Abstract
The characteristic geometry of the unicellular chlorophyte Chlamydomonas reinhardtii has contributed to its adoption as a model system for cellular asymmetry and organelle positioning. The eyespot, a photosensitive organelle, is localized asymmetrically in the cell at a precisely-defined position relative to the flagella and cytoskeletal microtubule rootlets. We have isolated a mutant, named pey1 for posterior eyespot, with variable microtubule rootlet lengths. The length of the acetylated daughter four-membered microtubule rootlet correlates with the position of the eyespot, which appears in a posterior position in the majority of cells. The correlation of rootlet length with eyespot positioning was also observed in the cmu1 mutant, which has longer acetylated microtubules, and the mlt1 mutant, in which the rootlet microtubules are shorter. Observation of eyespot positioning after depolymerization of rootlet microtubules indicated that eyespot position is fixed early in eyespot development and becomes independent of the rootlet. Our data demonstrate that the length of the daughter four-membered rootlet is the major determinant of eyespot positioning on the anterior-posterior axis and are suggestive that the gene product of the PEY1 locus is a novel regulator of acetylated microtubule length.
Keywords: Chlamydomonas, eyespot, microtubule rootlet, organelle positioning, pey1
INTRODUCTION
Asymmetry is a fundamental property of cellular organization across the spectrum of life. Apical-basal asymmetries define cell polarity in a variety of tissue types [Eaton and Simons, 1995] and the polarity of organelles contributes to the geometric organization of cells [Bornens, 2008]. The intrinsic asymmetry of the centrioles (basal bodies) establishes cell polarity by directing the positioning of organelles and localizing cortical polarization cues via interactions with components of the cytoskeleton [Beisson and Jerka-Dziadosz, 1999; Geimer and Melkonian, 2004]. The unicellular green alga Chlamydomonas reinhardtii has a well-defined and characteristic geometry which has contributed to its use as a model system for investigating the mechanisms controlling the asymmetric placement of organelles (Figure 1A). The basal bodies, acting as centrioles during mitosis, control the placement of the nucleus through connecting structures and thus transmit polarity cues for the anterior-posterior axis of the cell [Feldman et al., 2007]. The flagellar apparatus, nucleated by the basal bodies, marks the apical (anterior) pole of the cell. A single cup-shaped chloroplast fills the posterior two-thirds of the cell and contains a conspicuous pyrenoid situated at the posterior pole.
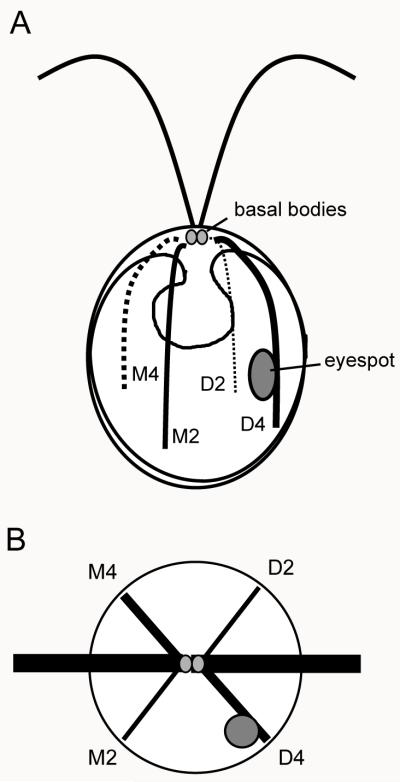
Figure 1.
Diagram of eyespot positioning and cytoskeletal organization in Chlamydomonas reinhardtii. A: The basal bodies (gray circles) nucleate the two flagella and four microtubule rootlets. Two rootlets (D2 and D4) extend from the region of the daughter basal body and two (M2 and M4) are inherited from the mother cell. The eyespot (large gray oval) is associated with the D4 rootlet. B: View from the anterior pole of a cell showing the cruciate arrangement of the rootlets. The eyespot is situated 45° from the plane of the flagella (dark bars).
A prominent feature of the C. reinhardtii cytoskeleton is the system of microtubule rootlets which originate from the basal bodies at the anterior pole of the cell (Figure 1A). These structures are highly acetylated, a modification that confers enhanced stability concurrent with increased resistance to microtubule-depolymerizing drugs [LeDizet and Piperno, 1986]. Data are indicative that the acetylation track of these rootlets extends the full length of the rootlet in the majority of cells [Mittelmeier et al., 2011]. The rootlets consist of four- and two-membered bundles of microtubules: a two-membered and four-membered rootlet originate near the striated fiber at the proximal end of the mother basal body, while a new two- and four-membered rootlet grow from the area near the daughter basal body during mitosis [Goodenough and Weiss, 1978]. The four-membered rootlets exist in a three-over-one arrangement [Melkonian, 1984] and the four-membered rootlets mark the plane of the cleavage furrow during cytokinesis [Ehler and Dutcher, 1998]. Like the basal bodies, each of which has an intrinsic asymmetry, the rootlet system exhibits asymmetric characteristics, as the daughter four-membered (D4) rootlet is noticeably longer than the others [Mittelmeier et al., 2011].
