Abstract
All organisms, including humans, possess a huge number of uncharacterized enzymes. Here, we describe a general cell-based screen for enzyme substrate discovery by untargeted metabolomics and its application to identify α/β-hydrolase domain-containing 3 (ABHD3) as a lipase that selectively cleaves medium-chain and oxidatively-truncated phospholipids. Abhd3−/− mice possess elevated myristoyl (C14)-phospholipids, including the bioactive lipid C14-lysophosphatidylcholine, confirming the physiological relevance of our substrate assignments.
Genome sequences have proven valuable for enumerating the full complement of enzymes that belong to a particular mechanistic class across a wide range of organisms1. Despite this basic knowledge, the physiologic substrates and products for many of these enzymes remain unknown2. This question has historically been addressed by biochemically screening with a restricted set of candidate substrates, but such targeted in vitro assays often require purified enzyme, which can limit their utility for challenging protein classes (e.g., integral membrane enzymes), and create non-physiologic environments that may impede the discovery of natural substrates. Computational approaches can predict substrates of enzymes, but these in silico methods require structural information for the target enzymes3,4. Metabolomic methods have also been introduced to discover endogenous substrates by comparatively profiling wild type versus enzyme-disrupted or enzyme-overexpressing cells and organisms5–9. While fruitful, these metabolomic studies have, to date, focused on individual enzymes, at least when applied to mammalian systems. Considering the vast number of mammalian enzymes in need of annotation, we asked whether a metabolomic strategy could be adapted for higher-throughput assessment of enzyme-substrate relationships in living systems.
Our approach involved the transient overexpression of a panel of uncharacterized enzymes in a common cellular system (HEK293T cells) followed by untargeted liquid chromatography-mass spectrometry (LC-MS)-based metabolomics, wherein the chromatograms for multiple transfections at a given m/z value are displayed together for comparison of peaks that are selectively changed by overexpression of one enzyme but not others (see Supplementary Fig. 1 in Supplementary Results and Supplementary Methods). We anticipated that directly measuring metabolomic perturbations from living cells would provide a more physiological setting for enzyme-substrate discovery compared to in vitro biochemical assays and also obviate the need to arbitrarily select candidate substrates for analysis. Furthermore, the transfected enzymes should each act as counter-controls for one another (working under the assumption that, in most circumstances, these enzymes would use different substrates), such that an inherent “specificity” filter is built into the screen. We selected for initial analysis a dozen uncharacterized enzymes from the metabolic serine hydrolase (SH) class (Fig. 1a), at least six of which are integral membrane proteins based on sequence predictions and activity-based proteomic data10. There are more than 110 predicted metabolic SHs in humans, and these enzymes can hydrolyze a diverse range of substrates, including polar small molecules, lipids, peptides, glycans, and proteins11. Many SHs, however, remain unannotated with respect to endogenous substrates and functions.
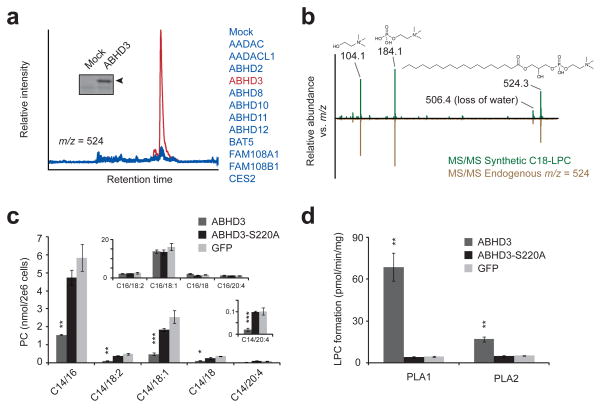
Fig. 1.
