wild sheep occurred 8000–9000 years ago. The first appearance of the remains of domestic sheep in the western part of Mediterranean Europe, dating approximately 5400 BC, is believed to reflect a rapid spread by sea [7, 8]. The Mediterranean Sea also had a key role in the history of livestock in postneolithic times, when peoples like Phoenicians, Greeks, Romans, and Berbers probably introduced new species of animals and new breeds of livestock into southwest Europe by sea. Some settlers may have improved local livestock by importing stock from overseas [8], which explained the unexpectedly high within-breed diversity in domestic goats [9, 10], the differential cattle migration along the Mediterranean coast [11], and the close genetic relationship between Tuscan and Near Eastern cattle breeds [12]. The role of the Mediterranean Sea as a natural corridor connecting the South West Europe to the Near East and North Africa is particularly plausible for domestic sheep and goats that were adaptable to various environments and easy to transport due to their size [8]. Subsequently, sheep breeds developed after selective breeding for desirable traits (wool, milk, and meat production) and environmental tolerance. Since domestication, sheep have established a wide geographic range due to their adaptability to poor diets and extreme climatic conditions as well as their manageable size.
The genetic history of sheep has been investigated using three major sources of genomic variation: autosomes, Y chromosome, and mitochondrial genome. Analysis of the nonrecombining region of the Y chromosome has revealed patterns of male-mediated introgression during breed development [13, 14]. Recent surveys have tested collections of animals from southern and northern Europe [15] or Europe and the Middle East [16] using microsatellites and enabled the analysis of genetic partitioning at a continental scale. Interestingly, southern European breeds displayed increased genetic diversity and decreased genetic differentiation compared to their northern European counterparts. This is consistent with the expectation that genetic diversity will be maintained high in populations close to the centre of domestication but decreases with increasing geographic distances. Kijas et al. [17] used a SNP panel to analyse sheep nuclear genome, providing the indication that breeds cluster into large groups based on geographic origin and that SNPs can successfully identify population substructures within individual breeds.
A recent study on retrovirus integrations [18] has provided additional information on the introduction of sheep into Europe, indicating an early arrival of the primitive sheep populations (European mouflons, North-Atlantic Island breeds) and a subsequent advent of wool producing sheep.
However, most of the information about history and domestication of the species have been gathered using mtDNA. The existence of multiple mtDNA lineages and their admixture within breeds [8, 19–22] could be due to multiple domestication events and subsequent human selection or introgression by domestic and wild species.
Mitochondrial DNA analyses in sheep identified an increasing number of maternal lineages: two [23–25], three [20, 26], and then five [22]. The main haplogroups A and B are both found in Asia, while B dominates in Europe. Haplogroup C has been found in Portugal, Turkey, the Caucasus, and China [7]. Haplogroup D, present in Rumanian Karachai and Caucasian animals, is possibly related to the haplogroup A. In contrast to taurine cattle, the sheep haplogroups hardly correlate with geography.
Because of their mode of inheritance, mitochondrial markers are more likely to lead to biased estimates of species phylogeny [27]. Combining nuclear and mitochondrial markers may help in avoiding this problem. Nuclear genome evolves five-to-ten times slower than mtDNA; it is contributed by both parents and its variability is less affected by demographic forces such as bottleneck. Therefore, nuclear markers can detect more recent genetic events that influence the extant divergence of domestic breeds. Several studies have demonstrated that the combination of nuclear and mtDNA markers can increase the information obtained [27–30]. The use of both markers might provide a more accurate and comprehensive understanding of a species' history [31]. SNP markers could help in understanding the recent evolutionary history of domestic animals [10, 32].
We aimed at investigating the geographic distribution of the genetic diversity of sheep breeds in Albania, Greece, and Italy and to gather information on the migration history of the species. To accomplish that, we employed sequence data from the mitochondrial D-loop and 27 nuclear loci (SNPs).
2. MATERIALS AND METHODS
2.1. Sampling and DNA Extraction
We focused on sheep breeds of Albania, Greece, and Italy. Samples of the European mouflon were also included. About twenty unrelated samples per breed were selected. Three animals per flock from 11 farms spread over the traditional rearing area were sampled. A total of 313 animals from 18 sheep breeds were analyzed. Breeds, acronyms used, and country of origin of each breed are reported in Table 1. Part of the samples were obtained from a previous project (Econogene, http://www.econogene.eu/). Blood samples were collected in EDTA tubes and frozen at −20°C until extraction. Genomic DNA was isolated using standard procedures, checked for DNA quality on agarose gel and quantified using a DTX microplate reader (Beckman Coulter) after staining with PicoGreen (Invitrogen).
Table 1.
Country of origin, breeds, and acronyms used in computations.
| Country | Breed | Acronym |
|---|---|---|
| Albania | Bardhoka Ruda Shkordane |
BAR RUD SHK |
|
| ||
| Greece | Kalarritiko Orino Pilioritiko Kefalleneas Lesvos Kymi Karagouniko Skopelos Anogeiano Sfakia |
KAL ORI PIL KEF LES KIM KAR SKO ANO SFA |
|
| ||
| Italy | Bergamasca Delle Langhe Laticauda Altamurana Gentile di Puglia |
BER LAN LAT ALT GDP |
2.2. Amplification and Sequencing of the Mitochondrial D-Loop
To amplify the partial D-loop of 721 bp, primers described by Tapio et al. [7] were used from 15,541 to 16,261 of the complete sequence described by Hiendleder et al. [33] available in GenBank (NC_001941.1). Polymerase chain reaction (PCR) was performed in a total volume of 50 μL containing 20 ng of genomic DNA, 40 pMol of each primer (Sigma-Aldrich), 200 μM dNTPs, 5X PCR buffer, and 5 units of Taq DNA polymerase (Promega) on a PCR Thermo Cycler (MJ Research). A 5 minutes denaturation step at 95°C was followed by 14 cycles of denaturation at 95°C for 30 sec, annealing for 30 sec starting at 62°C and decreasing 0.5°C per cycle and extension at 72°C for 120 sec, then by 20 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec and extension at 72°C for 120 sec; the final extension step was carried out at 72°C for 5 minutes. PCR products were purified through ExoSap-IT (USB Corporation) to remove residual primers and dNTPs and used as templates for forward and reverse sequencing reactions.
Sequencing was performed using the primers described by Tapio et al. [7] with a CEQ 8800 sequencer using DTCS QuickStart Kit and purifying with Agencourt CleanSEQ 96 (Beckman Coulter), according to the manufacturer's instructions. After the optimization of the sequencing protocol, sequencing was outsourced to Macrogen (http://www.macrogen.com/). The sequences of D-loop were submitted to GenBank (accession numbers: JN184789–JN184999).
2.3. Mitochondrial Sequence Analysis
A fragment of 435 bp, running from 15,541 to 16,261 bp (NC_0019041.1), was selected excluding a central region rich in tandem repeats (from 15,644 to 15,932 bp). mtDNA variations were identified on a total of 313 sequences of 18 breeds analyzed and aligned with BioEdit software [34]. DnaSP 5.00 software [35] was used to calculate haplotype, sequence variation, average number of nucleotide differences (D), and average number of nucleotide substitutions (Dxy) per site between breeds. Neighbour-joining tree for all haplotypes was constructed using Mega version 5 [36]. Analysis of molecular variance (AMOVA) was performed with Arlequin version 3.11 [37]. Sequences of the same D-loop fragment in wild sheep, published by Hiendleder et al. [33], were obtained from GenBank, Ovis vignei arkal (AY091489.1), Ovis vignei bochariensis (AY091490.1, AY091491.1, and AF039580.1), Ovis ammon collium (AY091492.1), Ovis ammon nigrimontana (AY091493.1 and AY091494.1), and used as outgroups in phylogenetic analysis.
