Abstract
Stop codons have been exploited for genetic incorporation of unnatural amino acids (Uaas) in live cells, but the efficiency is low possibly due to competition from release factors, limiting the power and scope of this technology. Here we show that the reportedly essential release factor 1 can be knocked out from Escherichia coli by fixing release factor 2. The resultant strain JX33 is stable and independent, and reassigns UAG from a stop signal to an amino acid when a UAG-decoding tRNA/synthetase pair is introduced. Uaas were efficiently incorporated at multiple UAG sites in the same gene without translational termination in JX33. We also found that amino acid incorporation at endogenous UAG codons is dependent on RF1 and mRNA context, which explains why E. coli tolerates apparent global suppression of UAG. JX33 affords a unique autonomous host for synthesizing and evolving novel protein functions by enabling Uaa incorporation at multiple sites.
INTRODUCTION
The canonical genetic code specifies 61 sense codons for amino acids and 3 nonsense codons for stop signals in protein translation. Although the canonical code is preserved in virtually every organism on earth, small deviations in codon assignments have been discovered in the mitochondrial and nuclear codes of an increasing number of organisms1,2. These include the reassignment of sense codons from one amino acid to another and the reassignment between nonsense and sense codons. In the laboratory, stop codons have been exploited for the incorporation of both natural and unnatural amino acids (Uaas) into proteins. Natural suppressor tRNAs decoding stop codons as common amino acids have been identified in E. coli and other organisms3,4. Orthogonal tRNA/synthetase pairs have been engineered to incorporate various Uaas into proteins in response to a stop codon5,6,7.
A major limitation of using a stop codon to encode Uaas is that the incorporation efficiency is low; this low efficiency can be inherent because the suppressor tRNA has to compete with endogenous release factors (RFs), whose native function is to recognize stop codons and terminate translation. The assignment of the stop codon is thus ambiguous, being a stop signal and an Uaa simultaneously, which severely limits the full exploitation and potential of this technology. Besides decreasing the Uaa incorporation efficiency, RF competition results in truncated protein products, which may interfere with target protein function or be deleterious to the host cell. Low incorporation efficiency also prevents the synthesis of proteins containing Uaas at multiple sites. Protein yields drop precipitously with the addition of even a second stop codon. Therefore, it is currently infeasible to efficiently synthesize proteins with Uaa modifications at multiple sites and to explore novel protein and organism functions through experimental evolution involving Uaas.
Another important yet unaddressed question is related to stop codons used by endogenous genes for translational termination in host cells. When a tRNA/synthetase pair is introduced to suppress a stop codon in exogenous genes, it is unclear whether and to what extent the legitimate stop codon in endogenous genes is suppressed. Would the extended proteins create pressure to host cells, and would host cells tolerate or adapt to such a challenge? These questions are not only important for overcoming the restrictions currently imposed on the Uaa incorporation methodology, but also for understanding how an organism copes with and eventually fixes codon reassignments during evolution.
To begin addressing these questions, we aim to fully reassign the amber codon UAG from the stop signal to an amino acid in E. coli. In prokaryotes, stop codons are recognized by two RFs, RF1 for UAA/UAG and RF2 for UAA/UGA8. To achieve full reassignment of UAG, RF1 must be removed from the system. However, the prfA gene encoding the RF1 is reportedly essential for E. coli survival9,10. Here we show that RF1 gene can be knocked out of the E. coli genome by fixing the expression of RF2. The RF1 knockout strain has been stable and sustainable for over 3 years. This new autonomous strain enables the genetic incorporation of various natural and unnatural amino acids into proteins at numerous UAG sites without being terminated. Moreover, we found whether an amino acid is incorporated by an orthogonal tRNA/synthetase pair at a legitimate UAG codon of endogenous genes is strongly dependent on RF1, and that mRNA context of the UAG codon determines the translation outcome.
RESULTS
Generation of an autonomous RF1 knockout strain
UAG is the least used stop codon in E. coli, terminating only ~7% of E. coli genes11. Although RF1 recognizes UAA and UAG, UAA can also be read by RF2. However, RF1 is considered essential in E. coli10, and only conditionally lethal knockouts have been described9. This indispensability suggests that either accurate stoppage at UAG is essential for E. coli, or stoppage of the numerous UAA is impossible with RF2 alone. Consistently, our initial attempts to directly knock out prfA, the RF1-encoding gene, in the common DH10β strain using a chloramphenicol resistance (CmR) knock-in cassette failed. We then tried a two-step strategy: prfA was expressed in DH10β exogenously using a plasmid curable at 37 °C or higher temperature. In the presence of this plasmid, the genomic prfA of DH10β was successfully knocked out. However, when these cells were transferred to higher temperatures to cure the prfA expressing plasmid, none of them survived. These experiments indicate that it is unfeasible to directly knock out the prfA gene from the DH10β genome.
The tmRNA surveillance system for translational stalling could cause lethality after prfA knockout. In the absence of RF1-mediated peptide release, ribosome will be stalled on UAG-ending mRNAs. The alanyl-tmRNA can recognize and enter the stalled ribosome, transfer the nascent peptide onto itself, and resume translation to append a degradation tag12. Tagged polypeptides are subsequently degraded. Degradation of an overwhelming number of proteins could induce cell death. We reasoned that a tmRNA-deficient E. coli strain would bypass degradation of too many proteins. However, our attempts to knock out prfA in the tmRNA-deficient strain (X90 ssrA1::cat)13 also failed.