Metabolomic profiling of an enzyme library identifies metabolites altered by ABHD3 overexpression. (a) Representative overlaid extracted ion chromatograms at m/z = 523.5–524.5 from HEK293T cells transfected with ABHD3 (red trace) versus other enzymes (blue traces). Insert: Activity-based labeling using the serine hydrolase-directed probe fluorophosphonate rhodamine (FP-Rh) in mock versus ABHD3-transfected cells. Arrowhead designates FP-Rh-labeled ABHD3 protein. (b) Representative MS/MS fragmentation of synthetic C18-LPC (top, in green) and endogenous m/z = 524 (bottom, in brown) gave identical daughter ions of 104.1 (choline), 184.1 (phosphocholine), and 506.1 (dehydro-C18-LPC). (c) Targeted MRM measurements of phosphocholines (PCs) from C8161 cells stably overexpressing epitope-tagged ABHD3 (dark grey), the catalytically dead ABHD3-S220A mutant (black), or GFP (light grey). PC species are indicated by C#/##, where the # indicates the sn-1 acyl chain and ## indicates the sn-2 acyl chain. Top insert: C16-containing PC species; bottom insert: an enlarged graph showing C14/20:4-PC. (d) PLA1 and PLA2 hydrolysis activities for ABHD3 using C14/18:2-PC as a substrate and lysates from stably transfected C8161 cells as the protein source. Data are presented as mean ± standard error; n = 4/group; * P < 0.05, ** P < 0.01, *** P < 0.001 for ABHD3 versus ABHD3-S220A groups.
Comparison of the LC-MS profiles for organic-soluble metabolites (m/z range of 200–1200 Da) from 12 enzyme-transfected and 1 mock-transfected cell preparations identified two cases where a metabolite peak was altered in a single enzyme profile (Fig. 1a and Supplementary Figs. 2 and 3). We focused our attention on one peak in positive polarity mode with m/z = 524 and a retention time ~22 min, which was selectively elevated in cells transfected with the integral membrane enzyme α/β-hydrolase domain-containing 3 (ABHD3) (Fig. 1a). Stable overexpression of epitope-tagged ABHD3, but not a catalytically dead mutant (ABHD3-S220A) or GFP, in two additional human cell lines (C8161 and MUM2C) recapitulated the elevation in m/z = 524 (Supplementary Fig. 4), demonstrating that this metabolite change is observed across multiple cell types expressing ABHD3 and requires the catalytic activity of this enzyme. The metabolite was identified by tandem MS fragmentation (Fig. 1b) and coelution with a synthetic standard (Supplementary Fig. 5) as C18-lysophosphatidylcholine (C18-LPC, 1). Subsequent targeted multiple reaction monitoring12 (MRM) analyses revealed increases in other LPCs (Supplementary Fig. 6) and large decreases in PCs containing a myristoyl acyl chain (C14-PCs) in ABHD3-overexpressing cells, but no changes in PCs with acyl chains of other lengths (Fig. 1c). Because myristic acid (2) itself was unchanged in ABHD3-overexpressing cells (Supplementary Fig. 7), these data together suggested that ABHD3 acts as a direct phospholipase for myristoyl-PCs. Consistent with this premise, lysates from C8161 cells stably overexpressing ABHD3 showed 17-fold and 3-fold higher phospholipase A1 (PLA1; cleavage of the sn-1 acyl chain on a phospholipid) and phospholipase A2 (PLA2; cleavage of the sn-2 acyl chain on a phospholipid) activities, respectively, when using sn-1 C14, sn-2 C18:2-PC (C14/18:2-PC, 3) as a substrate, compared to either ABHD3-S220A or GFP-expressing cell lysates (Fig. 1d).
We next addressed whether ABHD3 regulates C14-phospholipids (C14-PLs) in vivo. Abhd3−/− mice were obtained from Deltagen and found to be viable, fertile, and normal in their general cage behavior. Selective loss of ABHD3 protein in Abhd3−/− mice was confirmed by untargeted, MS-based proteomics and activity-based protein profiling (ABPP) using the probe fluorophosphonate (FP)-rhodamine13, which covalently labels the conserved serine nucleophile in the active sites of SH enzymes (Supplementary Fig. 8, Supplementary Table 1). Previous ABPP10 and gene expression profiling14 studies established that ABHD3 is highly expressed in brain, liver, and kidney (Supplementary Fig. 9). We therefore analyzed these tissue metabolomes from Abhd3+/+ and Abhd3−/− mice by untargeted LC-MS, which identified a single peak in positive polarity with an m/z value of 468 and a retention time ~20.0 min that was elevated in all three tissues from Abhd3−/− mice. Three additional metabolites with m/z values of 730.5, 754.5, and 778.5, all nearly coeluting at ~23.2 min, were also elevated in kidneys from Abhd3−/− mice (Fig. 2a).