Geographic distribution of eigenvectors was performed to investigate population genetic differences on the basis of their geographic distances. This approach permitted the generation of a synthetic configuration of locations based on the pairwise genetic distances that matched the real geographic configuration. Principal component analysis (PCA) scores for the first two components, obtained using Nei's 1973 genetic distance, were plotted on a geographic map. As breeds are scattered among several farms, a virtual geographic entity representing the centroid of each breed on geographic maps was created using WGS84 geographical coordinates [38]. For a given component, it is a measure of the variance accounted for by that component. On thematic maps produced with the geographic information system (GIS) Manifold software package (Manifold System, Version 7, Manifold Net Ltd., Carson City, USA, http://www.manifold.net/), all breeds are thus represented according to a geometric distribution (see Figures 3(a) and 3(b)). Breeds showing high eigenvectors contribute sensibly to the explanation of the variance related to the component displayed. Classes were elaborated on the basis of the criterion of the natural breaks (Jenks optimization method). This algorithm reduces the variance within classes and maximizes the variance between classes. Colour classes were chosen in order to support the distinction between the different categories of behaviours observed: green: positive contribution; yellow: intermediary values; red: negative contribution to the component displayed.
Figure 3.
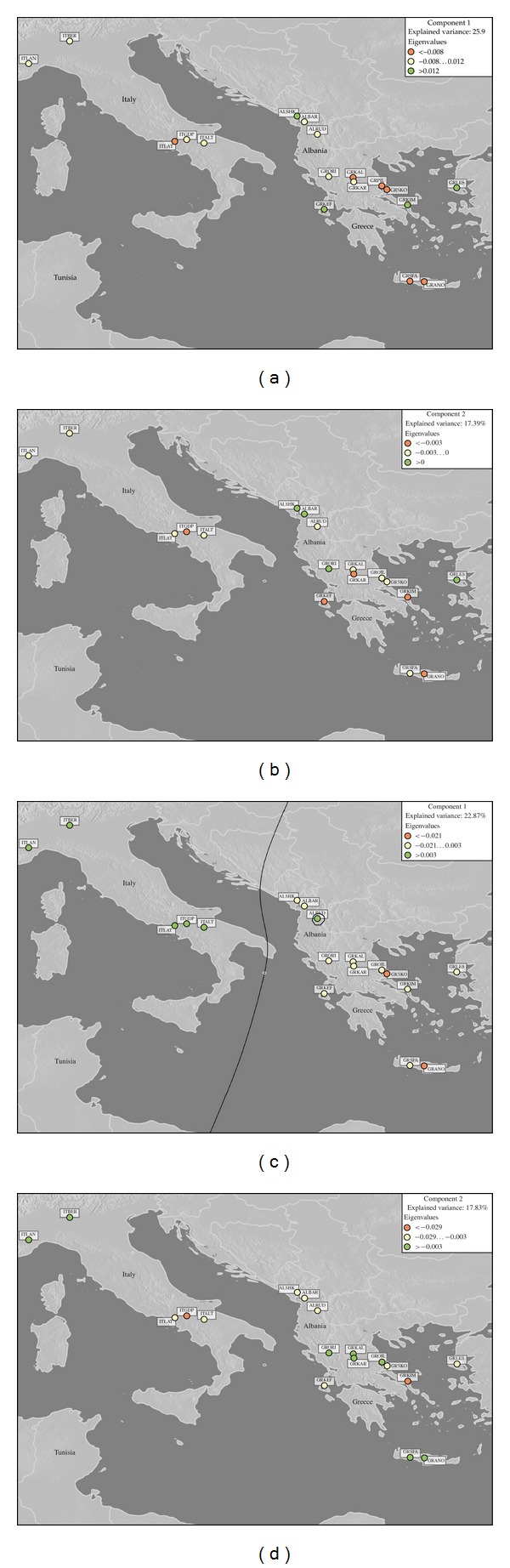
First (a) and second (b) components of eigenvectors spatial distribution calculated on mtDNA marker and first (c) and second (d) components calculated on SNPs markers. Background image is GTOPO30, a global digital elevation model (DEM) with a horizontal grid spacing of 30 arc seconds (approximately 1 kilometer) produced by the U.S. Geological Survey's Center for Earth Resources Observation and Science (EROS). The line in the map (c) shows the isoline for an eigenvalue of 0. It appears on panel C only because this is the only geographical configuration for which a limit is so obvious. The circle around the RUDA breed shows an isoline for the eigenvalue of 0.03.
2.4. Nuclear Polymorphism Analysis
The same 313 sheep belonging to 18 breeds sequenced at D-loop were genotyped with 37 previously described SNPs [39]. SNP ascertainment bias was minimised by sequencing target DNA in at least 8 individuals from different populations. Large-scale genotyping of all animals was performed by outsourcing to a commercial genotyping company (http://www.Kbioscience.co.uk/).
Allele frequencies, Nei's estimation of observed and expected heterozygosities (Ho and He, resp.), were calculated using Fstat 2.93 [40]. Weir and Cockerham's [41] estimates of Fis per population, Fst per locus, and population pairs were calculated for each locus using Genalex 4.0 [42]. The same software was used to test deviations from Hardy-Weinberg equilibrium (HWE) for each locus and population and for locus over all populations; test for conformity with HWE expectations was assessed by calculating the Chi-squared value.
Correlation between geographic and Nei's 1973 pairwise genetic distances was tested using Mantel tests (999 permutation) implemented in Genalex 4.0 software [42].
A PCA was performed on the covariance matrix of SNP frequency data to investigate spatial patterns of genetic variation using GENETIX software [43].
Nei [44] and Reynolds [45] genetic distances between population pairs were calculated using PowerMarker v3.25 [46].
Geographic distribution of eigenvectors was performed as described above using pairwise genetic distances [47] calculated on the basis of the selected SNP markers.
3. RESULTS
3.1. Mitochondrial Haplotypes
Ninety-three polymorphic sites and 154 haplotypes were identified from 313 sequences. Relatively high haplotype diversity was found in all three sampled geographic regions; the largest nucleotide diversity is present in Albania (0.02107) while the highest number of haplotypes observed is recorded in Greece (83) (Table 2).
Table 2.
Sample size per country (n), number of haplotypes observed (Haplotypes), number of polymorphic sites, mean number of pairwise differences among sequences (pairwise diff.), haplotype diversity (h), and nucleotide diversity (π) are shown.
| Country | n | Haplotypes | Polymorphic sites | Pairwise diff. | h | π |
|---|---|---|---|---|---|---|
| Italy | 93 | 62 | 58 | 4.180 | 0.978 | 0.01007 |
| Greece | 167 | 83 | 73 | 5.934 | 0.934 | 0.01469 |
| Albania | 53 | 37 | 57 | 8.704 | 0.979 | 0.02107 |
The average number of nucleotide differences and the average number of nucleotide substitutions per site were used to calculate the genetic distance between breeds. The lowest distance was observed between Laticauda and Anogeiano (D: 2.357—Dxy: 0.006), while the highest distance was observed between Bardhoka and Kymi (D: 12.450—Dxy: 0.03) (Table 3).
Table 3.
Average number of nucleotide differences, D (below), and average number of nucleotide substitutions per site between populations, Dxy [48] (above).