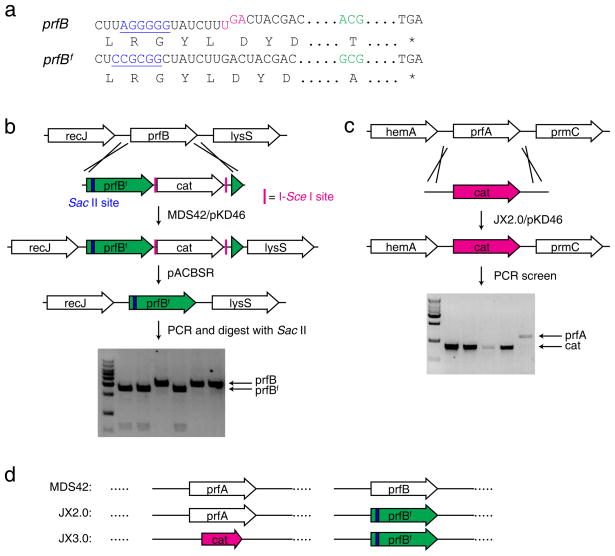
RF2 expression in E. coli is tightly autoregulated by an in-frame UGA codon in its mRNA that requires a +1 frameshift to generate full-length RF214. Interestingly, E. coli strains derived from K-12 contain a peculiar mutation (Ala246Thr) in RF2 that lowers its release activity for the UAA codon 5-fold15. Once RF1 is removed in K-12 derivatives, RF2 expressed at the endogenous level may be unable to efficiently terminate all UAA codons. To relieve the potentially increasing burden imposed on RF2, we removed the in-frame UAG autoregulation element and mutated residue 246 back to Ala to generate the “fixed” prfBf gene (Fig. 1a). We used E. coli MDS42 as the parental strain16, because the deletion of nearly 700 nonessential genes in MDS42 may alleviate the termination load imposed on RF2. The RF2-encoding gene prfB in MDS42 cells was first replaced by the prfBf gene coupled to a CmR cassette using the λ-red recombination system17 (Fig. 1b). The CmR cassette was subsequently excised from Cm resistant clones using the pACBSR plasmid for markerless insertions18 (see Supplementary Methods). The resultant strain, JX2.0, has the prfB replaced by prfBf. Knockout of prfA was then attempted in JX2.0 by electroporating the linear CmR knock-in cassette with flanking sequence identical to those of prfA (Fig. 1c). Genomic PCR screening of Cm resistant colonies showed that they contained the CmR cassette at the endogenous prfA locus, indicating that RF1 was successfully knocked out (Fig. 1c). The RF1-knocked out strain was named as JX3.0 (Fig. 1d).
Figure 1. RF1 can be knocked out from E. coli after RF2 is fixed.
(a) Features of the RF2-encoding prfB gene in K-12 E. coli strains: an in-frame UGA stop codon (magenta) for autoregulation of RF2 expression and the Ala246Thr mutation (green) impairing RF2’s release activity for the UAA codon. In prfBf, the UGA regulation was removed and residue 246 reverted to Ala. A Shine-Dalgarno like sequence (blue) in prfB was silently mutated to a Sac II site (blue) in prfBf to facilitate the screening of prfBf knock-in. (b) Generation of the JX2.0 strain. The prfB gene in MDS42 was first replaced with prfBf followed by a Cm resistant cat cassette. The cat cassette was subsequently removed by pACBSR (see Supplementary Methods). (c) Generation of the JX3.0 strain. The RF1-encoding prfA gene was successfully knocked out with a cat cassette in JX2.0 to afford JX3.0. (d) Illustration of main features of the three strains, MDS42, JX2.0 and JX3.0.
We measured the growth rates of JX2.0 and JX3.0 cells (Supplementary Results, Supplementary Fig. 1). JX2.0 grew with a doubling time of 27.1 minutes in the Luria-Bertani media, similar to the parental DH10β. JX3.0 doubled every 74.4 minutes, significantly slower than JX2.0 and DH10β, indicating that RF1 knockout puts pressure on cell growth. However, JX3.0 cells are viable and sustainable, suggesting that RF1 is not essential for their survival. Interestingly, out of numerous JX3.0 colonies we found a single colony, JX33, growing faster than others with a doubling rate (27.5 minutes) close to JX2.0.
Genomic sequencing of JX2.0 and JX3.0 strains
To confirm the introduced genomic changes and to detect any potential mutations compensatory for RF1 knockout, we performed full genomic sequencing on JX2.0 and JX3.0, and compared to E. coli K-12 MG1655. For JX3.0, colonies with slow (JX31) and fast (JX33) growth rate were both sequenced. Both JX2.0 and JX3.0 showed gene deletions identical to the parental MDS42 strain, a multiple-deletion descendent of MG165516. They also both contained the exact changes we made in prfBf. The knockout of prfA by the CmR cassette in JX3.0 was confirmed, and this is the only deletion difference between JX2.0 and JX3.0. No other differences and mutations were found between JX2.0, JX31 and MG1655. These results clearly indicate that RF1 can be knocked out from JX2.0 without incurring compensatory mutations in other genes, indicating that RF1 is nonessential in JX2.0.
Two single nucleotide polymorphisms (SNPs) were found between JX2.0 and JX33 (Supplementary Fig. 2a). One is a silent mutation in the coding region of ypdE and the other results in an amino acid change (A293E) in RF2. The A293E mutation has not been discovered in any previous complementation screens for RF1 deficiency19–22. We then determined if the A293E mutation in RF2 is sufficient for rescuing RF1 function in E. coli. We replaced the RF2 gene of a temperature sensitive RF1 (tsRF1) strain (MRA8)9 with the RF2(A293E) gene from JX33 using established procedures17 to create strain MRA8 A293E. However, no difference in growth phenotype was observed between the parental (MRA8) and mutant (MRA8 A293E) strains (Supplementary Fig. 2b), indicating that the A293E mutation is not able to rescue the RF1 temperature sensitive phenotype in E. coli. As the only difference between JX31 and JX33 is the A293E mutation, this mutation is likely responsible for the fast growth phenotype of JX33.
Incorporation of tyrosine at multiple UAG sites in JX33
Deletion of RF1 from E. coli presumably changes the UAG codon from the stop signal to a blank codon. Introduction of an orthogonal tRNA/synthetase pair to recognize the UAG codon in JX3.0 would translate UAG with the amino acid cognate for the synthetase, essentially reassigning UAG to a sense codon. In the absence of RF1 competition, incorporation efficiency at UAG should be significantly increased, and multiple UAGs should be suppressible simultaneously. We tested these hypotheses using the fast-growing JX33 strain.
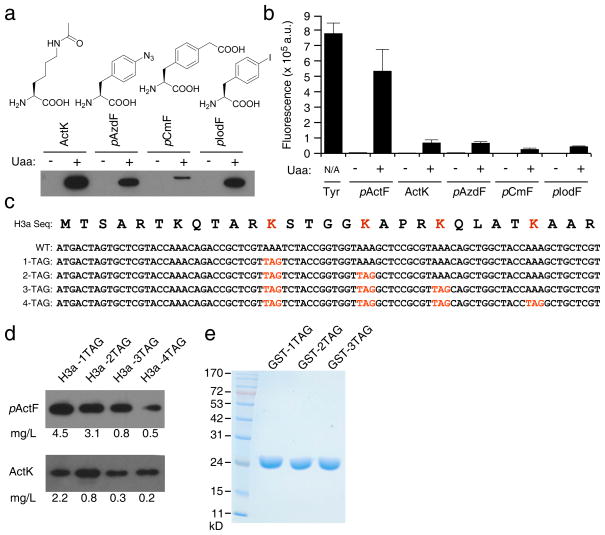
A single All-in-One expression plasmid (pAIO) was constructed to contain an orthogonal amber suppressor tRNA, an orthogonal aminoacyl-tRNA synthetase, and an EGFP reporter with an N-terminal hexahistidine (His6) tag (Fig. 2a). TAG mutations were introduced into tyrosyl sites in the EGFP gene to create 1-, 2-, 3-, and 6-TAG EGFP reporters. We first tested incorporation efficiency at UAG sites using the orthogonal derived from archaebacterium Methanococcus jannaschii5, which inserts tyrosine at UAG codons. In JX2.0, this system showed a reduction of EGFP protein yields with each additional UAG, and no full-length EGFP in the 6-TAG reporter was detected on Western blot (Fig. 2b). In stark contrast, JX33 showed increased levels of protein and no reduction in EGFP protein yields across all TAG mutants. For the 1-TAG reporter, the protein expression level in JX33 increased to 254% of in JX2.0. For EGFP with 6 TAG sites, JX2.0 yielded no protein whereas JX33 afforded 6.8 mg/L, which reached 46% of wild-type EGFP without any TAG. In-cell fluorescence intensity was measured for each mutant using fluorometry. In JX2.0, fluorescence intensity decreased with each additional TAG, while JX33 fluorescence was similar among all mutants and much higher than in JX2.0 (Fig. 2c). These results demonstrate that JX33 has markedly higher incorporation efficiency for tyrosine at UAG sites than the parental JX2.0. In addition, the knockout of RF1 allows multiple UAG sites to be efficiently suppressed with tyrosine in JX33, but not in the RF1-containing JX2.0.