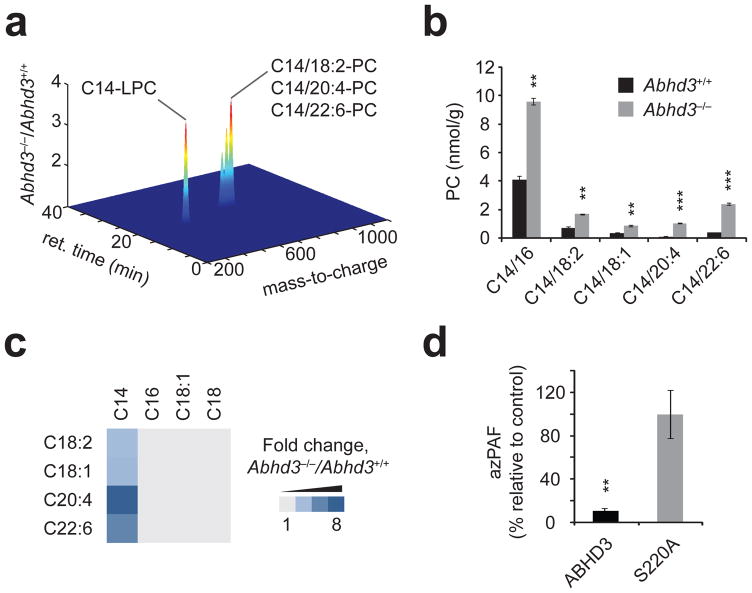
Fig. 2.
Abhd3−/− mice possess elevated C14-PCs. (a) Untargeted LC-MS profiling of kidney metabolomes from Abhd3+/+ or Abhd3−/− mice (plot is shown in the positive ionization mode). Only those metabolites with P < 0.05 and fold change > 1.5 are shown. Data are presented as mean fold changes; n = 6/group. (b) Targeted MRM measurements of PCs from kidney tissue of Abhd3+/+ (black bars) or Abhd3−/− (grey bars) mice. Similar changes in PCs were observed in brain, liver, and plasma from Abhd3−/− mice (see Supplementary Table 3). (c) Fold change of PC species from kidney tissue of Abhd3−/− versus Abhd3+/+ mice. The PC acyl chains for a given species are indicated by the top and left axis. Lighty grey indicates no change between genotypes whereas the darkest blue indicates a 8.5-fold change. Data are presented as means; n = 4/group. (d) Relative levels of azPAF in C8161 cells following live-cell incubation with azPAF (10 μM, 4 h, 37°C). For (b) and (d), data are presented as mean ± standard error; n = 3–4/group; * P < 0.05, ** P < 0.01, *** P < 0.001 for Abhd3+/+ versus Abhd3−/− or ABHD3 versus ABHD3-S220A.
Tandem MS experiments showed loss of an m/z species of 184, corresponding to phosphocholine, from all four metabolites (Supplementary Fig. 10). From this, m/z = 468 was deduced to be C14-LPC (4), and, by extrapolation, the m/z values of 730.5, 754.5, and 778.5 corresponded to PCs with acyl chains of C14/18:2, C14/20:4 (5), and C14/22:6 (6), respectively. These metabolic changes were confirmed and quantified by MRM (Fig. 2b), which also revealed their additional presence in brain and liver of Abhd3−/− mice and showed that other LPC species, myristic acid, and C14-CoA (7) were unchanged (Supplementary Table 2). For all three tissues, the greatest fold-increases in Abhd3−/− mice were observed for C14-PCs bearing polyunsaturated acyl chains, while non-C14-containing PCs were unaltered (Fig. 2c and Supplementary Table 2). Finally, other C14-PLs were also elevated in tissues from Abhd3−/− mice (Supplementary Table 2).
We found that the striking specificity displayed by ABHD3 for C14-PLs in vivo was reflected in its inherent substrate preferences in vitro. As noted previously, cell lysates overexpressing ABHD3 showed highly elevated phospholipase activity with C14-PCs compared to ABHD3-S220A cell lysates (Fig. 1d), but no such differences were observed for C15- or C16-PCs (Supplementary Table 3). ABHD3 displayed some head group tolerance and could hydrolyze C14/14-phosphatidylglycerol (PG, 8), but not other PLs under our assay conditions (Supplementary Table 3). Since ABHD3 appears capable of directly hydrolyzing some, but not all C14-PLs in vitro, we speculate that a portion of the C14-PL changes in tissues from Abhd3−/− mice may reflect the indirect action of other enzymes that interconvert PL head-groups using accumulated PC as a substrate. An analogous argument could explain elevations in C14-LPC in Abhd3−/− mice, where ABHD3 would exert indirect control over C14-LPC by regulating the availability of C14-PC pools that constitute substrates for other phospholipase enzymes.