Like many chlorophytes, C. reinhardtii has a photosensory organelle, the eyespot, which forms de novo following each cell division and is localized asymmetrically at the equator of the cell. The eyespot is invariably positioned 45° from the flagellar plane (Figure 1B) [Holmes and Dutcher, 1989] and is associated with the D4 rootlet [Ringo, 1967; Moestrup, 1978; Melkonian, 1984]. The precise positioning of the eyespot is essential for coordination with flagellar beat frequency and proper photoorientation of the cell in response to light signals [Rüffer and Nultsch, 1991]. The eyespot comprises layers of carotenoid-rich pigment granules in the chloroplast that are overlayed by an elliptical patch of rhodopsin photoreceptors in the plasma membrane [Melkonian and Robenek, 1980; Berthold et al., 2008]. Evidence is suggestive that the tubulin cytoskeleton guides localization of the rhodopsin photoreceptors to the daughter side of the cell [Mittelmeier et al., 2011] and that this positioning cue plays a role in initiating the coordinated assembly of the eyespot and association of its chloroplastic components [Boyd et al., 2011].
Despite the established role of the D4 rootlet in the asymmetric positioning of the eyespot in C. reinhardtii, the factors that define the positioning of this organelle on the anterior-posterior axis of the cell have not been thoroughly investigated. In addition to mutants affecting assembly of the basal bodies and flagellar apparatus [Preble et al., 2001; McVittie, 1972; Barsel et al., 1988; Tam et al., 2007; Berman et al., 2003], previous genetic studies have identified mutants defective in various aspects of cytoskeletal structure that have supplied clues in the pursuit of this question. The cmu1 (cytoplasmic microtubules unorganized) mutant has supernumerary acetylated and non-acetylated microtubules that extend and curl around the posterior of the cell [Horst et al., 1999]. The mlt1 (multiple eyespot) mutant is characterized by supernumerary eyespots [Lamb et al., 1999] and acetylated rootlets that are significantly shorter than wild-type lengths [Mittelmeier et al., 2011]. In this study we describe the identification and characterization of a novel mutant, pey1 (posterior eyespot), which has variable microtubule rootlet lengths that correlate with the position of the eyespot in the cell. We demonstrate that the D4 microtubule rootlet is the major determining factor for establishing eyespot placement in C. reinhardtii, and that positioning of this multi-compartmental organelle becomes independent of the rootlet early after eyespot assembly.
RESULTS
Microtubule rootlet length varies in eyespot-position mutants
Wild-type Chlamydomonas reinhardtii cells have a single asymmetrically-localized eyespot at or near the cell equator (Figure 2A). Cells of the previously-described cmu1-1 mutant are characterized by defects in microtubule organization and altered cell shape [Horst et al., 1999] and, when examined by bright field microscopy, often exhibit posteriorly-positioned eyespots (Figure 2B). Cells of the mlt1 mutant have multiple eyespots that are frequently more anteriorly positioned than wild-type eyespots and can form on either side of the cell [Lamb et al., 1999] (Figure 2E). To identify additional mutants with defects in eyespot assembly or positioning, we conducted a forward genetic screen of 600 insertion mutants (gift of Patrice Hamel, The Ohio State University, Columbus, OH) using a simple phototaxis assay [Lamb et al., 1999]. One strain was initially characterized by erratic, jerky swimming behavior. When scored by bright field light microscopy, a majority of cells within a population of this strain had an eyespot at or near the posterior end of the cell (Figure 2C and D). We therefore named the mutant pey1 for posterior eyespot. Subsequent out-crosses of this strain indicated that the swimming defect did not segregate with the eyespot-positioning phenotype. Cells of an out-crossed pey1 strain were able to swim smoothly and undergo positive phototaxis. Although we mapped the location of the APHVII insertion in pey1, we found that the posterior-eyespot phenotype did not segregate with the insertion, hindering identification of the gene product of the PEY1 locus. Southern blotting confirmed that hygromycin-resistance was due to a single insertion event unlinked to the PEY1 locus. Genetic analysis confirmed that the pey1 mutation is unlinked to the cmu1 mutation.
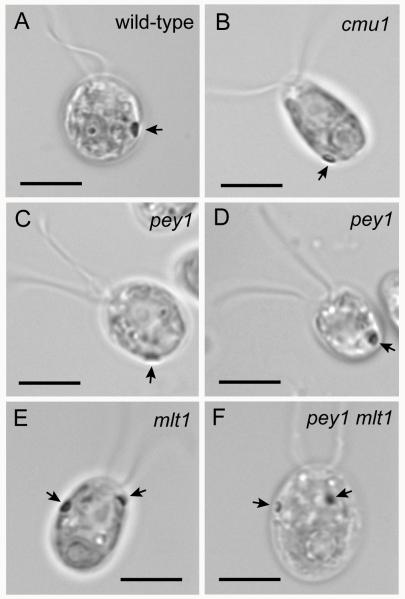
Figure 2.
Bright field micrographs of wild-type C. reinhardtii cell and eyespot-position mutants. Arrows indicate eyespots. A: Wild-type cell with an equatorially-positioned eyespot. B: cmu1 mutant cell with a posteriorly-positioned eyespot near the pyrenoid. C,D: pey1 mutant cells with posterior eyespots. E: mlt1 mutant cell with two eyespots, one of which is positioned anteriorly in the chloroplast lobe. F: The pey1 mlt1 double mutant exhibits the mlt1 phenotype. Bars, 5 μm.