Figure 2. RF1 knockout enables incorporation of natural or unnatural amino acids at multiple UAG sites in JX33.
(a) Features of the All-in-One plasmid and structures of Tyr and pActF. (b) Western analysis of EGFP expression when UAG codons were decoded as Tyr by pAIO-TyrRS. The same number of cells were used for each sample, and the blot was probed with a penta-His antibody. (c) In-cell fluorescence assay of EGFP intensity when UAG codons were decoded as Tyr by pAIO-TyrRS. The same number of cells was used for each sample. Measurement was performed on 3 independent batches of cells and error bars represent s.e.m.. (d) Western analysis of EGFP expression when UAG codons were decoded as the Uaa pActF by pAIO-LW1RS. Conditions are the same in (b). For each sample, a duplicate of cultures were grown in the presence or absence of pActF in the growth media. (e) In-cell fluorescence assay of EGFP intensity when UAG codons were decoded as pActF by pAIO-LW1RS. Measurements were performed as in (c), n = 3, error bars represent s.e.m. (f) Fluorescence images of cells when UAG was decoded as Tyr or pActF. (g) SDS-PAGE analysis of EGFP proteins with pActF incorporated in JX33 or in BL21(DE3) coexpressing plasmid pET-L11C26. The same pAIO-LW1RS plasmids containing 1- or 3-TAG EGFP mutant gene were used in both cells. EGFP was purified with Ni-NTA chromatography. Loading was normalized to the same number of cells, and the gel was stained with Coomassie blue.
Incorporation of Uaa pActF at multiple UAG sites in JX33
To determine if RF1-deletion permits an Uaa to be incorporated at multiple UAG sites, we used the orthogonal pair23 to incorporate Uaa p-acetylphenylalanine (pActF) into EGFP. In JX2.0, only the 1-TAG EGFP reporter produced full-length EGFP protein; no full-length EGFP was detected in reporters containing 2-, 3-, or 6-TAGs by Western blot (Fig. 2d). The evolved LW1RS is less active than wt TyrRS in aminoacylation23, consistent with the observation that small amounts of EGFP were detected in the 2- and 3-TAG reporters in JX2.0 with but not with . In JX33, EGFP expression with pActF incorporated at a single TAG site was doubled compared to in JX2.0. Large amounts of full-length EGFP were also produced in the 2- and 3-TAG reporters using the pair. Notably, protein yield did not diminish when the number of UAG codons increased from 1 to 3 (Supplementary Table 1). Even for EGFP with 6 TAG sites, 0.5 mg/L of pActF containing EGFP was purified from JX33.
In-cell fluorescence measurement confirmed that pActF was incorporated into EGFP at multiple sites only in JX33 and not in JX2.0 (Fig. 2e and 2f). Green fluorescence was detected for 1-TAG reporter only in JX2.0, but for 1-, 2-, and 3-TAG reporters in JX33. A decrease in fluorescence intensity was observed when pActF was incorporated at the second UAG site but no further decrease at the third UAG site. This observation is consistent with previous studies that the change of GFP fluorescence depends on the amino acid and the mutation site24,25. The 6-TAG mutant also produced full-length protein in JX33 (Fig. 2d), but with a lower yield than other TAG mutants (Supplementary Table 1) and exhibited no green fluorescence (Fig. 2e and 2f). The introduction of 6 pActF into EGFP may affect its folding and stability, thus reducing protein yields and abolishing fluorescence.
Overexpression of the C-terminus of ribosomal protein L11 (L11C) has been reported to enable Uaa incorporation into GFP at 1–3 TAG sites26. For comparison, we incorporated pActF into EGFP at identical 1-TAG and 3-TAG sites with the same pAIO plasmids using the L11C method and our approach, respectively (Fig. 2g). After Ni-NTA purification, JX33 afforded 27 mg/L of EGFP for 1-TAG mutant and 23 mg/L for 3-TAG mutant; while coexpression of the L11C in BL21(DE3) afforded 4.6 mg/L for 1-TAG mutant and 1.2 mg/L for 3-TAG mutant. JX33 provides 5.9-fold protein for the 1-TAG mutant and 19-fold for the 3-TAG mutant, showing a marked increase for 1 UAG site and more dramatic increase at 3 UAG sites. With RF1 intact, the L11C overexpression method still suffers quick reduction of incorporation efficiency when more than one UAG codon is introduced. These results further demonstrate that RF1 is the primary competitor preventing high incorporation efficiency at the UAG codons.