ABHD3 also cleaved PCs with shorter acyl chains (< C14), including C12/12-PC (9, Supplementary Table 3), a lipid that has been recently shown to be an agonist ligand for the nuclear receptor LRH-115. ABHD3 displayed highest activity with C5-C8 substrates and oxidatively truncated PCs (oxPCs), such as azPAF (10, 1-hexadecyl-2-azelaoyl-glycerol-3-phosphocholine) (Supplementary Table 3 and Supplementary Fig. 11). ABHD3 showed a similar substrate selectivity profile (azPAF > C14/22:6 PC ≥ C14/18:2 PC) in crude lysates from multiple cell lines or as a partially purified protein enriched via a polyhistidine tag on metal affinity resin (Supplementary Fig. 11), indicating that this substrate profile is likely an inherent property of the enzyme.
oxPCs are derived from polyunsaturated PCs under conditions of cellular oxidative stress16 and possess a range of biological activities, including promoting cell death and atherosclerosis17,18. Their enzymatic hydrolysis may thus ensure efficient removal of damaged lipids from cellular membranes. We were unable to detect oxPCs in tissues from Abhd3+/+ or Abhd3−/− mice, likely owing to their low abundance and selective production under conditions of oxidative stress. We therefore performed two additional studies to test whether ABHD3 is likely to play a role in metabolizing oxPCs in biological systems. First, we exposed the polyunsaturated lipid C16/18:2-PC (11) to CuSO4 oxidation protocols that generate a mixture of oxPCs19 and then incubated this mixture with C8161 cell lysates overexpressing ABHD3 or ABHD3-S220A and monitored the generation of C16-LPC (12). ABHD3-overexpressing proteomes produced much greater quantities of C16-LPC than ABHD3-S220A-overexpressing cell proteomes (Supplementary Fig. 12). Second, we treated ABHD3- and ABHD3-S220A-overexpressing C8161 cells with azPAF and found that ABHD3-overexpressing cells showed much greater capacity to metabolize this azPAF in situ (Fig. 2d). These data thus indicate that ABHD3 can regulate oxPC metabolism in complex biochemical and cellular contexts.
Taken together, our results reveal a multi-faceted role for ABHD3 in the catabolism of medium-chain phospholipids that is distinct from other known phospholipases. ABHD3 shares closest sequence similarity with ABHD1 (47% identity) and more distant relatedness to ABHD2 (25%) (Supplementary Fig. 13), two poorly characterized serine hydrolases that lack known substrates. While additional phospholipases, such as PLA2G7 and PAFAH2, have been proposed to play roles in regulating oxPCs in vivo20, these enzymes do not, to our knowledge, endogenously regulate medium-chain PLs such as C14-PCs. We speculate that the unusual substrate profile displayed by ABHD3, which shows a relaxed specificity among shorter-chain PCs, but a strict upper limit for PCs with a C14 acyl chain, might serve as an enzyme-intrinsic regulatory mechanism to allow ABHD3 to exert control over specific pools of lipids under both physiologic (C14-PLs) and pathologic (oxPC) conditions. Future studies using Abhd3−/− mice or ABHD3 inhibitors, once they become available, should illuminate the effects of perturbing ABHD3 and its medium-chain PL substrates in mammalian physiology and disease, where these lipids have been postulated to serve a range of signaling functions21–23.
Projecting forward, we are encouraged by our discovery of a physiologically relevant biochemical pathway from a screen containing only a dozen mammalian enzymes. Indeed, traditional biochemical assays may have failed to uncover the unusual connectivity between ABHD3 and medium-chain PLs, if, for instance, more common longer-chain PLs were tested as general candidate phospholipase substrates. We recognize, however, potential limitations to our screen, such as the absence of relevant endogenous substrates in heterologous cellular systems, the observation of non-physiologic metabolic changes due to enzyme overexpression, or the failure to detect a biochemical change due to sensitivity limits or incomplete profiling and annotation of the metabolome. These issues could be addressed, in part, by expanding the analysis to other fractions of the metabolome (e.g., polar metabolites24) or by coupling metabolomics with RNA-interference-mediated knockdowns of endogenously expressed enzymes. Such potential limitations aside, our data indicate that metabolomic screening of enzyme libraries, by enabling the de novo discovery of substrates in living cells, may prove generally applicable for mapping natural enzyme-substrate relationships and assigning functions to the myriad uncharacterized enzymes in mammalian proteomes.