| BER | ALT | LAT | LAN | GDP | BAR | RUD | SHK | KAL | ORI | PIL | KEF | LES | KIM | KAR | SKO | ANO | SFA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BER | ∗ | 0.009 | 0.009 | 0.012 | 0.011 | 0.018 | 0.011 | 0.021 | 0.009 | 0.017 | 0.009 | 0.018 | 0.018 | 0.025 | 0.012 | 0.011 | 0.008 | 0.009 |
| ALT | 3.908 | ∗ | 0.007 | 0.011 | 0.009 | 0.017 | 0.009 | 0.02 | 0.007 | 0.016 | 0.008 | 0.016 | 0.017 | 0.024 | 0.011 | 0.009 | 0.006 | 0.008 |
| LAT | 3.728 | 2.93 | ∗ | 0.01 | 0.009 | 0.016 | 0.009 | 0.02 | 0.007 | 0.015 | 0.007 | 0.016 | 0.017 | 0.025 | 0.01 | 0.008 | 0.006 | 0.007 |
| LAN | 5.079 | 4.425 | 4.279 | ∗ | 0.012 | 0.019 | 0.013 | 0.022 | 0.01 | 0.018 | 0.01 | 0.019 | 0.019 | 0.026 | 0.013 | 0.012 | 0.01 | 0.01 |
| GDP | 4.632 | 3.906 | 3.756 | 5.134 | ∗ | 0.018 | 0.011 | 0.021 | 0.009 | 0.017 | 0.01 | 0.017 | 0.018 | 0.024 | 0.012 | 0.011 | 0.009 | 0.009 |
| BAR | 7.447 | 6.933 | 6.856 | 8.107 | 7.579 | ∗ | 0.018 | 0.026 | 0.016 | 0.023 | 0.017 | 0.023 | 0.024 | 0.03 | 0.02 | 0.018 | 0.016 | 0.016 |
| RUD | 4.612 | 3.95 | 3.7 | 5.245 | 4.711 | 7.647 | ∗ | 0.021 | 0.009 | 0.017 | 0.01 | 0.018 | 0.018 | 0.025 | 0.012 | 0.011 | 0.008 | 0.009 |
| SHK | 8.778 | 8.272 | 8.345 | 9.269 | 8.696 | 10.567 | 8.789 | ∗ | 0.02 | 0.025 | 0.02 | 0.025 | 0.025 | 0.029 | 0.023 | 0.021 | 0.02 | 0.02 |
| KAL | 3.712 | 3 | 2.708 | 4.183 | 3.754 | 6.741 | 3.778 | 8.241 | ∗ | 0.015 | 0.007 | 0.016 | 0.017 | 0.024 | 0.01 | 0.008 | 0.006 | 0.007 |
| ORI | 7.108 | 6.488 | 6.395 | 7.536 | 7.132 | 9.4 | 7.122 | 10.395 | 6.352 | ∗ | 0.016 | 0.023 | 0.023 | 0.029 | 0.018 | 0.017 | 0.015 | 0.015 |
| PIL | 3.91 | 3.289 | 3.011 | 4.5 | 4.021 | 7.049 | 4.067 | 8.419 | 2.978 | 6.556 | ∗ | 0.017 | 0.017 | 0.025 | 0.011 | 0.009 | 0.007 | 0.007 |
| KEF | 7.349 | 6.861 | 6.832 | 7.853 | 7.319 | 9.804 | 7.475 | 10.306 | 6.757 | 9.486 | 6.971 | ∗ | 0.022 | 0.026 | 0.019 | 0.018 | 0.016 | 0.016 |
| LES | 7.515 | 7.042 | 6.941 | 8.053 | 7.507 | 10.1 | 7.588 | 10.396 | 6.896 | 9.576 | 7.125 | 9.367 | ∗ | 0.027 | 0.019 | 0.018 | 0.016 | 0.016 |
| KIM | 10.426 | 10.083 | 10.276 | 10.994 | 10.211 | 12.45 | 10.438 | 11.931 | 10.194 | 12.028 | 10.383 | 11.141 | 11.375 | ∗ | 0.026 | 0.026 | 0.024 | 0.024 |
| KAR | 5.144 | 4.478 | 4.197 | 5.595 | 5.139 | 8.217 | 5.185 | 9.378 | 4.294 | 7.65 | 4.503 | 7.894 | 7.975 | 10.975 | ∗ | 0.012 | 0.01 | 0.01 |
| SKO | 4.497 | 3.852 | 3.538 | 4.989 | 4.544 | 7.585 | 4.611 | 8.883 | 3.472 | 7.102 | 3.719 | 7.455 | 7.618 | 10.813 | 4.936 | ∗ | 0.008 | 0.009 |
| ANO | 3.514 | 2.678 | 2.357 | 4.053 | 3.526 | 6.614 | 3.505 | 8.123 | 2.439 | 6.132 | 2.712 | 6.655 | 6.73 | 10.079 | 4.003 | 3.301 | ∗ | 0.006 |
| SFA | 3.768 | 3.164 | 2.878 | 4.266 | 3.801 | 6.698 | 3.834 | 8.015 | 2.906 | 6.266 | 3.182 | 6.658 | 6.77 | 9.757 | 4.284 | 3.596 | 2.643 | ∗ |
AMOVA revealed that mitochondrial diversity is mainly distributed within breeds (95.04%) and only in part among regions (0.90%); low variability was also found among breeds/within regions (4.06%) (Table 4).
Table 4.
Hierarchical analysis of molecular variance (AMOVA) with 10,000 permutations.
| Source of variation | Variation (%) | Fixation indices [49] | P value |
|---|---|---|---|
| Among regions | 0.90 | FSC: 0.0495 | <0.01 |
| Among breeds/within regions | 4.06 | F st: 0.04960 | <0.01 |
| Within breeds | 95.04 | FCT: 0.00903 | <0.5 |
3.2. Phylogenetic Analysis and Haplogroups
The NJ tree obtained from mtDNA haplotypes and wild sheep sequences, used as out-group, revealed three of the five haplogroups described in the literature: A, B, and C (Figure 1). Haplogroup B is the most frequent among the analyzed samples (89%), while A and C are less common (8% and 3%, resp.). Greek and Albanian breeds are present in all three haplogroups, while Italian breeds are present only in haplogroups B and A (Table 5). This is shown also in Figure 2, representing the percentage of occurrence of each haplogroup in Albania, Greece, and Italy.
Figure 1.

Neighbour-joining tree based on the 154 mtDNA haplotypes, showing differences among haplogroups and wild sheep (outgroup). Numbers indicate the percentage bootstrap support (10,000 resamplings).
Table 5.
Sequences analysed, number of variable sites (# Var), number of haplotypes (Hap), and percentage of haplotypes in the haplogroups A, B, or C per breed.
| Breed | Sequences | # Var | Hap | A | B | C |
|---|---|---|---|---|---|---|
| BAR | 15 | 43 | 14 | 0.07 | 0.79 | 0.14 |
| RUD | 20 | 35 | 15 | 0.07 | 0.93 | 0 |
| SHK | 18 | 42 | 11 | 0.09 | 0.73 | 0.18 |
| ANO | 19 | 12 | 7 | 0 | 1 | 0 |
| KAL | 18 | 18 | 11 | 0 | 1 | 0 |
| KAR | 20 | 38 | 14 | 0.07 | 0.93 | 0 |
| KEF | 16 | 31 | 11 | 0.18 | 0.82 | 0 |
| KIM | 8 | 27 | 6 | 0.33 | 0.77 | 0 |
| LES | 16 | 38 | 13 | 0.15 | 0.85 | 0 |
| ORI | 18 | 43 | 13 | 0.08 | 0.84 | 0.08 |
| PIL | 15 | 17 | 11 | 0 | 1 | 0 |
| SFA | 19 | 15 | 11 | 0 | 1 | 0 |
| SKO | 18 | 27 | 13 | 0 | 1 | 0 |
| ALT | 18 | 19 | 11 | 0 | 1 | 0 |
| BER | 17 | 30 | 12 | 0.08 | 0.92 | 0 |
| GDP | 19 | 32 | 16 | 0.06 | 0.94 | 0 |
| LAN | 20 | 41 | 17 | 0.06 | 0.94 | 0 |
| LAT | 19 | 17 | 13 | 0 | 1 | 0 |
Figure 2.
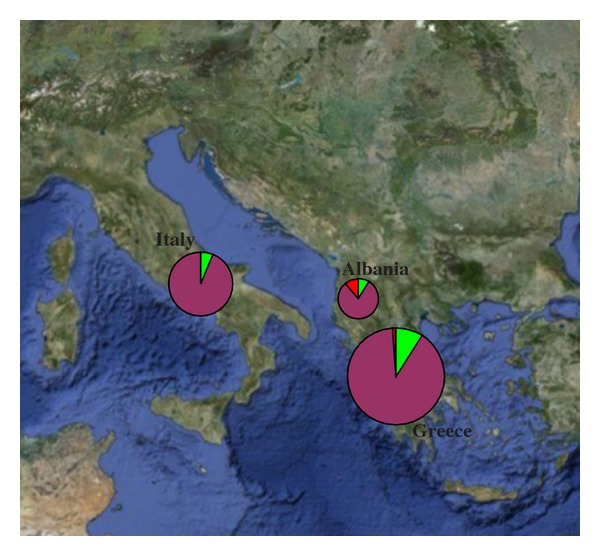
Geographical distribution of haplogroups, circles are proportional to the number of samples. (green: haplogroup A; purple: haplogroup B; red: haplogroup C).