We examined microtubule rootlets in the mlt1 and pey1 mutants using an antibody to acetylated-α-tubulin (clone 6-11-B-1, Sigma, St. Louis, MO). Interestingly, mlt1 cells have significantly shorter microtubule rootlets than wild-type, with the ratio of longest rootlet to cell length (R1/L) approximately one quarter less than the wild-type average (Figure 3B). The lengths of microtubule rootlets in the pey1 mutant are more variable and often longer than wild-type (Figure 3C). These observations led to the hypothesis that D4 rootlet length correlates with eyespot position in C. reinhardtii.
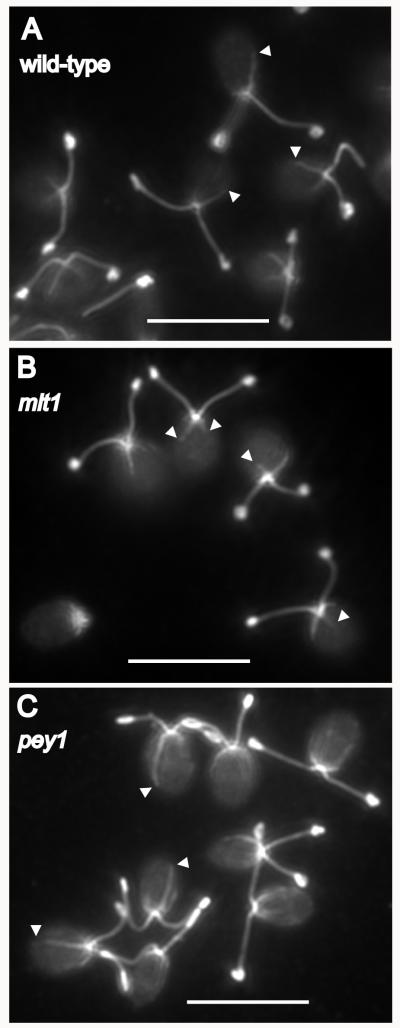
Figure 3.
Acetylated rootlet length is perturbed in eyespot-position mutants. Immunofluorescence micrographs of fields of methanol-fixed C. reinhardtii cells stained with anti-acetylated tubulin. A: Wild-type cells. Ends of the acetylated D4 rootlets are marked by arrowheads. B: The mlt1 mutant has shorter acetylated rootlets. C: Rootlets in the pey1 mutant vary in length and are often longer than those of wild-type cells. Bars, 10 μm.
Eyespot position corresponds to acetylated microtubule rootlet length
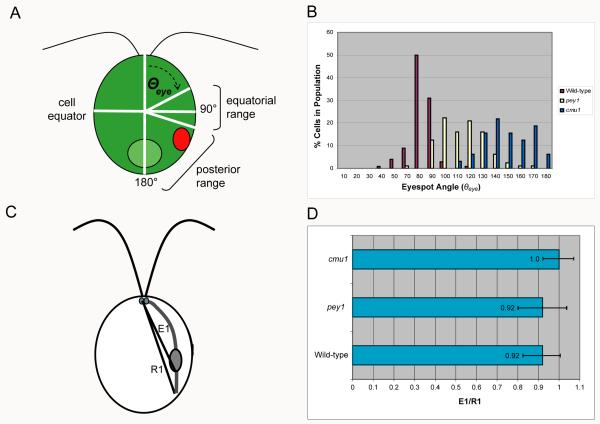
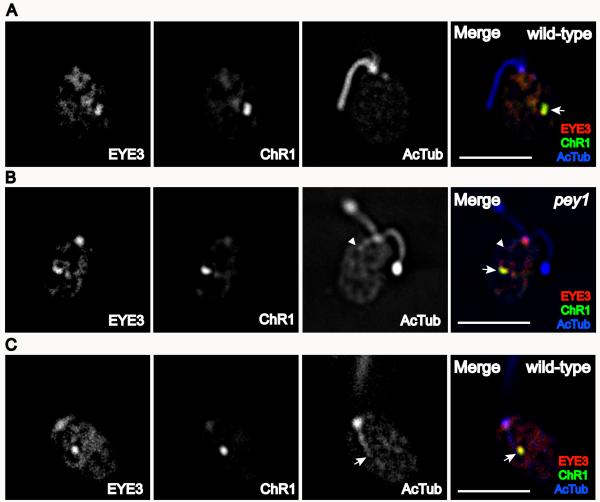
To quantify eyespot position in the pey1 and cmu1 mutants, we defined Θeye as the angle of the eyespot along the anterior-posterior axis from the center of the cell body. The anterior flagellar pole lies at an angle of 0° and the posterior pole at an angle of 180° (Figure 4A). Θeye varied widely in pey1 cells, in contrast to the wild-type range, and eyespots of cells of a cmu1 population were all in a posterior range (Figure 4B). The mean Θeye in wild-type cells was 89 ± 9° (n=116) while pey1 cells had a mean Θeye of 110 ± 18° (n=81) and cmu1 a mean of 144 ± 19° (Table I). The mean eyespot angle of pey1 cells was significantly different than the means of both wild-type and cmu1 cells (Table II). We also quantified the position of the eyespot in relation to the highly-acetylated D4 rootlet, defining E1 as the distance from the anterior pole to the posterior of the eyespot and R1 as the distance from the anterior pole to the end of rootlet acetylation (Figure 4C). Immunofluorescence microscopy of pey1 and cmu1 cells stained with antibodies against acetylated α-tubulin and the photoreceptor channelrhodopsin-1 (ChR1) revealed that the position of the eyespot strongly correlated with the length of the acetylated track of the D4 rootlet (Table I and Figure 4D). The E1/R1 ratio did not differ significantly between wild-type and pey1 cells, but the E1/R1 ratio of cmu1 was significantly higher than either wild-type and pey1 (Table II). The lengths of pey1 and cmu1 flagella did not differ markedly from wild-type (Table I).