Mass spectrometry (MS) was used to confirm the incorporation of pActF at UAG sites in JX33. Electrospray ionization MS (ESI-MS) of intact EGFP protein expressed by the 1-TAG reporter in JX33 showed two peaks (27801 and 27897 Da), corresponding to the mature pActF-containing EGFP minus the N-terminal methionine (theoretical mass 27799.2 Da) and the pActF-containing EGFP with an immature chromophore (theoretical mass 27899.2 Da), respectively. ESI-MS analysis of EGFP expressed by the 2- and 3-TAG reporters showed a single peak at 27898 and 27924 Da, respectively. These peaks lie within ±2 Da of the theoretical masses of the 2 and 3 pActF-containing mature EGFP minus the N-terminal methionine (27896.3 and 27922.3 Da, respectively). No peaks were observed in any sample corresponding to mutant EGFP containing any natural amino acid at the UAG position. These results corroborate our Western blot and in-cell fluorescence data showing no significant EGFP expression in the absence of pActF (Fig. 2d and 2e). Liquid chromatography tandem MS (LC-MS/MS) of chymotrypsin-digested protein samples was performed to identify the amino acid incorporated at the UAG sites. The fragment ion masses were unambiguously assigned confirming the site-specific incorporation of pActF at the UAG site for all 1-, 2-, and 3-TAG EGFP mutants (Fig. 3a). Extracted ion chromatograms (EIC) showed that the peptide with pActF incorporated at the UAG site was the dominant species, with only trace amounts of Gln-containing peptide (Fig. 3b). Because both pActF and Gln have neutral side chains, we expect the two peptides containing pActF or Gln at the UAG site to be similar in ionization efficiency27. Therefore, we used the peptide intensity calculated from peak area in EIC to determine the incorporation fidelity of pActF. Incorporation of pActF in JX33 was found >99.81% at all UAG sites (Supplementary Table 2), consistent with the reported incorporation fidelity (>99.8%) of pActF at a single UAG site in DH10β23. Taken together, these results demonstrate that the JX33 strain enables the efficient and specific incorporation of the Uaa pActF into a protein at multiple UAG sites.
Figure 3. Mass spectrometric analyses of EGFP expressed in JX33 show that pActF was selectively incorporated at multiple UAG sites with high fidelity.
(a) MS/MS spectrum of EGFP peptide ADHUQQNTPIGDGPVLLPDNHY. U represents the UAG codon at residue 182. Star (*) in the spectrum denotes peptide fragments containing pActF, which unambiguously indicate that pActF was incorporated at the UAG site. (b) Extracted ion chromatograms (EIC) of the above peptide containing pActF (top) or Gln (bottom) at the UAG 182 position. The peak areas are indicated.
Simultaneous suppression of ten UAG sites in JX33
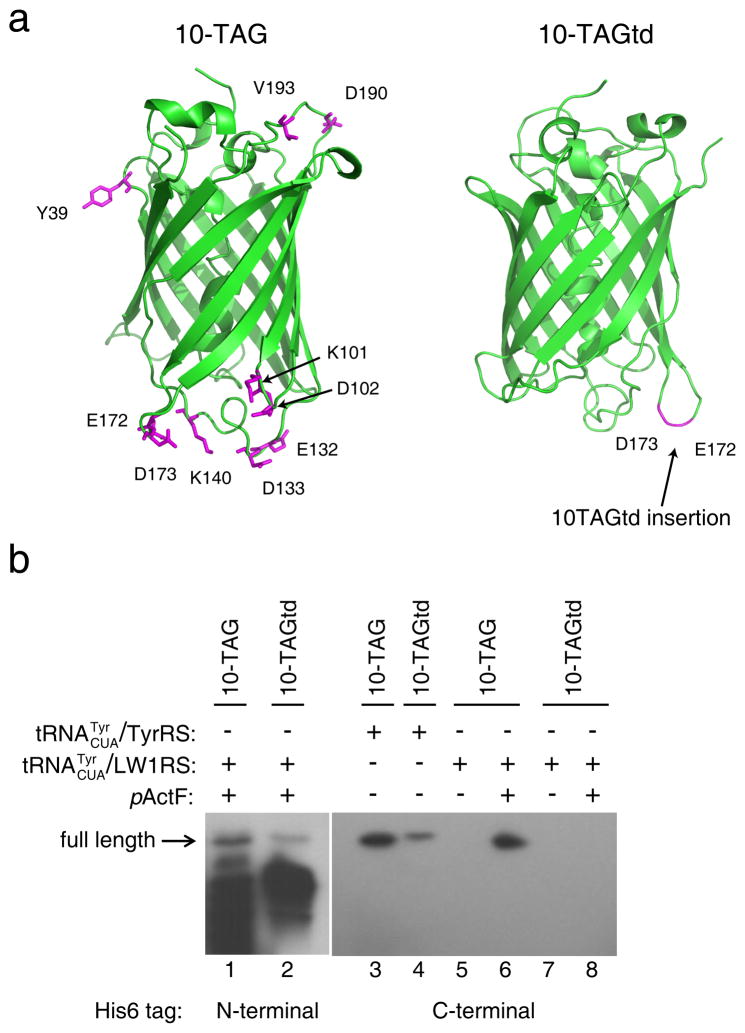
To assess if more UAG sites can be suppressed simultaneously for amino acid incorporation in JX33, we synthesized two EGFP reporters: one contains 10 TAGs across various loops (10-TAG) and the other has 10 tandem TAGs inserted in one loop (10-TAGtd, Fig. 4a). Using the to incorporate pActF into these mutants, we found that despite a substantial increase in truncated products full-length EGFP was still produced (Fig. 4b). Expression of 10-TAGtd reporter was lower than the 10-TAG reporter, possibly because 10 consecutive pActFs would more negatively affect EGFP folding and stability. To facilitate protein yield quantification, the His6 tag was moved to the C-terminus, and proteins were purified by Ni-NTA chromatography followed by FPLC. Incorporation of tyrosine using the yielded full-length EGFP for both 10 TAG mutants, with a similar decrease in expression of the 10-TAGtd reporter (Fig. 4b). For the 10-TAG mutant, protein yields were 0.4 mg/L for tyrosine incorporation and 0.5 mg/L for pActF incorporation. Fluorescence was abolished in all 10-TAG mutants regardless of identity of the amino acid incorporated. Expression level of the 10-TAG reporter was reduced compared to the 1-, 2-, and 3-TAG reporters, presumably due to folding and stability issues caused by the large number of mutations. Nonetheless, the ability to produce any level of protein with 10 Uaas selectively incorporated is a novel property of JX33, and has never been accomplished before in any cells. So long as chosen incorporation sites do not negatively affect protein folding and stability, JX33 should permit Uaa incorporation at more than 10 UAG sites.
Figure 4. Ten UAG sites are simultaneously suppressed with natural or unnatural amino acids in JX33.
(a) GFP structure (PDB 1GFL) illustrating the sites where 10 UAG codons were introduced. (b) Western blot analysis of the expression of 10-TAG and 10-TAGtd EGFP in JX33. The UAG codons were decoded as Tyr by or as pActF by . Cell lysates from same number of cells were separated by SDS-PAGE and probe with a penta-His antibody.
Multi-site incorporation of various Uaas in JX33
To ascertain if incorporation at multiple UAG sites in JX33 was generally applicable, we assayed the expression of the 3-TAG EGFP reporter with various Uaas. EGFP was efficiently expressed with Nε-acetyl-L-lysine (ActK)28, p-azido-L-phenylalanine (pAzdF), p-carboxymethyl-phenylalanine (pCmF)29, and p-iodo-phenylalanine (pIodF)30 incorporated as shown by Western blot (Fig. 5a) and in-cell fluorescence (Fig. 5b). No full-length EGFP was detected by Western blot in the absence of the Uaa. We obtained about 1 mg/L of purified protein containing ActK, pAzdF, and pIodF at 3 UAG positions (Supplementary Table 1). JX33 was compatible for use with orthogonal tRNA/aaRS pairs derived from the M. barkeri tRNAPyl/PylRS (for ActK) and the M. jannaschii tRNATyr/TyrRS (for other 3 Uaas).