3.3. SNP Analysis
A total of 37 SNPs identified as polymorphic on eighteen sheep breeds selected throughout Europe [39] were applied to genotype 313 individuals of three Albanian, ten Greek, and five Italian sheep breeds. After removing those found monomorphic in the selected breeds, 27 SNPs were used for the analysis.
The frequencies of the major alleles ranged from 0.99 for the locus LEP1 to 0.538 for the locus IL2_1. Except for CAST_1, LEP1, LEP2, GDF8, and PRNP_1, which show frequencies of the rare alleles of 0.035, 0.010, 0.020, 0.021, and 0.036, respectively, the remaining SNPs have rare allele frequency of greater than 5%. Observed heterozygosity of all loci ranged from 0.019 (LEP1) to 0.489 (IL2_1), with a mean of 0.250. Expected heterozygosity of the loci ranged from 0.018 (LEP1) to 0.474 (IL2_1), with a mean of 0.259.
Significant deviations from Hardy-Weinberg equilibrium (HWE) for each locus and population (P value < 0.01) were observed for nine loci. Locus BMPR was not in HWE in Skopelos breed. Locus ACVR2B_2 was not in HWE in Bardhoka population, and locus SERPINA3 was not in HWE in Karagouniko population. Loci ZP2 and KRTAP6 were not in HWE in Altamurana. PRNP_1 was not in HWE in four Italian breeds: Bergamasca, Delle Langhe, Gentile di Puglia, and Laticauda. CSN3 was not in HWE in Ruda, Skopelos, and Bergamasca populations; KRT1 in Delle Langhe, Laticauda, and Karagouniko; GHR in Karagouniko and Laticauda.
F is per population ranged from −0.102 (Shkordane) to 0.276 (Altamurana) with a mean of 0.033 (P value = 0.00010 on 9720 randomisations).
The analysis of population differentiation revealed an overall Fst of 0.048, that is, 4.8% of allelic variation accounted across breeds and 95.2% within breeds. Weir and Cockerham's [41] estimate of Fst per locus ranged from 0.0132 (IL2_2) to 0.098 (DESMIN_2), with a mean of 0.047.
Both Nei's [44] and Reynolds' [45] genetic distances were calculated (Table 6). The distances range from 0.022 (Ruda-Karagouniko) to 0.253 (Sfakia-Gentile di Puglia) using Reynolds' distances and from 0.008 (Ruda-Karagouniko) to 0.096 (Gentile di Puglia-Anogeiano) using Nei's distances. Both indices indicate Ruda and Karagouniko as the breeds with the minimum pairwise distance and show the maximum distances between the Italian breed Gentile di Puglia and Anogeiano, Karagouniko, Kefalleneas, Kymi, Lesvos, Pilioritiko, Sfakia, Skopelos, and Bergamasca breeds. The Mantel test showed correspondence between geographic and genetic distances with a P = 0.04.
Table 6.
Pairwise population matrix of Nei's [44], below, and Reynolds' [45], above, genetic distances. Breed codes are as in Table 1.
| BAR | RUD | SHK | ANO | KAL | KAR | KEF | KIM | LES | ORI | PIL | SFA | SKO | ALT | BER | GDP | LAN | LAT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BAR | ∗ | 0.060 | 0.046 | 0.065 | 0.052 | 0.054 | 0.055 | 0.104 | 0.110 | 0.117 | 0.083 | 0.061 | 0.070 | 0.107 | 0.103 | 0.167 | 0.121 | 0.055 |
| RUD | 0.018 | ∗ | 0.036 | 0.072 | 0.082 | 0.022 | 0.046 | 0.067 | 0.059 | 0.148 | 0.104 | 0.062 | 0.062 | 0.079 | 0.049 | 0.203 | 0.100 | 0.051 |
| SHK | 0.017 | 0.009 | ∗ | 0.072 | 0.072 | 0.039 | 0.058 | 0.078 | 0.087 | 0.112 | 0.063 | 0.080 | 0.042 | 0.101 | 0.062 | 0.174 | 0.123 | 0.058 |
| ANO | 0.019 | 0.029 | 0.025 | ∗ | 0.086 | 0.055 | 0.075 | 0.109 | 0.096 | 0.147 | 0.103 | 0.045 | 0.098 | 0.128 | 0.085 | 0.247 | 0.122 | 0.073 |
| KAL | 0.014 | 0.019 | 0.023 | 0.023 | ∗ | 0.078 | 0.071 | 0.127 | 0.124 | 0.076 | 0.062 | 0.107 | 0.112 | 0.142 | 0.104 | 0.159 | 0.119 | 0.092 |
| KAR | 0.013 | 0.008 | 0.009 | 0.020 | 0.013 | ∗ | 0.047 | 0.071 | 0.072 | 0.142 | 0.103 | 0.057 | 0.060 | 0.087 | 0.048 | 0.216 | 0.102 | 0.054 |
| KEF | 0.009 | 0.015 | 0.014 | 0.025 | 0.013 | 0.014 | ∗ | 0.095 | 0.091 | 0.141 | 0.106 | 0.074 | 0.076 | 0.089 | 0.091 | 0.248 | 0.124 | 0.058 |
| KIM | 0.034 | 0.024 | 0.025 | 0.039 | 0.033 | 0.021 | 0.035 | ∗ | 0.081 | 0.154 | 0.134 | 0.107 | 0.115 | 0.102 | 0.100 | 0.241 | 0.157 | 0.075 |
| LES | 0.029 | 0.017 | 0.025 | 0.029 | 0.028 | 0.021 | 0.030 | 0.028 | ∗ | 0.180 | 0.148 | 0.095 | 0.110 | 0.116 | 0.086 | 0.249 | 0.134 | 0.070 |
| ORI | 0.016 | 0.018 | 0.018 | 0.017 | 0.010 | 0.009 | 0.011 | 0.027 | 0.027 | ∗ | 0.090 | 0.169 | 0.161 | 0.182 | 0.168 | 0.152 | 0.163 | 0.140 |
| PIL | 0.024 | 0.020 | 0.013 | 0.021 | 0.025 | 0.012 | 0.020 | 0.028 | 0.025 | 0.021 | ∗ | 0.124 | 0.104 | 0.161 | 0.125 | 0.152 | 0.161 | 0.107 |
| SFA | 0.014 | 0.026 | 0.025 | 0.015 | 0.026 | 0.020 | 0.024 | 0.040 | 0.028 | 0.020 | 0.021 | ∗ | 0.098 | 0.140 | 0.097 | 0.253 | 0.122 | 0.067 |
| SKO | 0.022 | 0.021 | 0.013 | 0.040 | 0.034 | 0.020 | 0.021 | 0.039 | 0.037 | 0.031 | 0.021 | 0.034 | ∗ | 0.128 | 0.083 | 0.232 | 0.152 | 0.098 |
| ALT | 0.031 | 0.024 | 0.031 | 0.047 | 0.041 | 0.026 | 0.025 | 0.037 | 0.040 | 0.030 | 0.042 | 0.051 | 0.041 | ∗ | 0.119 | 0.226 | 0.132 | 0.073 |
| BER | 0.039 | 0.021 | 0.021 | 0.035 | 0.028 | 0.019 | 0.036 | 0.042 | 0.032 | 0.026 | 0.024 | 0.040 | 0.029 | 0.049 | ∗ | 0.233 | 0.070 | 0.083 |
| GDP | 0.077 | 0.062 | 0.077 | 0.096 | 0.086 | 0.068 | 0.079 | 0.089 | 0.083 | 0.076 | 0.083 | 0.081 | 0.094 | 0.067 | 0.082 | ∗ | 0.212 | 0.173 |
| LAN | 0.053 | 0.045 | 0.058 | 0.054 | 0.047 | 0.044 | 0.053 | 0.076 | 0.061 | 0.037 | 0.058 | 0.051 | 0.069 | 0.057 | 0.032 | 0.078 | ∗ | 0.099 |
| LAT | 0.017 | 0.017 | 0.019 | 0.025 | 0.025 | 0.016 | 0.017 | 0.030 | 0.024 | 0.022 | 0.023 | 0.021 | 0.034 | 0.026 | 0.035 | 0.064 | 0.048 | ∗ |
3.4. PCA on SNPs
Genetic relationships were also explored by means of PCA. To examine the overall pattern of population differentiation, we considered the first three axes, which cumulatively explained 48.87% of the total inertia contained in the data set (Figure 4). From PCA, it can be seen that the some breeds are quite differentiated, with good correspondence to geographical locations, even if SNPs were few (4 of the 5 Italian breeds are well separated from the main cluster). Particularly, a differentiation between northern Italian (Bergamasca and Delle Langhe) and southern Italian (Gentile di Puglia, Altamurana, and Laticauda) breeds can be seen. The projection of loci in the space formed by the first three principal components (data not shown) shows that the differentiation of outlying breeds is caused by a small number of SNPs: the Delle Langhe population is mainly affected by alleles in the MSTN gene along the second component.; the Skopelos breed position in the graph is affected by alleles in the PRP gene and the Laticauda breed (lesser) differentiation is mainly due to alleles in the CALPA and LEP genes.