Figure 4.
Eyespot position varies over a wider, longer range in the pey1 and cmu1 mutants compared to wild-type, and eyespot position correlates with the length of the D4 rootlet. A: Diagram of a C. reinhardtii cell showing basis for positioning measurements. Eyespot angle (Θeye) along the anterior-posterior axis was defined from the center of the cell. For wild-type cells, the distribution of Θeye centers on the cell equator. B: Histogram showing the distribution of Θeye in populations of wild-type, pey1, and cmu1 cells. The distribution of eyespot angles in the pey1 mutant is skewed toward the posterior of the cell, and eyespots in cmu1 cells are always observed in the posterior range. C: Diagram showing basis for distance measurements. Distances from the anterior pole to the distal ends of the eyespot (E1) and D4 rootlet (R1) were measured as straight lines. Distance between end of eyespot staining and rootlet staining was calculated from E1/R1. D: Graph plotting the mean E1/R1 ratio (± S.D.) between the eyespot and end of the D4 rootlet in populations of wild-type, pey1, and cmu1 cells scored by indirect immunofluorescence staining of ChR1 photoreceptor and acetylated tubulin.
Table I.
Measurements (mean ± standard deviation) of wild-type, pey1, and cmu1 cells. pey1 and cmu1 cells are characterized by posteriorly-positioned eyespots and more variable D4 rootlet lengths.
| Strain | Θeyea (°) | (n) | E1/R1b | (n) | Flagellar Length (μm) |
(n) | Tag Lengthc (μm) | (n) |
|---|---|---|---|---|---|---|---|---|
| wild-type | 89 ± 9 | (116) | 0.92 ± 0.09 | (63) | 8.9 ± 1.2 | (100) | 0.34 ± 0.33 | (20) |
| pey1 | 110 ± 18 | (81) | 0.89 ± 0.12 | (84) | 8.5 ± 1.6 | (100) | 0.54 ± 0.45 | (20) |
| cmu1 | 144 ± 19 | (32) | 1.00 ± 0.07 | (32) | 9.3 ± 1.5 | (100) | 0.26 ± 0.26 | (20) |
eyespot angle
ratio of eyespot position to D4 rootlet length
Table II.
Statistical analysis of measurements of wild-type, pey1, and cmu1 cells. P-values were calculated using one-tail Student’s T tests assuming equal variance.
| Pairwise Comparison | P(0.05) Θeye | P(0.05) E1/R1 | P(0.05) Tag Length |
|---|---|---|---|
|
| |||
| wild-type vs. pey1 | 4.3 × 10−22 | 0.45 | 0.07 |
| wild-type vs. cmu1 | 3.8 × 10−50 | 3.2 × 10−5 | 0.2 |
| pey1 vs. cmu1 | 4.1 × 10−15 | 4.0 × 10−4 | 0.01 |
The pey1 and cmu1 mutations do not affect eyespot morphology
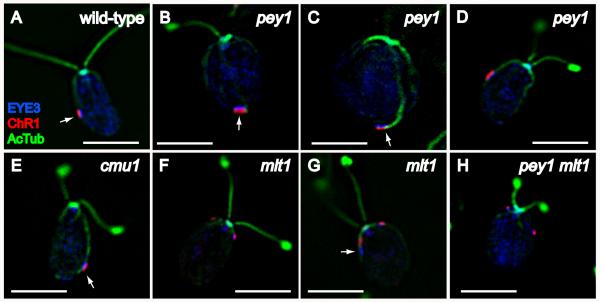
As assessed by triple-staining for acetylated tubulin, ChR1, and the eyespot pigment granule marker EYE3, eyespot morphology appeared normal in pey1 and cmu1 cells (Figure 5B-E), indicating that the cytoskeletal defects of these mutants do not affect eyespot assembly. Examination of the mlt1 mutant by immunofluorescence staining with EYE3 and ChR1 antisera confirmed that the anteriorly-positioned eyespots correlated with the short-rootlet phenotype of this mutant (Figure 5F), in agreement with previously-reported results [Mittelmeier et al., 2011]. mlt1 mutant cells frequently exhibit defects in eyespot morphology, with loss of association between the pigment granules and photoreceptor evident in 54% of cells scored in an asynchronously-dividing population in stationary phase (n=160) (Figure 5G). A double mutant of pey1 and mlt1 exhibited the mlt1 phenotype, indicating that mlt1 is epistatic to pey1 (Figure 5H).