Supplementary Material
Acknowledgments
We gratefully acknowledge Daniel Bachovchin and Shasha Ji for providing the serine hydrolase expression plasmids, Alan Saghatelian for the fatty acyl-CoA extraction protocol, and Stephanie Palmer for the Matlab script used to generate Fig. 2a. This work was supported by the US National Institutes of Health (CA132630) and the Skaggs Institute for Chemical Biology.
Footnotes
Author contributions.
J.Z.L. and B.F.C. designed the experiments, analyzed the data, and wrote the manuscript. J.Z.L. and J.S.C. synthesized C14 PCs. J.Z.L. and D.M. performed the experiments. S.N. generated and analyzed the untargeted proteomic data. C.W. generated the R script used for screening chromatograms from cellular metabolomes. S.A.T. and G.S. assisted in metabolite identification and MS/MS experiments.
Competing financial interests.
The authors declare no competing financial interests.
References
- 1.Lander ES. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–46. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 3.Hermann JC, et al. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–9. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rottig M, Rausch C, Kohlbacher O. Combining structure and sequence information allows automated prediction of substrate specificities within enzyme families. PLoS Comput Biol. 2010;6:e1000636. doi: 10.1371/journal.pcbi.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raamsdonk LM, et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nature Biotechnology. 2001;19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- 6.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–50. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Saghatelian A, et al. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–9. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 8.Tagore DM, et al. Peptidase substrates via global peptide profiling. Nat Chem Biol. 2009;5:23–5. doi: 10.1038/nchembio.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachovchin DA, et al. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20941–6. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. Journal of Biological Chemistry. 2010;285:11051–5. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakanishi H, Iida Y, Shimizu T, Taguchi R. Analysis of oxidized phosphatidylcholines as markers for oxidative stress, using multiple reaction monitoring with theoretically expanded data sets with reversed-phase liquid chromatography/tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1366–74. doi: 10.1016/j.jchromb.2008.09.041. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14694–9. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JM, et al. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 2011 doi: 10.1038/nature10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson AD, et al. Structural identification by mass spectrometry of oxidized phospholipids in minimally oxidized low density lipoprotein that induce monocyte/endothelial interactions and evidence for their presence in vivo. Journal of Biological Chemistry. 1997;272:13597–607. doi: 10.1074/jbc.272.21.13597. [DOI] [PubMed] [Google Scholar]
- 17.Chen R, Yang L, McIntyre TM. Cytotoxic phospholipid oxidation products. Cell death from mitochondrial damage and the intrinsic caspase cascade. Journal of Biological Chemistry. 2007;282:24842–50. doi: 10.1074/jbc.M702865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navab M, et al. The oxidation hypothesis of atherogenesis: the role of oxidized phospholipids and HDL. Journal of Lipid Research. 2004;45:993–1007. doi: 10.1194/jlr.R400001-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Davies SS, et al. Oxidized alkyl phospholipids are specific, high affinity peroxisome proliferator-activated receptor gamma ligands and agonists. Journal of Biological Chemistry. 2001;276:16015–23. doi: 10.1074/jbc.M100878200. [DOI] [PubMed] [Google Scholar]
- 20.McIntyre TM, Prescott SM, Stafforini DM. The emerging roles of PAF acetylhydrolase. Journal of Lipid Research. 2009;50 (Suppl):S255–9. doi: 10.1194/jlr.R800024-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemming PK, et al. Sensing of lysophospholipids by TRPC5 calcium channel. Journal of Biological Chemistry. 2006;281:4977–82. doi: 10.1074/jbc.M510301200. [DOI] [PubMed] [Google Scholar]
- 22.Murakami N, Yokomizo T, Okuno T, Shimizu T. G2A is a proton-sensing G-protein-coupled receptor antagonized by lysophosphatidylcholine. Journal of Biological Chemistry. 2004;279:42484–91. doi: 10.1074/jbc.M406561200. [DOI] [PubMed] [Google Scholar]
- 23.Yea K, et al. Lysophosphatidylcholine activates adipocyte glucose uptake and lowers blood glucose levels in murine models of diabetes. Journal of Biological Chemistry. 2009;284:33833–40. doi: 10.1074/jbc.M109.024869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bajad SU, et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. Journal of Chromatography A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.