Figure 4.
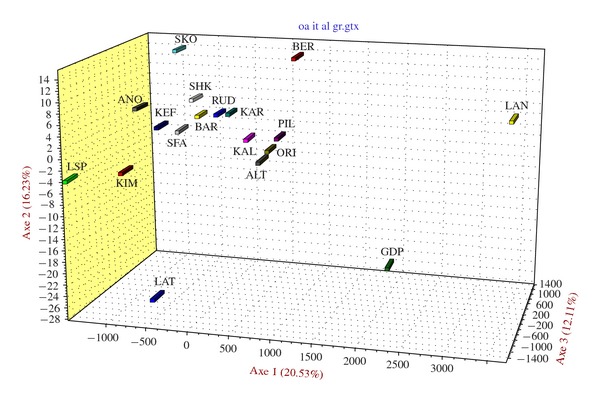
PCA analysis of the breeds assessed by SNP markers. The first three axes, cumulatively explaining 48.87% of the total inertia, were considered.
PCA scores calculated on mtDNA marker for the first 2 components were plotted on a geographic map, using the centroids of the sampling area of each breed (Figures 3(a) and 3(b)). The highest eigenvector contribution (coloured in green) was observed for Albanian and Greek breeds, as expected, even if four breeds show unexpectedly low diversity.
As for mtDNA analyses, the results of the PCA based on SNPs data were also used for making inferences about population genetic differences on the basis of their geographic distances. PCA scores for the first two principal components were plotted on a geographic map, using the centroids of the sampling area of each breed (Figures 3(c) and 3(d)). The line separating the map in two regions shows the isoline for an eigenvalue of 0. RUDA breed is an exception showing an eigenvalue above 0 but located in an area where all other breeds show lower eigenvalues. In accordance with the domestication history of the species, genetic diversity was higher in south-eastern populations than in north-western populations. The first component of Figure 3(c), in fact, shows a regular loss of genetic diversity towards North West.
4. DISCUSSION
Agriculture arose mainly within the distribution range of the wild ancestors of the most valuable livestock, such as the Fertile Crescent of southwest Asia, where early farmers were able to outcompete local hunter-gatherers. Once livestock slowly spread northwest across Europe, farming also shifted northwest from the Fertile Crescent to areas where farming had never arisen independently—first to Greece, then to Italy, and finally to northwest Europe [50]. Therefore, today the most productive farming zones do not correspond to those most productive in the past. Then, founder effect, genetic drift, and natural or artificial selection led to the formation of distinct breeds or varieties [51].
We therefore focused on the analysis of sheep of three countries aligned on this route, to evidence signs of migration. Geographical isolation, natural and artificial selection for physical or productive characters, genetic drift, mutations, and interpopulation gene flows have altered gene frequencies over many generations. The genetic diversity within and across breeds and species forms the basis of our current animal genetic resources for food production and other purposes.
The nature of the markers used for the analysis can affect the detection of geographical structuring, as suggested by Naderi et al. [52]. In fact, mtDNA informativeness is limited because it does not detect male-mediated gene flow and does not predict the nuclear genomic diversity [53]. Moreover, results may be affected by phenomena such as homoplasy, incomplete lineage sorting, effective population sizes, and sex-biased dispersal [27]. By combining markers with different modes of inheritance and rates of evolution this bias can be minimized [54].
Our mtDNA analysis shows higher levels of sheep nucleotide diversity in the South-East, which is congruent with data reported in the literature [22] and congruent with the proximity to the domestication centre. This is confirmed by eigenvector analysis, which showed high contribution to variance by Albanian and Greek breeds, even if four breeds show unexpectedly low diversity. However, this behaviour can be explained by recent isolation or selection for some traits that reduced the overall genetic diversity through bottleneck.
Very high haplotypes diversity was found in all three regions analysed (greater than 0.9), in agreement with previously published works on Portuguese breeds [9], Indian breeds [55], and Balkan sheep [56]. The major mitochondrial variation is distributed within breeds (95.04%), while it is lower among regions (0.90%) and among breeds within regions (4.06%). Phylogenetic methods were employed to examine the evolutionary history of the 18 breeds. Neighbour-joining and median-joining network revealed three of the described haplogroups, A, B, and C. The mouflon shares a haplotype with domestic sheep, as previously reported by Hiendleder et al. [33].
The SNP analysis revealed a rare allele frequency <5% for LEP1 and LEP2 loci, in agreement with those observed on a different European breed panel by Pariset et al. [39]. Observed and expected mean heterozygosity also showed similar values to those reported in the same paper. Expected heterozygosity values, which can indicate response to selection, are higher than observed heterozygosity values (Hs 0.063, 0.07, and 0.042; Ho 0.052, 0.06, and 0.038 in CALPA, PRNP-1, and GDF8, resp.).
Among the breeds tested, Altamurana showed the highest Fis value suggesting the inbreeding in this population.
Regarding the phylogeographic structure we found that the 95.2% of variation occurred within breeds indicating the weak phylogeographic structure in sheep. These data are consistent with those previously published by Kijas et al. using a different SNP panel [17]. Sheep generally do not have a strong geographic structure and show a high genetic variability within breeds.
Anyway, Mantel test analysis using SNPs revealed a correlation between genetic and geographic distance. The possibility to assess the presence of a geographic component in genetic diversity using SNPs was already reported in previous studies on sheep [39] and goats [10, 57, 58].
In the PCA, the breeds appear differentiated with 48.9% of the variance explained by the first three principal components. Also this analysis shows a good correspondence to geographical locations: the breeds remaining separated by the main group are all Italian. PCA supports therefore a westward route to Italy that could indicate that transport of animals made by sea as already proposed for cattle [11, 12] and goats [8, 59]. This is particularly plausible because small sized species as sheep are easy to transport during migration and commercial trade and can adapt easily to various environments [53, 60, 61]. A similar decrease in genetic diversity as well as an increase in the level of differentiation at the breed level from South-East to North-West in European sheep breeds, supporting the hypothesis of livestock migration from the Middle East towards western and northern Europe, was found by Peter et al. [16] and Lawson Handley et al. [15], using other nuclear markers.
The formulation of the modern breed concept during mid-1800s has caused remarkable changes in the livestock sector: large-scale production expanded and its application to breeding and husbandry practices led to the formation of well-defined breeds that were exposed to intense anthropogenic selection. The differentiation of three breeds observed using PCA analysis could be related to a recent selection, which appears to be linked to CALPA (Laticauda), PRNP_1 (Skopelos), and GDF8 (Delle Langhe) SNP markers. Gentile di Puglia breed seems influenced by both CALPA and GDF8. In particular, these two genes have an effect on conformation and therefore are an easy target of selection. Other SNPs related to meat traits were found potentially under the effect of selection and apparently not associated with production attitude of the breeds [58].