Figure 5. JX33 enables multisite incorporation of various Uaas and in different proteins.
(a) Western analysis of the expression of 3-TAG EGFP reporter in JX33 with the UAG codon decoded as different Uaas. The same number of cells was used for each sample. After cell lysis, proteins were separated on SDS-PAGE and probed with a penta-His antibody. Densitometric analysis of Western blot bands and purified protein yields (Supplementary Table 1) were consistent on incorporation efficiency. (b) In-cell fluorescence assay of these mutant EGFP proteins containing different Uaas at 3 UAG sites. Measurements were performed using 3 independent batches of cells. Error bars represent s.e.m.. (c) The N-terminal sequence of human histone H3a and UAG codons introduced at the known acetylation sites. (d) Western analysis showing that full-length histone H3a was expressed for 1-, 2-, 3-, and 4-TAG H3a constructs in the presence of pActF or ActK in JX33 cells. For pActF, the sample loading ratio was 1:1:3:3; for ActK, the sample loading ratio was 1:3:7:9. Yields of H3a proteins were calculated after purification and shown at the bottom. (e) SDS-PAGE analysis of GST proteins expressed in JX33 with pActF incorporated at 1, 2, and 3 UAG sites. GST was purified with Ni-NTA chromatography, separated on the gel and stained with Coomassie blue. Loading was normalized to the same number of cells. Measurements of GST yields were performed on 3 independent batches of cells and errors represent s.e.m..
Multi-site incorporation of Uaa into various proteins
To demonstrate the use of JX33 in expressing other proteins, we expressed human histone H3a in JX33 with ActK and pActF incorporated at 1, 2, 3, and 4 UAG codons placed at known acetylation sites (Fig. 5c). In JX2.0, no expression of H3a protein was detected when more than 1 UAG sites were introduced. In JX33, all H3a mutants were successfully expressed in full length in the presence of pActF or ActK (Fig. 5d). Protein yields were sufficient for most in vitro studies.
We also incorporated pActF into glutathione S-transferase (GST) at 1, 2, and 3 UAG sites in JX33 (Fig. 5e). The protein yields were 67 (±11), 57 (±12), and 68 (±9) mg/L for 1-, 2-, and 3-TAG mutants, respectively. Similar to EGFP, the GST expression yield did not decrease when the number of UAG sites increased from 1 to 3. Taken together, these results indicate that JX33 can encode Uaas at multiple UAG sites into different proteins.
Suppression of endogenous UAG codons is RF1 dependent
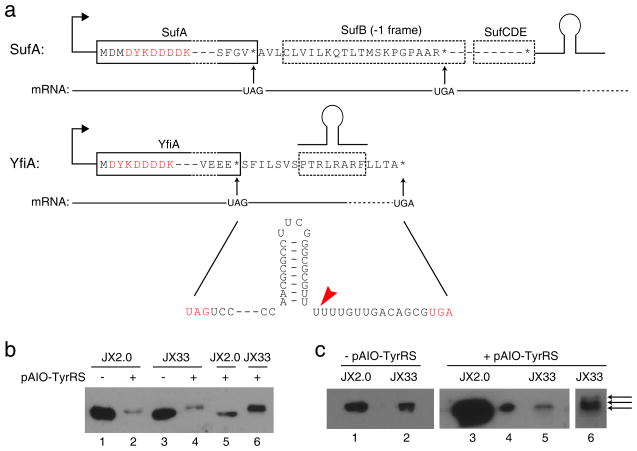
Over 300 endogenous genes end with TAG in the E. coli genome. What would happen to these legitimate TAG sites upon the introduction of an orthogonal amber suppressor tRNA/synthetase pair and upon the removal of RF1? We divided these genes into two categories defined by their downstream context (Fig. 6a). The majority of the TAG-ending genes have a secondary in-frame stop codon (UAA or UGA) downstream before a transcriptional terminator, as represented by sufA. To facilitate detection, we scarlessly appended a FLAG-tag to the N-terminus of the sufA gene in the genomes of JX2.0 and JX33. Surprisingly, Western analysis showed no detectable extension of SufA protein in JX2.0 harboring pAIO-TyrRS (Fig. 6b), suggesting that suppression of its UAG codon is inefficient in the presence of RF1. In contrast, SufA protein purified from JX33 harboring pAIO-TyrRS showed an increase in size corresponding to the expected molecular weight increase for extension to the next stop codon (Fig. 6b); LC-MS/MS analysis of this SufA protein identified numerous extended peptide fragments (Supplementary Table 3), confirming that translation extended to the next stop codon and that the UAG codon was suppressed with Tyr.
Figure 6. Legitimate UAG codons of endogenous genes are suppressed efficiently after RF1 knockout, and protein extension beyond the UAG codon is dependent on mRNA context.
(a) Two categories of endogenous genes ending in TAG: sufA representing those with an in-frame, non-UAG stop codon downstream before a transcription terminator (shown as a hairpin); yfiA representing those with a terminator before the next in-frame, non-UAG stop codon. A FLAG-tag (DYKDDDDK) was inserted scarlessly to the N-terminus of these two proteins in the genome to facilitate protein detection. Red arrow indicates the end of the mRNA as defined by the terminator. (b) Western analysis of SufA purified from cells. Extension of SufA to its next stop codon occurred only in lanes 4 and 6; all other lanes showed a band corresponding to the wild type SufA. Loading was normalized to the same number of cells in lane 1–4. Lane 5 and 6 were duplicates of lane 2 and 4, respectively, with increasing amounts. An anti-FLAG antibody was used for detection. (c) Western analysis of YfiA purified from cells. Loading in lane 1, 2, 3 and 5 were normalized to the same number of cells. Loading in lane 4 is 1/50 of lane 3, which showed it is a single band. Lane 6 is a re-run of lane 5 sample in a higher percentage gel (20% vs. 15%) for longer time to achieve better separation. Lane 5 and 6 showed 3 predominant bands, while other samples showed only one band.