Figure 5.
Characterization of eyespot morphology and rootlet associations in wild-type C. reinhardtii and eyespot-positioning mutants. Combined immunofluorescence micrographs of methanol-fixed cells stained with anti-EYE3 (blue), anti-channelrhodopsin-1 (ChR1) (red), and anti-acetylated tubulin (green). A: Wild-type cell with equatorially-positioned eyespot, visible by the co-positioned layers of photoreceptor (red) and eyespot pigment granules (EYE3, blue). Note association of eyespot with the D4 rootlet (AcTub, blue). B-D: pey1 mutant cells exhibit a range of eyespot positions including posterior (arrows in B and C) and equatorial (D). Arrow in C highlights rootlet association. Eyespot morphology appears normal in pey1 cells. E: cmu1 mutant cell. The posteriorly-positioned eyespot is associated with the rootlet (arrow). F and G: Eyespots in mlt1 mutant cells are often anteriorly-positioned (F) and exhibit loss of co-positioning of photoreceptor with pigment granules (arrow in G). H: The pey1 mlt1 double mutant exhibits the multiple-eyespot phenotype of mlt1 including anterior eyespots. Bars, 5 μm.
Extension of D4 rootlet acetylation beyond the eyespot is retained in pey1 and cmu1 mutants
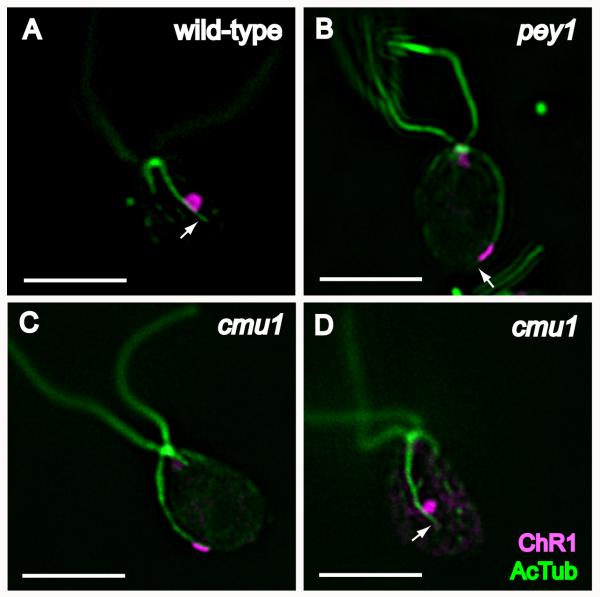
As previously reported, rootlet acetylation often extends a short distance beyond the eyespot in wild-type cells [Mittelmeier et al., 2011]. We examined whether mutations affecting rootlets and eyespot position altered this “tag” of acetylation. Observation of strains double-stained for ChR1 and acetylated tubulin indicated that a tag was present in 75% of both wild-type and cmu1 cells and 95% of pey1 cells (n=20); (Figure 6). This tag could be observed even in pey1 cells with a far-posterior eyespot (Figure 6B). The average tag length in pey1 cells was not significantly different than wild-type (Tables I and II). These data imply that the mechanism for determining the extent of acetylation beyond the eyespot is distinct from the regulation of overall steady-state length of the microtubule rootlet.
Figure 6.
Extension of D4 rootlet acetylation beyond the eyespot is retained in pey1 and cmu1 mutants. Combined immunofluorescence micrographs of individual cells stained for ChR1 (magenta) and acetylated tubulin (green). A: wild-type cell showing “tag” of acetylation on D4 rootlet beyond posterior edge of ChR1 staining (arrow), present in 75% of wild-type cells. B: pey1 cell with acetylated tag (arrow) beyond posterior-positioned eyespot. Tags were present in 95% of pey1 cells scored. C: cmu1 cell showing ChR1 staining co-extensive with end of rootlet. D: Face-on view of a cmu1 cell showing tag (arrow). Acetylated tags were present in 75% of cmu1 cells scored.
mlt1 exhibits enhanced sensitivity to colchicine
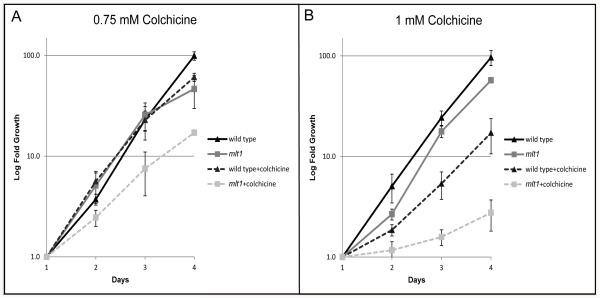
To further characterize effects of eyespot-positioning mutations on the cytoskeleton, cultures were treated with the microtubule-destabilizing drug colchicine and assayed for growth over several days. While the growth of colchicine-treated pey1 and cmu1 strains did not differ observably from wild-type, the growth rate of mlt1 cultures was significantly attenuated. At a concentration of 0.75 mM colchicine, the doubling time of mlt1 cells slowed by four hours compared to one hour for wild-type cells. At a slightly higher dose of 1 mM colchicine, the doubling time of wild-type cells slowed by six hours whereas mlt1 exhibited substantially increased sensitivity to the drug, the doubling time slowing by 38 hours (Figure 7; see Tables S2 and S3 in supplementary material). These results provide additional evidence that the MLT1 locus affects the stability or dynamics of the microtubule cytoskeleton.