5. CONCLUSIONS
We employed mtDNA and nuclear SNPs to investigate the genetic diversity of sheep breeds of three countries of the Mediterranean basin: Albania, Greece, and Italy. Our results showed significant genetic differentiation among the sheep breeds, supported by mtDNA and by SNP. The differentiation identified by nuclear markers could indicate a reduced gene flow due to geographical isolation, associated with different flocks management, or an effect of the introduction of different stocks centuries ago (cf. Figure 3(d), showing the 2nd dimension geographic distribution of eigenvectors). In general, D-loop sequence analysis shows a pattern reflecting migrations that occurred in postneolithic and historical times, with the most divergent mtDNA lineage occurring in the southern breeds, as shown in Figure 2 and Table 5. PCA on SNP data differentiated breeds with good correspondence to geographical locations. It is interesting that the correlation between genetic and geographic distances revealed using nuclear SNPs was not confirmed by mtDNA, for which Mantel test was not significant. Our results seem to indicate a better correlation between geographic distances and autosomal markers.
ACKNOWLEDGMENTS
This work has been supported by a PRIN 2007 (prot. 2007XRBAEN_003) of Italian Ministry of University and Research. Part of the sampling was operated in the frame of the EU ECONOGENE project (QLK5-CT2001-02461). The content of the publication does not necessarily represent the views of the commission or its services. The authors wish to thank Gabriella Porcai and Paolo Ciorba for technical assistance.
APPENDIX
Members of the Econogene Consortium
Abo-Shehada Mahamoud, Parasitology Resesarch Laboratory, Faculty of Veterinary Medicine, Jordan University of Science and Technology, Ibid, Jordan; Ajmone Marsan Paolo, Istituto di Zootecnica, Università Cattolica del S. Cuore, Piacenza, Italy; Al Tarrayrah Jamil, Department of Animal Production, Aristotle Faculty of Agriculture, University of Thessaloniki, Thessaloniki, Greece; Angiolillo Antonella, Dipartimento di Scienze Animali Vegetali e dell'Ambiente, Università del Molise, Campobasso, Italy; Baret Philip, Department of Animal Genetics, Catholic University of Louvain, Louvain-la-Neuve, Belgium; Baumung Roswitha, University of Natural Resources and Applied Life Sciences, Vienna, Vienna, Austria; Beja-Pereira Albano, Laboratoire de Biologie des Populations d'Altitude, Université Joseph Fourier et Centre National de la Recherche Scientifique, Grenoble, France; Bertaglia Marco, Department of Food Economics and Consumption Studies, University of Kiel, Kiel, Germany; Bordonaro Salvatore, Dipartimento di Scienze Agronomiche e delle Produzioni Animali, Università degli Studi di Catania, Catania, Italy; Bruford Mike, School of Biosciences, Cardiff University, Cardiff, UK; Caloz Régis, Laboratoire de Systémes d'Information Géographique (LASIG), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland; Canali Gabriele, Istituto di Economia Agro-Alimentare, Università Cattolica del S. Cuore, Piacenza, Italy; Canon Javier, Universidad Computense de Madrid, Madrid, Spain; Cappuccio Irene, Dipartimento di Produzioni Animali, Università della Tuscia, Viterbo, Italy; Carta Antonello, Istituto Zootecnico e Caseario per la Sardegna, Sassari, Italy; Cicogna Mario, Istituto di Zootecnia Generale, Università degli Studi di Milano, Milano; Italy, Crepaldi Paola, Istituto di Zootecnia Generale, Università degli Studi di Milano, Milano, Italy; Dalamitra Stella, Biological Sciences, University of East Anglia, Norwich, UK; Daniela Krugmann, Institut für Tierzucht und Haustiergenetik, Justus Liebig-Universität of Gießen, Gießen, Germany; Dobi Petrit, Department of Animal Production, Faculty of Agriculture, Tirana, Albania; Dominik Popielarczyk, Department of Sheep and Goat Breeding, Warsaw Agricultural University, Warsaw, Poland; Dunner Susana, Universidad Computense de Madrid, Madrid, Spain; D'Urso Giuseppe, Dipartimento di Scienze Agronomiche e delle Produzioni Animali, Università degli Studi di Catania, Catania, Italy; El-Barody M. A. A., Animal Production Department, Faculty of Agriculture, Minia University, Minia, Egypt; England Phillip, Laboratoire de Biologie des Populations d'Altitude, Université Joseph Fourier et CNRS, Grenoble, France; Erhardt Georg, Institut für Tierzucht und Haustiergenetik, Justus Liebig-Universität of Gießen, Gießen, Germany; Ertugrul Okan, Veterinary Faculty, Ankara University, Ankara, Turkey; Eva-Maria Prinzenberg, Institut für Tierzucht und Haustiergenetik, Justus Liebig-Universität of Gießen, Gießen, Germany; Eveline Ibeagha-Awemu, Institut für Tierzucht und Haustiergenetik, Justus Liebig-Universität of Gießen, Gießen, Germany; Ewa Strzelec, Department of Sheep and Goat Breeding, Warsaw Agricultural University, Warsaw, Poland; Fadlaoui Aziz, Department of Animal Genetics, Catholic University of Louvain, Louvain-la-Neuve, Belgium; Fornarelli Francesca, Istituto di Zootecnia Generale, Università degli Studi di Milano, Milano, Italy; Garcia David, Universidad Computense de Madrid, Madrid, Spain; Georgoudis Andreas, Department of Animal Production, Faculty of Agriculture, Aristotle University of Thessaloniki, Thessaloniki, Greece; Gesine Lühken, Institut für Tierzucht und Haustiergenetik, Justus Liebig-Universität of Gießen, Gießen, Germany; Giovenzana Stefano, Istituto di Zootecnia Generale, Università degli Studi di Milano, Milano, Italy; Gutscher Katja, Institut für Tierzucht und Haustiergenetik, Justus Liebig-Universität of Gießen, Gießen, Germany; Hewitt Godfrey, Biological Sciences, University of East Anglia, Norwich, UK; Hoda Anila, Department of Animal Production, Faculty of Agriculture, Tirana, Albania; Horst Brandt, Institut für Tierzucht und Haustiergenetik, Justus Liebig-Universität of Gießen, Gießen, Germany; Istvan Anton, Research Institute for Animal Breeding and Nutrition, Department of Genetics, Herceghalom, Hungary; Juma Gabriela, School of Biosciences, Cardiff University, Cardiff, UK; Joost Stéphane, Laboratoire de Systèmes d'Information Géographique (LASIG), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland; Jones Sam, The Sheep Trust, Biology Area-8, University of York, York, England; Karetsou Katerina, Department of Animal Production, Faculty of Agriculture, Aristotle University of Thessaloniki, Thessaloniki, Greece; Kliambas Georgios, Department of Animal Production, Faculty of Agriculture, Aristotle University of Thessaloniki, Thessaloniki, Greece; Koban Evren, Department of Biology, Middle East Technical University, Ankara, Turkey; Kutita Olga, Department of Animal Production, Faculty of Agriculture, Aristotle University of Thessaloniki, Thessaloniki, Greece; Lazlo Fesus, Research Institute for Animal Breeding and Nutrition, Department of Genetics, Herceghalom, Hungary; Lenstra Johannes A., Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands; Ligda Christina, National Agricultural Research Foundation, Thessaloniki, Greece; Lipsky Shirin, Institut für Tierzucht und Haustiergenetik, Justus Liebig-Universität of Gießen, Gießen, Germany; Luikart Gordon, Laboratoire de Biologie des Populations d'Altitude, Université Joseph Fourier et CNRS, Grenoble, France; Marie-Louise Glowatzki, Institut für Genetik Ernährung und Haltung von Haustieren, Universität Bern, Bern, Switzerland; Marilli Marta, Istituto di Zootecnia Generale, Università degli Studi di Milano, Milano, Italy; Marletta Donata, Dipartimento di Scienze Agronomiche e delle Produzioni Animali, Università degli Studi di Catania, Catania, Italy; Milanesi