The second category of genes has a transcriptional terminator between the UAG and the next in frame UAA or UGA, as exemplified by yfiA (Fig. 6a). The terminator will generate the 3′-end of the mRNA at the distal poly-U portion of the terminator hairpin31 (Fig. 6a). We appended a scarless N-terminal FLAG tag to the yfiA gene in the genomes of JX2.0 and JX33. In JX2.0 harboring pAIO-TyrRS, YfiA expression showed a single band on Western without extension to the next stop codon, suggesting that its UAG codon was also inefficiently suppressed. In JX33 harboring pAIO-TyrRS, however, YfiA expression showed three bands on Western as well as a dramatic protein reduction compared to JX2.0 (Fig. 6c). This difference can be explained by efficient UAG suppression in JX33. Suppression of the UAG in yfiA causes the ribosome to stall at the mRNA end as defined by the terminator. tmRNA recognizes stalled ribosomes and induces degradation of the extended polypeptide12. In JX2.0, RF1 allows stoppage at the UAG to produce wild-type YfiA. Therefore, JX33 had much less YfiA protein than JX2.0. To verify this, we analyzed YfiA purified from JX33 using LC-MS/MS (Supplementary Table 4). Both Western blot and MS did not reveal peptides extended to the next in-frame UGA stop codon, consistent with that the mRNA is ended at the terminator hairpin without the UGA codon. No proteins extended to the end of mRNA were detected either, suggesting that they were efficiently degraded by the tmRNA12. The three bands resolved in Fig. 6c correspond to extensions of 0, 2, or 6 amino acids away from the UAG site, respectively. Terminator hairpin structure and ribosome stalling at the mRNA end can result in early release of ribosomes, yielding small amount of proteins containing these peptides, which are not tagged by tmRNA for degradation. Our results corroborate a previous report on tmRNA-induced degradation of non-stop mRNA from plasmid-borne exogenous genes32, except that we studied endogenous genes. In all extended YfiA proteins expressed in JX33, the TAG site was incorporated with tyrosine by the orthogonal .
In short, legitimate UAG codons of endogenous genes were not efficiently suppressed in RF1-containing JX2.0 but were efficiently suppressed in the RF1-knockout JX33. This difference was also reflected on cell growth. When the orthogonal was expressed to incorporate Tyr at UAG sites, JX2.0 cells showed slight decrease in growth rate (doubling time 27.1 vs. 30.7 min) whereas the growth of JX33 was significantly retarded (doubling time 27.5 vs. 36.6 min) (Supplementary Fig. 1).
DISCUSSION
JX33 is a novel RF1 knockout strain with unique properties for Uaa incorporation. Generated 3.5 years ago, JX33 has been stable and autonomous without major growth or deleterious defects. Without RF1 competition, JX33 increases the amino acid incorporation efficiency at a single TAG site more than 100% than the parental RF1-containing JX2.0. More important, JX33 enables amino acids to be incorporated at multiple TAG sites without decreasing efficiency. This ability has not been achieved in any cells before, and is an essential trait for UAG being completely reassigned as a sense codon.
Although RF1 competition no longer exists in JX33, the efficiency of UAG functioning as a sense codon depends on multiple factors. Orthogonal synthetases evolved for Uaas are often less active than wild type synthetases5,6, and thus may generate less aminoacylated orthogonal tRNAs. In addition, the binding of natural aminoacyl-tRNAs to elongation factor Tu and to ribosome has been evolutionary tuned for optimal decoding33,34, yet the Uaa loaded orthogonal tRNA has not been fully optimized toward either. Moreover, many tRNAs are subjected to post-transcriptional modifications for specific and efficient decoding of cognate codons35. The orthogonal tRNA, with its anticodon artificially mutated to CUA, has not evolutionary optimized for UAG decoding. All these factors could make the UAG codon less efficient than canonical sense codons in encoding amino acids. For these reasons, the four UAG codons placed closely in the N-terminus of the H3a may behave as a cluster of “rare” codons. In E. coli, a rare-codon cluster lowers protein expression level36, which may account for the drop-off in yields with additional UAG observed for H3a expression. These non-optimal factors would also explain why Uaa-containing mutant proteins have not yet reached the same expression level as the wt protein.
A long-standing question for genetically encoding Uaas with a stop codon is how endogenous genes ending with the stop codon are affected. By studying two representative endogenous genes, we found that an amber suppressor tRNA/synthetase did not efficiently incorporate its cognate amino acid at the legitimate TAG site in the presence of RF1. This surprising finding suggests that an unknown mechanism may prevent these legitimate stop codons from being suppressed. The inefficient suppression of endogenous TAGs in JX2.0 explains why no significant adverse effect to E. coli is observed when orthogonal amber suppressor tRNA/synthetase pairs are used to incorporate Uaas. However, we discovered that upon RF1 removal in JX33 the TAG codon of endogenous genes was efficiently suppressed by the tRNA/synthetase pair. Translation then extended to the next in-frame different stop codon when there is no transcription terminator before the next stop codon; when there is a transcription terminator, translation was terminated between the TAG and the mRNA end defined by the terminator hairpin. Endogenous UAG suppression in JX33 also led to a slower growth phenotype. Studying UAG suppression in more endogenous genes would confirm whether the above observations are general.
RF1 is reported to be essential for E. coli, but our results argue against this paradigm. We showed that RF1 can be knocked out when wild type RF2 expression is not auto-regulated. The resultant JX31 strain has no compensatory mutations anywhere in the genome. Although JX31 has a slower growth rate, it is an independent and stable strain with RF1 deleted, which suggests that RF1 is nonessential for E. coli. Interestingly, the A293E mutation of RF2 found in JX33 restores the growth rate of JX33 to the same level of the parental JX2.0. However, the RF2(A293E) is unable to substitute RF1 in terminating UAG codons because it could not rescue the RF1 temperature sensitive phenotype in MRA8 cells. We note that non-stop incorporation of pActF into EGFP was also observed in JX31, which has no RF2(A293E) mutation; the protein yields for 1-, 2- and 3-TAG EGFP mutants from JX31 were 5.7 (±0.4), 6.1 (±0.5) and 7.0 (±0.5) mg/L, respectively. This result suggests that the RF2(A293E) mutation is not required for efficient incorporation of Uaa at multiple TAG sites. Nonetheless, how A293E mutation contributes to the fast growth of JX33 warrants further studies.
When this paper was being prepared, it was reported that RF1 can be knocked out after supplying 7 essential genes and a suppressor tRNA on a plasmid37. The dependence on simultaneous change of many essential genes is consistent with the previous conclusion that RF1 is essential. A major difference of our work is that the knockout of RF1 is independent of supplying any other genes or a suppressor tRNA. More important, the generation of JX31 suggests that RF1 is nonessential. To our best knowledge, this work represents the first unconditional knockout of RF1 and the generation of an autonomous stable RF1 deletion strain.