Figure 7.
mlt1 cells exhibit enhanced sensitivity to colchicine. Graphs plotting the average logarithmic-fold concentrations of triplicate samples of wild-type and mlt1 cells ± standard deviation grown with and without colchicine at concentrations of 0.75 and 1 mM. Data points were normalized to the first 24-hour counts. (A) With treatment at 0.75 mM colchicine, only mlt1 showed a significant retardation in growth at all time points (p value < 0.05). At day 4 both wild-type and mlt1 strains showed significant growth retardation with drug treatment (p value < 0.05), with wild-type doubling time reduced by one hour and mlt1 doubling time reduced by four hours relative to the non-treated controls. (B) At 1 mM colchicine, both strains showed a significant retardation of growth at all time points (p value < 0.05). At day 4 wild-type doubling time was reduced by six hours, whereas mlt1 doubling time was reduced by 38 hours.
Eyespot position is established and becomes independent of the D4 rootlet prior to interphase
Our investigation of the role of the D4 rootlet in eyespot placement along the anterior-posterior axis led us to ask whether the positioning of the eyespot is labile and could be experimentally manipulated by altering microtubule organization. If eyespot position in interphase cells is dependent upon the length of the D4 rootlet, shortening or lengthening the rootlet would be expected to cause a proportional change in eyespot position on the anterior-posterior axis. As microtubule stabilizing and destabilizing drugs have limited effects on the rootlets of living cells, we assessed eyespot position in asynchronous populations of wild-type and pey1 cells after an extended incubation at 0°C, which depolymerizes all cytoplasmic microtubules [LeDizet and Piperno, 1986]. Cells were fixed and stained with anti-acetylated tubulin, EYE3, and ChR1 after 90 minutes at 0°C. After treatment, acetylated tubulin staining was mostly confined to the basal bodies and flagella, indicating that the rootlet microtubules had been depolymerized almost completely; however, in cells with a depolymerized D4 rootlet, the eyespot retained its structure, observed by the continued co-positioning of pigment granules and ChR1 photoreceptor, and did not move to an anterior position (Figure 8A). Figure 8B shows a pey1 cell with an incompletely depolymerized D4 rootlet and the eyespot remaining in a fixed position. In many cells, the D4 rootlet remained after treatment even though all other microtubule rootlets had depolymerized (Figure 8C), demonstrating that the D4 rootlet is especially stable and resistant to depolymerization relative to other members of the tubulin cytoskeleton. These data indicate that eyespot positioning becomes independent of the D4 rootlet after being established, and that the structural organization of the eyespot is not dependent on rootlet association in interphase.
Figure 8.
Eyespot positioning in cold-treated wild-type and pey1 cells. A: Immunofluorescence micrographs of a wild-type cell treated for 90 minutes at 0°C to depolymerize microtubules. Eyespot structure remains intact and retains approximately equatorial position as indicated by EYE3 and ChR1 staining (arrow in merge). B: Immunofluorescence micrographs of a pey1 cell showing incompletely depolymerized rootlet (arrowhead) while the eyespot retains its position. C: Immunofluorescence micrographs of a wild-type cell after 90 minutes at 0°C showing D4 rootlet remaining after depolymerization of other microtubule rootlets (arrow). Bars, 5 μm.
DISCUSSION
The organization and placement of cellular organelles is largely dependent on the guidance of cytoskeletal elements. The basal bodies (centrioles) of the unicellular alga Chlamydomonas reinhardtii instruct the positioning of the nucleus [Feldman et al., 2007] and direct the characteristic asymmetric placement of organelles in the cell, such as the chloroplast, by establishment of cortical polarity [Ehler and Dutcher, 1998]. The daughter four-membered (D4) microtubule rootlet, emanating from the vicinity of the daughter basal body, directs the asymmetric placement of the eyespot, a photosensory organelle located near the cell equator. We have determined that in addition to its role in positioning the eyespot on the daughter half of the cell, the D4 rootlet is the major determinant of eyespot positioning on the anterior-posterior axis, and that this positioning is to a large extent fixed following eyespot assembly.
The characteristic placement of the eyespot near the end of the acetylated track of the D4 rootlet in wild-type cells is retained in the pey1 mutant, which exhibits variable microtubule rootlet lengths, the cmu1 mutant, which has supernumerary microtubules of extended length, and the multiple-eyespot mutant mlt1, in which rootlet length is shorter than the wild-type average. This correlation is consistent with a role of the D4 rootlet in directing the placement of the eyespot on the anterior-posterior axis of the cell.