Elisabetta, Istituto di Zootecnica, Università Cattolica del S. Cuore, Piacenza, Italy; Negrini Riccardo, Istituto di Zootecnica, Università Cattolica del S. Cuore, Piacenza, Italy; Nijman, Isaäc J., Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands; Obexer-Ruff Gabriela, Institut für Genetik Ernährung und Haltung von Haustieren, Universität Bern, Bern, Switzerland; Papachristoforou Christos, Agricultural Research Institute, Nicosia, Cyprus; Pariset Lorraine, Dipartimento di Produzioni Animali, Università della Tuscia, Viterbo, Italy; Pellecchia Marco, Istituto di Zootecnica, Università Cattolica del S. Cuore, Piacenza, Italy; Peter Christina, Institut für Tierzucht und Haustiergenetik, Justus Liebig-Universität of Gießen, Gießen, Germany; Perez Trinidad, School of Biosciences, Cardiff University, Cardiff, UK; Pietrolà Emilio, Dipartimento di Scienze Animali, Vegetali e dell'Ambiente, Università del Molise, Campobasso, Italy; Pilla Fabio, Dipartimento di Scienze Animali, Vegetali e dell'Ambiente, Università del Molise, Campobasso, Italy; Roman Niznikowski, Department of Sheep and Goat Breeding, Warsaw Agricultural University, Warsaw, Poland; Roosen Jutta, Department of Food Economics and Consumption Studies, University of Kiel, Kiel, Germany; Scarpa Riccardo, Environment Department, University of York, York, UK; Sechi Tiziana, Istituto Zootecnico e Caseario per la Sardegna, Sassari, Italy; Taberlet Pierre, Laboratoire d'Ecologie Alpine (LECA), Université Joseph Fourier, Grenoble, France; Taylor Martin, Biological Sciences, University of East Anglia, Norwich, UK; Togan Inci, Department of Biology, Middle East Technical University, Ankara, Turkey; Trommetter Michel, Institut de recherché Agronomique, Unité d'Économie et Sociologie Rurales, Grenoble, France; Valentini Alessio, Dipartimento di Produzioni Animali, Università della Tuscia, Viterbo, Italy; Van Cann, Lisette M., Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands; Vlaic Augustin, Department of Animal Genetics, Faculty of Zootechnics, University of Cluj-Napoca, Cluj-Napoca, Romania; Wiskin Louise, Biological Sciences, University of East Anglia, Norwich, UK; Zundel Stephanie, Laboratoire d'Ecologie Alpine (LECA), Université Joseph Fourier, Grenoble, France.
References
- 1.Vigne JD, Carrère I, Guilaine J. Unstable status of early domestic ungulates in the Near East: the example of Shillourokambos (Cyprus, IX-VIIIth millennia Cal. B.C.) Bulletin de Correspondance Hellenique. 2003;43, supplement:239–251. [Google Scholar]
- 2.Nadler CF, Korobitsina KV, Hoffmann RS, Vorontsov NN. Cytogenetic differentiation, geographic distribution, and domestication in Palaearctic sheep (Ovis) Zeitschrift für Säugetierkunde. 1973;38:109–125. [Google Scholar]
- 3.Bunch TD, Foote WC, Spillett JJ. Translocations of acrocentric chromosomes and their implications in the evolution of sheep (Ovis) Cytogenetics and Cell Genetics. 1976;17(3):122–136. doi: 10.1159/000130704. [DOI] [PubMed] [Google Scholar]
- 4.Dobney K, Larson G. Genetics and animal domestication: new windows on an elusive process. Journal of Zoology. 2006;269(2):261–271. [Google Scholar]
- 5.Zeder MA. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(33):11597–11604. doi: 10.1073/pnas.0801317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zilhão J. Radiocarbon evidence for maritime pioneer colonization at the origins of farming in west Mediterranean Europe. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):14180–14185. doi: 10.1073/pnas.241522898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tapio M, Marzanov N, Ozerov M, et al. Sheep mitochondrial DNA variation in European, Caucasian, and Central Asian areas. Molecular Biology and Evolution. 2006;23(9):1776–1783. doi: 10.1093/molbev/msl043. [DOI] [PubMed] [Google Scholar]
- 8.Pereira F, Davis SJM, Pereira L, McEvoy B, Bradley DG, Amorim A. Genetic signatures of a Mediterranean influence in Iberian Peninsula sheep husbandry. Molecular Biology and Evolution. 2006;23(7):1420–1426. doi: 10.1093/molbev/msl007. [DOI] [PubMed] [Google Scholar]
- 9.Pereira F, Pereira L, Van Asch B, Bradley DG, Amorim A. The mtDNA catalogue of all Portuguese autochthonous goat (Capra hircus) breeds: high diversity of female lineages at the western fringe of European distribution. Molecular Ecology. 2005;14(8):2313–2318. doi: 10.1111/j.1365-294X.2005.02594.x. [DOI] [PubMed] [Google Scholar]
- 10.Pariset L, Cuteri A, Ligda C, Ajmone-Marsan P, Valentini A. Geographical patterning of sixteen goat breeds from Italy, Albania and Greece assessed by Single Nucleotide Polymorphisms. BMC Ecology. 2009;9, article 20 doi: 10.1186/1472-6785-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cymbron T, Freeman AR, Malheiro MI, Vigne JD, Bradley DG. Microsatellite diversity suggests different histories for Mediterranean and Northern European cattle populations. Proceedings of the Royal Society B. 2005;272(1574):1837–1843. doi: 10.1098/rspb.2005.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellecchia M, Negrini R, Colli L, et al. The mystery of Etruscan origins: novel clues from Bos taurus mitochondrial DNA. Proceedings of the Royal Society B. 2007;274(1614):1175–1179. doi: 10.1098/rspb.2006.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meadows JRS, Hanotte O, Drögemüller C, et al. Globally dispersed Y chromosomal haplotypes in wild and domestic sheep. Animal Genetics. 2006;37(5):444–453. doi: 10.1111/j.1365-2052.2006.01496.x. [DOI] [PubMed] [Google Scholar]
- 14.Meadows JRS, Kijas JW. Re-sequencing regions of the ovine Y chromosome in domestic and wild sheep reveals novel paternal haplotypes. Animal Genetics. 2009;40(1):119–123. doi: 10.1111/j.1365-2052.2008.01799.x. [DOI] [PubMed] [Google Scholar]
- 15.Lawson Handley LJ, Byrne K, Santucci F, et al. Genetic structure of European sheep breeds. Heredity. 2007;99(6):620–631. doi: 10.1038/sj.hdy.6801039. [DOI] [PubMed] [Google Scholar]
- 16.Peter C, Bruford M, Perez T, Dalamitra S, Hewitt G, Erhardt G. Genetic diversity and subdivision of 57 European and Middle-Eastern sheep breeds. Animal Genetics. 2007;38(1):37–44. doi: 10.1111/j.1365-2052.2007.01561.x. [DOI] [PubMed] [Google Scholar]
- 17.Kijas JW, Townley D, Dalrymple BP, et al. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE. 2009;4(3) doi: 10.1371/journal.pone.0004668. Article ID e4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chessa B, Pereira F, Arnaud F, et al. Revealing the history of sheep domestication using retrovirus integrations. Science. 2009;324(5926):532–536. doi: 10.1126/science.1170587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen SY, Duan ZY, Sha T, Xiangyu J, Wu SF, Zhang YP. Origin, genetic diversity, and population structure of Chinese domestic sheep. Gene. 2006;376(2):216–223. doi: 10.1016/j.gene.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Pedrosa S, Uzun M, Arranz JJ, Gutiérrez-Gil B, San Primitivo F, Bayón Y. Evidence of three maternal lineages in near eastern sheep supporting multiple domestication events. Proceedings of the Royal Society B. 2005;272(1577):2211–2217. doi: 10.1098/rspb.2005.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meadows JRS, Li K, Kantanen J, et al. Mitochondrial sequence reveals high levels of gene flow between breeds of domestic sheep from Asia and Europe. Journal of Heredity. 2005;96(5):494–501. doi: 10.1093/jhered/esi100. [DOI] [PubMed] [Google Scholar]