Selective incorporation of Uaas at multiple sites will open up new possibilities in protein research and laboratory evolution. For instance, multisite incorporation of posttranslational modification mimics (e.g., ActK and pCmF) will be valuable for studying epigenetics and signal transductions. JX33 may enable laboratory evolution of bacteria in search for new protein properties or organismal functions by exploiting the novel properties of Uaas. Such experiments were not feasible or effective before, as the Uaa is incorporated at a single site with low efficiency; novel functions may require an Uaa at multiple positions simultaneously. Moreover, the presence of RF1 generates truncated protein products, which may interfere with selection and evolution. JX33 resolves all these problems and should prove valuable in harnessing the expanding Uaa repertoire for directed evolution.
RF1 knockout strains can also be valuable for investigating the evolution of the genetic code. Organisms in different taxa have been found to reassign stop codons to sense codons1,2, yet they represent the reassignment endpoint and provide limited information on the reassignment process and organismal adaptation. This study demonstrates the feasibility of reassigning the UAG stop codon to a sense codon in the extant organism E. coli, providing empirical evidence in support of such codon reassignment events. E. coli is tolerant of codon reassignment and unexpectedly flexible in adapting to a new code, suggesting that the code can evolve in modern organisms. JX3.0 affords a previously unavailable model system for experimentally studying the physiological change and adaptation of a living organism to codon reassignment on a laboratory time scale.
METHODS
Strain construction
All strains in this study were created using the λ-red recombinase system17,18, and are described in detail in the Supplementary Methods.
Growth assay
A colony was picked for each E. coli strain and grown overnight with appropriate antibiotic. Cells were normalized to an OD600 of 1 and diluted 1:50 in fresh media without antibiotic. OD600 was then measured every 20 minutes for 10 hours.
Plasmid construction
All plasmids were assembled by standard cloning methods and confirmed by DNA sequencing. pAIO plasmids containing EGFP gene with different TAG codons were synthesized as the following: EGFP cassettes with an N-terminal His6 tag, containing TAG’s at various positions were created using overlapping PCRs. The following sites were used: Y182 for 1-TAG; Y39 and Y182 for 2-TAG; Y39, Y182 and Y151 for 3-TAG; Y39, Y74, Y143, Y151, Y182 and Y200 for 6-TAG; Y39, K101, D102, E132, D133, K140, E172, D173, D190 and V193 for 10-TAG; a 10 tandem TAG codons in place of E172 and D173 for 10-TAGtd. These cassettes were first cloned into pBP-Blunt (Biopioneer, San Diego, CA), and then digested and ligated into pBK-AIO vectors containing the orthogonal and the M. jannaschii TyrRS5 or the LW1RS23 using Spe I and Bgl II.
Human histone H3a was expressed using two plasmids: pTak-tRNA-H3 and pBKt-ActKRS. The pTak plasmid contained the M. barkeri tRNAPyl and the human histone H3a gene with a His6 tag appended at the C-terminus. tRNAPyl was driven by the lpp promoter and terminated with the rrnC terminator. The gene for human histone H3a was codon-optimized using Gene Design38, and synthesized by overlapping PCRs using multiple 40 bp primers. The optimized gene was cloned into pTak using Spe I and Bgl II sites, and was driven by the T5 promoter. Various mutant forms of histone H3a were then synthesized and also cloned into the pTak plasmid. The following histone H3a mutants were cloned: 1TAG – K9; 2TAG, K9 and K14; 3TAG – K9, K14, and K18; 4TAG – K9, K14, K18, and K23. The second plasmid pBKt-ActKRS expresses the ActK-specific synthetase. Six mutations (D76G, L266V, L270I, Y271F, L273A, and C313F) were introduced into the wild-type M. barkeri PylRS using overlapping PCR to generate the ActKRS. This cassette was digested with Nde I and Pst I and ligated into the precut pBK-JYRS vector 5. The GlnRS promoter originally in pBK-JYRS was replaced with the trc promoter from pTrc (Invitrogen, Carlsbad, CA) to drive the expression of ActKRS.
GST was expressed with plasmids pVL-GST and pBK-LW1RS23. TAG codons were introduced at residue Y58 (1-TAG), Y58 and Y111 (2-TAG), Y58, Y111 and Y164 (3-TAG) of the Schistosoma japonicum GST gene. TAG-containing GST genes were cloned into pLEIZ23 using Spe I and Bgl II to afford pVL-GST. pVL-GST encodes the orthogonal under the control of the lpp promoter and the rrnC terminator, and the GST(TAG) gene driven by the T5 promoter with a His6 tag appended at the C-terminus.
Western analyses
E. coli cells expressing EGFP or histone H3a were lysed, and proteins were separated on 12% or 15% SDS polyacrylamide gel, respectively. After transfer, EGFP and histone H3a were detected using the HRP-conjugate penta-His antibody (Qiagen, Valencia, CA). Protein purification of YfiA and SufA was performed using established procedures39,40 with minor modifications. Purified YfiA and SufA were run on 15% or 20% SDS polyacrylamide gel, and detected by using a monoclonal FLAG M2 antibody (Sigma, St. Louis, MO). Blots were developed using the pico chemiluminescence kit (Thermo Scientific, Rockford, IL) according to manufacturer’s specifications. See Supplementary Methods for details.
In-cell fluorescence assay
In-cell fluorescence intensity was determined using a FluoroLog-3 (Horiba Jobin Yvon). E. coli colonies were picked and grown 16 hours with or without Uaas. Cells were washed two times in PBS buffer, and diluted in PBS to an OD600 of 0.1. The emission spectrum of EGFP was measured using an excitation wavelength of 488 nm scanning emission from 503 to 560 nm. Fluorescence intensity of each sample was compared using the intensity at the maximal emission at 511 nm. Slit widths and integration times remained constant between all readings.
Protein purification
EGFP extracted from E. coli cells was first purified by Ni-NTA affinity chromatography (see Supplementary Methods), and further purified using 1 mL Resource Q anion exchange column on a UPC-900 FPLC (GE healthcare, Piscataway, NJ). The column was equilibrated with a low salt buffer (20 mM Tris·HCl, pH 8.0), and proteins were eluted with a linear gradient of 0 – 0.5 M NaCl. Peak fractions were analyzed by SDS-PAGE and pooled for further analysis. Purification of human histone H3a was performed using established procedures41 and following the protocol of using Ni-NTA resin (Qiagen) under denaturing conditions (see Supplementary Methods). All protein concentrations and total yields were determined using the Bio-Rad protein assay kit (Hercules, CA) according to manufacturer’s specifications.
Mass spectrometry
Intact proteins were analyzed by ESI-MS using a LTQ Velos mass spectrometer (Thermo Scientific, Rockford, IL). Automated 2D nanoflow LC-MS/MS analysis was performed using LTQ tandem mass spectrometer (Thermo Electron Corporation, San Jose, CA). See Supplementary Methods for details.