The observation that depolymerization of microtubule rootlets in interphase wild-type cells had no effect on eyespot position demonstrates that eyespot position becomes independent of the D4 rootlet after a certain point in the cell cycle. The fact that the eyespot apparatus is a stable structure that can be isolated biochemically [Schmidt et al., 2006] may account for its relative immobility once fully assembled. It is possible that the cytoskeleton contributes to maintaining the position of the eyespot, but observation of the organelle at longer time points after loss of rootlet association would be required. Eyespots of the similar species C. eugametos were observed to move in the chloroplast toward the pyrenoid after cells had been cultured for several days in the microtubule-depolymerizing drug colchicine [Walne, 1967], but it is unclear what observed changes in ultrastructure may have been due to secondary effects of the drug. It is likely that cell-cycle control of the expression of eyespot components and kinetic factors regulating microtubule extension together constitute the defining elements in the final determination of eyespot placement.
Several mechanisms could account for the aberrant placement of eyespots in the pey1 mutant: (1) the “fixation time” of the eyespot is delayed, so that eyespot components would travel to a more posterior position as the rootlet extends before the organelle coalesces into a sufficiently stable structure; (2) transport and delivery of chloroplastic eyespot-assembly factors to (or their association with) the D4 rootlet could be delayed, resulting in a scenario similar to that described above; or (3) acetylation and thus stabilization of the rootlet occurs faster or is otherwise mis-regulated, resulting in placement of eyespots near the rootlet ends in more posterior positions. Distinguishing between these models would require development of markers for real-time observation of microtubule rootlet dynamics and localization of eyespot proteins during and immediately following cytokinesis, as well as the availability of drugs for manipulating microtubule acetylation.
The specific roles of the MLT1 and CMU1 loci in cytoskeletal organization remain unclear. The MLT1 gene is predicted to encode a high-molecular weight, low-complexity protein with no recognized functional domains (JGI C. reinhardtii v 4.0; Protein ID 188661; http://www.chlamy.org), and its subcellular localization has not yet been determined. The epistasis of mlt1 to pey1, along with the observation that the mlt1 mutation disrupts the normal daughter-directed localization of eyespot components, implies an upstream effect of MLT1 in the pathway of eyespot assembly involving interaction of MLT1 with cytoskeletal elements in the basal body region. The correspondence of the mlt1 mutation with increased sensitivity to colchicine is also supportive of interaction of MLT1 with the microtubule cytoskeleton. The gene product of the CMU1 locus has not yet been identified, but is postulated to function in microtubule +-end capture in a manner similar to interactions of motor proteins with microtubules previously characterized in yeast [Horst et al., 1999]. The rootlet phenotype observed in pey1 seems to imply a defect in microtubule regulation rather than assembly of eyespot proteins, but it is also possible that a transport defect, such as a mutation in a microtubule-associated motor protein, could affect both processes. It remains possible that the lesion in the PEY1 locus does not result in a null mutation, and some of the phenotypic variability is due to dosage-dependent effects. Identification of the gene products of the CMU1 and PEY1 loci should clarify the mechanisms that control the timing of eyespot placement and lend new insights into the regulation of microtubule length and cytoskeletal organization.
MATERIALS AND METHODS
Chlamydomonas strains and media
Strains used in this study are listed in Table S1 in supplementary material. Chlamydomonas reinhardtii wild-type strains 137c mt+ (CC-125) and mt− (CC-124) were obtained from the Chlamydomonas Stock Center (University of Minnesota, St. Paul, MN). Strain mlt1-1 (CC-4304) was originally obtained following UV-mutagenesis of strain 137c mt+ [Lamb et al., 1999]. The NIT1 insertional mutant strain cmu1-1 (CC-3945) was obtained from George Witman (University of Massachusetts Medical Center, Worcester, MA). Strain 4A (CC-4051) was obtained from Patrice Hamel (The Ohio State University, Columbus, OH). Strain pey1-1 arg7 was obtained from a genetic screen of clones of strain 3A (arg7, mt+) after transformation by electroporation with 100 ng of aphVII insert DNA amplified from plasmid Hyg3 by PCR with primers Aph7-F (5′-TCGATATCAAGCTTCTTTCTTGC-3′) and Aph7-R (5′-AAGCTTCCATGGGATGACG-3′). The Arg+ pey1-1 mt+ strain was derived from an out cross of pey1-1 arg7 mt+ to strain 137c mt−.
Strains were maintained on solid tris-acetate-phosphate (TAP) medium [Harris, 1989] or TAP supplemented with 200 μg/mL arginine. For liquid cultures, freshly-grown cells from plates (2 to 4 days) were inoculated into either modified Sager and Granick medium I with Hutner’s trace elements (M medium) or M medium lacking nitrogen (M-N medium) and grown overnight at 25°C under continuous light.
Genetic screen
Approximately 600 strains of Chlamydomonas reinhardtii strain 3A carrying insertions of either a paramomycin-resistance or AphVII (hygromycin-resistance) cassette were screened using a simple assay for phototactic ability. Strains were patched on solid TAP medium plus arginine and inoculated into 1.2 mL liquid M-N medium in test tubes. Cultures were grown overnight at 25°C and assayed for phototaxis by placement in a covered box with a narrow slit for illumination. Phototaxis-defective or motility-deficient strains were examined by bright field microscopy.