- 22.Meadows JRS, Cemal I, Karaca O, Gootwine E, Kijas JW. Five ovine mitochondrial lineages identified from sheep breeds of the near east. Genetics. 2007;175(3):1371–1379. doi: 10.1534/genetics.106.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood NJ, Phua SH. Variation in the control region sequence of the sheep mitochondrial genome. Animal Genetics. 1996;27(1):25–33. doi: 10.1111/j.1365-2052.1996.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 24.Hiendleder S, Lewalski H, Wassmuth R, Janke A. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. Journal of Molecular Evolution. 1998;47(4):441–448. doi: 10.1007/pl00006401. [DOI] [PubMed] [Google Scholar]
- 25.Hiendleder S, Kaupe B, Wassmuth R, Janke A. Molecular analysis of wild and domestic sheep questions current nomenclature and provides evidence for domestication from two different subspecies. Proceedings of the Royal Society B. 2002;269(1494):893–904. doi: 10.1098/rspb.2002.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo J, Du LX, Ma YH, et al. A novel maternal lineage revealed in sheep (Ovis aries) Animal Genetics. 2005;36(4):331–336. doi: 10.1111/j.1365-2052.2005.01310.x. [DOI] [PubMed] [Google Scholar]
- 27.Flanders J, Jones G, Benda P, et al. Phylogeography of the greater horseshoe bat, Rhinolophus ferrumequinum: contrasting results from mitochondrial and microsatellite data. Molecular Ecology. 2009;18(2):306–318. doi: 10.1111/j.1365-294X.2008.04021.x. [DOI] [PubMed] [Google Scholar]
- 28.Heckel G, Burri R, Fink S, Desmet JF, Excoffier L. Genetic structure and colonization processes in European populations of the common vole, Microtus arvalis. Evolution. 2005;59(10):2231–2242. [PubMed] [Google Scholar]
- 29.Bryja J, Smith C, Konečný A, Reichard M. Range-wide population genetic structure of the European bitterling (Rhodeus amarus) based on microsatellite and mitochondrial DNA analysis. Molecular Ecology. 2010;19(21):4708–4722. doi: 10.1111/j.1365-294X.2010.04844.x. [DOI] [PubMed] [Google Scholar]
- 30.Tollefsrud MM, Sønstebø JH, Brochmann C, Johnsen O, Skrøppa T, Vendramin GG. Combined analysis of nuclear and mitochondrial markers provide new insight into the genetic structure of North European Picea abies. Heredity. 2009;102(6):549–562. doi: 10.1038/hdy.2009.16. [DOI] [PubMed] [Google Scholar]
- 31.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Molecular Ecology. 2004;13(4):729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 32.Gonçalves GL, Moreira GRP, Freitas TRO, Hepp D, Passos DT, Weimer TA. Mitochondrial and nuclear DNA analyses reveal population differentiation in Brazilian Creole sheep. Animal Genetics. 2010;41(3):308–310. doi: 10.1111/j.1365-2052.2009.01986.x. [DOI] [PubMed] [Google Scholar]
- 33.Hiendleder S, Mainz K, Plante Y, Lewalski H. Analysis of mitochondrial DNA indicates that domestic sheep are derived from two different ancestral maternal sources: no evidence for contributions from urial and argali sheep. Journal of Heredity. 1998;89(2):113–120. doi: 10.1093/jhered/89.2.113. [DOI] [PubMed] [Google Scholar]
- 34.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 35.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. doi: 10.1093/molbev/msr121. Molecular Biology and Evolution. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 38.Joost S, Colli L, Baret PV, et al. Integrating geo-referenced multiscale and multidisciplinary data for the management of biodiversity in livestock genetic resources. Animal Genetics. 2010;41(1):47–63. doi: 10.1111/j.1365-2052.2010.02037.x. [DOI] [PubMed] [Google Scholar]
- 39.Pariset L, Cappuccio I, Ajmone-Marsan P, et al. Characterization of 37 breed-specific single-nucleotide polymorphisms in sheep. Journal of Heredity. 2006;97(5):531–534. doi: 10.1093/jhered/esl020. [DOI] [PubMed] [Google Scholar]
- 40.Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3) 2001, http://www2.unil.ch/popgen/softwares/fstat.htm.
- 41.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 42.Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6(1):288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. (1996–2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5171, Université de Montpellier II, Montpellier, France.
- 44.Nei M. Genetic distance between populations. American Naturalist. 1972;106(949):p. 283. [Google Scholar]
- 45.Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105(3):767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu K, Muse SV. PowerMaker: an integrated analysis environment for genetic maker analysis. Bioinformatics. 2005;21(9):2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 47.Nei M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the United States of America. 1973;70(12):3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nei M. Molecular Evolutionary Genetics. New York, NY, USA: Columbia University Press; 1987. [Google Scholar]
- 49.Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129(2):555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418(6898):700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 51.Berthouly C, Bed’Hom B, Tixier-Boichard M, et al. Using molecular and multivariate methods to study the genetic diversity on local European and Asian chickens breeds. Animal Genetics. 2008;39:121–129. doi: 10.1111/j.1365-2052.2008.01703.x. [DOI] [PubMed] [Google Scholar]
- 52.Naderi S, Rezaei HR, Taberlet P, et al. Large-scale mitochondrial DNA analysis of the domestic goat reveals six haplogroups with high diversity. PLoS ONE. 2007;2(10) doi: 10.1371/journal.pone.0001012. Article ID e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruford MW, Bradley DG, Luikart G. DNA markers reveal the complexity of livestock domestication. Nature Reviews Genetics. 2003;4(11):900–910. doi: 10.1038/nrg1203. [DOI] [PubMed] [Google Scholar]
- 54.Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society B. 2004;359(1442):183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pardeshi VC, Kadoo NY, Sainani MN, Meadows JRS, Kijas JW, Gupta VS. Mitochondrial haplotypes reveal a strong genetic structure for three Indian sheep breeds. Animal Genetics. 2007;38(5):460–466. doi: 10.1111/j.1365-2052.2007.01636.x. [DOI] [PubMed] [Google Scholar]
- 56.Ćinkulov M, Popovski Z, Porcu K, et al. Genetic diversity and structure of the West Balkan Pramenka sheep types as revealed by microsatellite and mitochondrial DNA analysis. Journal of Animal Breeding and Genetics. 2008;125(6):417–426. doi: 10.1111/j.1439-0388.2008.00742.x. [DOI] [PubMed] [Google Scholar]
- 57.Cappuccio I, Pariset L, Ajmone-Marsan P, et al. Allele frequencies and diversity parameters of 27 single nucleotide polymorphisms within and across goat breeds. Molecular Ecology Notes. 2006;6(4):992–997. [Google Scholar]
- 58.Pariset L, Cappuccio I, Joost S, et al. Characterization of single nucleotide polymorphisms (SNPs) in sheep and their variation as an evidence of selection. Animal Genetics. 2006;37:290–292. doi: 10.1111/j.1365-2052.2006.01441.x. [DOI] [PubMed] [Google Scholar]
- 59.Cañón J, García D, García-Atance MA, et al. Geographical partitioning of goat diversity in Europe and the Middle East. Animal Genetics. 2006;37(4):327–334. doi: 10.1111/j.1365-2052.2006.01461.x. [DOI] [PubMed] [Google Scholar]
- 60.Luikart G, Gielly L, Excoffier L, Vigne JD, Bouvet J, Taberlet P. Multiple maternal origins and weak phylogeographic structure in domestic goats. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(10):5927–5932. doi: 10.1073/pnas.091591198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernández H, Hughes S, Vigne JD, et al. Divergent mtDNA lineages of goats in an Early Neolithic site, far from the initial domestication areas. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(42):15375–15379. doi: 10.1073/pnas.0602753103. [DOI] [PMC free article] [PubMed] [Google Scholar]