Genomic sequencing of E. coli strains
Genomic DNA from JX2.0, JX31 and JX33 was harvested, purified, and prepared into libraries. These genomic DNA libraries were sequenced using the Illumina Genome Analyzer II (Illumina, San Diego, CA) as per manufacturer’s instructions. Sequence alignments and SNP analysis were performed using the SHORE package42 according to the documentation provided with the software. See Supplementary Methods for details.
Supplementary Material
Acknowledgments
We are very grateful to Dr. Robert Sauer (MIT) for providing strain X90 ssrA1::cat,to Dr. Leif Isaksson (Stockholms Universitet) for providing strain MRA8, to Dr. Wenshe Liu (Texas A&M University) for providing plasmid pET-L11C, and to Dr. Jim Kadonaga (UCSD) for suggestions on H3a purification. We thank Vanessa K. Lacey for proofreading the manuscript. J.X. was partially funded by the Pioneer Fellowship. M.D.S was supported by National Science Foundation IGERT training grant (DGE-0504645). R.J.S. was supported by an NIH NRSA postdoctoral fellowship (F32-HG004830). J.R.E. acknowledges support from the Mary K. Chapman Foundation. L.W. acknowledges support from the Ray Thomas Edwards Foundation, Searle Scholar Program (06-L-119), Beckman Young Investigator Program, March of Dimes Foundation (#5-FY08-110), California Institute for Regenerative Medicine (RN1-00577-1) and National Institutes of Health (1DP2OD004744-01).
Footnotes
Author contributions
D.B.F.J. incorporated Tyr and various Uaas into EGFP and histone H3a using JX33, characterized JX2.0 and JX3.0 strains with growth, western, fluorescence, and temperature-sensitive complementation assays, performed endogenous UAG suppression studies, analyzed the data, and wrote the manuscript; J.X. generated the RF1 knockout strains and analyzed the data; Z.S. and S.P.B. characterized amino acid and Uaa incorporation with mass spectrometry, analyzed the data, and wrote the mass spectrometry section; J.K.T. incorporated Uaa into GST, compared Uaa incorporation efficiency in JX33 and in BL21 expressing L11C, and analyzed the data; M.D.S., R.J.S., and J.R.E. sequenced all E. coli strains described, analyzed the data, and wrote the genomic sequencing section; L.W. conceived and directed the project, analyzed the data, and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Osawa S, Jukes T, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992;56:229–64. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 3.Benzer S, Champe SP. A change from nonsense to sense in the genetic code. Proc Natl Acad Sci U S A. 1962;48:1114–21. doi: 10.1073/pnas.48.7.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29:4767–82. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed Engl. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Parrish AR, Wang L. Expanding the genetic code for biological studies. Chem Biol. 2009;16:323–36. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scolnick E, Tompkins R, Caskey T, Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci U S A. 1968;61:768–74. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rydén S, Isaksson L. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet. 1984;193:38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes SY, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–84. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–24. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 13.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–3. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 14.Craigen WJ, Cook RG, Tate WP, Caskey CT. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985;82:3616–20. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uno M, Ito K, Nakamura Y. Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie. 1996;78:935–43. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 16.Posfai G, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–6. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 17.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40:191–7. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 19.Ito K, Uno M, Nakamura Y. Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc Natl Acad Sci U S A. 1998;95:8165–9. doi: 10.1073/pnas.95.14.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Ryden-Aulin M, Kirsebom LA, Isaksson LA. Genetic implication for an interaction between release factor one and ribosomal protein L7/L12 in vivo. J Mol Biol. 1994;242:614–8. doi: 10.1006/jmbi.1994.1611. [DOI] [PubMed] [Google Scholar]
- 21.Dahlgren A, Ryden-Aulin M. A novel mutation in ribosomal protein S4 that affects the function of a mutated RF1. Biochimie. 2000;82:683–91. doi: 10.1016/s0300-9084(00)01160-3. [DOI] [PubMed] [Google Scholar]
- 22.Kaczanowska M, Ryden-Aulin M. Temperature sensitivity caused by mutant release factor 1 is suppressed by mutations that affect 16S rRNA maturation. J Bacteriol. 2004;186:3046–55. doi: 10.1128/JB.186.10.3046-3055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–44. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Xie J, Deniz AA, Schultz PG. Unnatural amino acid mutagenesis of green fluorescent protein. J Org Chem. 2003;68:174–6. doi: 10.1021/jo026570u. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, et al. A convenient method for genetic incorporation of multiple noncanonical amino acids into one protein in Escherichia coli. Mol Biosyst. 2010;6:683–6. doi: 10.1039/b920120c. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Schultz PG, Brock A. An improved system for the generation and analysis of mutant proteins containing unnatural amino acids in Saccharomyces cerevisiae. J Mol Biol. 2007;371:112–22. doi: 10.1016/j.jmb.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–4. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 29.Xie J, Supekova L, Schultz PG. A genetically encoded metabolically stable analogue of phosphotyrosine in Escherichia coli. ACS Chem Biol. 2007;2:474–8. doi: 10.1021/cb700083w. [DOI] [PubMed] [Google Scholar]
- 30.Xie J, et al. The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat Biotechnol. 2004;22:1297–1301. doi: 10.1038/nbt1013. [DOI] [PubMed] [Google Scholar]
- 31.Abe H, Abo T, Aiba H. Regulation of intrinsic terminator by translation in Escherichia coli: transcription termination at a distance downstream. Genes Cells. 1999;4:87–97. doi: 10.1046/j.1365-2443.1999.00246.x. [DOI] [PubMed] [Google Scholar]
- 32.Ueda K, et al. Bacterial SsrA system plays a role in coping with unwanted translational readthrough caused by suppressor tRNAs. Genes Cells. 2002;7:509–19. doi: 10.1046/j.1365-2443.2002.00537.x. [DOI] [PubMed] [Google Scholar]
- 33.Schrader JM, Chapman SJ, Uhlenbeck OC. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc Natl Acad Sci U S A. 2011;108:5215–20. doi: 10.1073/pnas.1102128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ledoux S, Uhlenbeck OC. Different aa-tRNAs are selected uniformly on the ribosome. Mol Cell. 2008;31:114–23. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoyama S, Nishimura S. Modified nucleosides and codon recognition. In: Soll D, RajBhandary UL, editors. tRNA: Structure, Biosynthesis, and Function. ASM Press; Washington, DC: 1995. pp. 207–223. [Google Scholar]
- 36.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 37.Mukai T, et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson SM, Wheelan SJ, Yarrington RM, Boeke JD. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 2006;16:550–6. doi: 10.1101/gr.4431306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agafonov DE, Kolb VA, Spirin AS. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2001;2:399–402. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JH, Yeo WS, Roe JH. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol Microbiol. 2004;51:1745–55. doi: 10.1111/j.1365-2958.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- 41.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 42.Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–33. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.