Genetic analysis
Fresh cultures from plates were grown for two days on solid R medium containing 1/10 of the normal nitrogen source at 25°C under continuous illumination. Cells were inoculated into 1 mL M-N medium and incubated 4 hours at 25°C, and 200 μL of each culture were combined and allowed to mate for one hour under continuous illumination at 25°C. Mating mixtures were plated on solid R medium containing 4% agar and kept in the dark for at least 4 days. Dissection and tetrad analysis were conducted according to standard methods [Harris 1989].
Bright field microscopy
Cells from overnight liquid cultures were viewed according to the protocol described in Mittelmeier et al. (2008).
Immunofluorescence microscopy
Samples for observation by immunofluorescence were prepared according to the protocol described in Mittelmeier et al. (2008), with modifications described in Boyd et al. (2011). Images in figures 2 and 3 were captured on a Leica DMRXA microscope using a 100x planapochromat 1.4 numerical aperture oil immersion objective with 1.6 x optivar and a Chroma 71001A filter set (Chroma Technology Corp., Rockingham, VT). Exposures were captured using a QImaging (Burnaby, B.C. Canada) Retiga EX cooled CCD camera driven by Universal Imaging (Downingtown, PA) MetaMorph v.6.1.2 software. Images in figures 5, 6 and 8 were captured on a DeltaVision inverted epifluorescence microscope (Applied Precision, Issaquah, WA) using an Olympus TH4 100x objective with a 1.6 optivar and deconvolved using the SoftWorx imaging program (Applied Precision). Images were adjusted for brightness and contrast and combined in National Institutes of Health (NIH) ImageJ, then cropped in Adobe Photoshop (Adobe Systems, Palo Alto, CA).
Measurements
Wild-type (137c) and pey1 cells were stained for EYE3 detected with donkey anti-rabbit Alexa-488-conjugated secondary (Molecular Probes) as described previously, and eyespot position was measured in immunofluorescence micrographs of cell fields using the Angle tool in NIH ImageJ. The angle from the anterior pole of each cell to the approximate center of the fluorescent spot stained with anti-EYE3 was defined as Θeye using the center point of the cell as reference. Eyespot distance from the end of the D4 rootlet was measured as a straight line from the anterior pole to the end of staining in cells double-stained for ChR1 and acetylated tubulin by immunofluorescence using the Straight Line Segment tool in ImageJ. For measurements of flagellar length, cells of strains 4A mt+ (wild-type), pey1 mt+, and cmu1 mt+ were stained with anti-acetylated α-tubulin and detected with a goat anti-mouse Alexa-488-conjugated secondary antibody (Molecular Probes) according to the protocol described previously, and individual flagella were measured using the Straight Line Segment tool in ImageJ. Lengths of acetylated tags were measured similarly from images double-stained for ChR1 and acetylated tubulin. Measurements were converted to micrometers in Microsoft Excel.
Drug treatment
Chlamydomonas reinhardtii wild-type strain 137c mt+ and mutant strain mlt1 mt+ cells were grown on solid TAP medium and cells were inoculated into 2 ml of M medium. After four hours of incubation at 25° C under constant illumination, cells were counted using a hemocytometer, and 1.5×105 cells were transferred from the medium to a 50 ml flask containing 5 ml of fresh M medium plus or minus either 0.75 mM or 1 mM colchicine (Sigma, St. Louis, MO). Cultures were incubated at 25° C with constant light and agitation. Every 24 hours for four days 200μl of medium was removed and cells were counted using a hemocytometer.
Cold treatment
Cultures were grown overnight in liquid M medium and cells were pelleted by centrifugation. Cells were resuspended in autolysin and incubated for 30 minutes at room temperature with gentle shaking, pelleted by centrifugation, and resuspended in small volumes of PBS (approximately 50 μL/culture). Cell suspensions were then incubated on ice for 90 minutes, spotted on poly-lysine-coated slides, and fixed and stained for immunofluorescence microscopy as described above.
Construction of figures
Images were cropped with Adobe Photoshop and figures prepared with Adobe Illustrator (Adobe Systems, Palo Alto, CA). Data were compiled and statistically analyzed in Microsoft Excel and graphs were exported to Adobe Illustrator.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Patrice Hamel (The Ohio State University, Columbus, OH) for donating the insertional library, Peter Hegemann (Humboldt-Universität Berlin, Germany) for the gift of ChR1 antiserum, Telsa Mittelmeier, Kylie Swisher, and Ximin Du for research support and assistance, and Matthew Laudon at the Chlamydomonas Stock Center for reliable supply of strains. This work was supported by National Institutes of Health Graduate Training Grant in Biochemistry and Molecular Biology T32GM008659 (to J. S. B. and M. D. T.) and National Science Foundation Grant MCB-0843094 (to C. L. D.). M. M. G. was supported by Research Experience for Undergraduates (REU) Grant DBI-0551115 from the National Science Foundation.
REFERENCES
- Barsel S-E, Wexler DE, Lefebvre PA. Genetic analysis of long flagella mutants of Chlamydomonas reinhardtii. Genetics. 1988;118:637–648. doi: 10.1093/genetics/118.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson J, Jerka-Dziadosz M. Polarities of the centriolar structure: morphogenetic consequences. Biol. Cell. 1999;91:367–378. [PubMed] [Google Scholar]
- Berman SA, Wilson NF, Haas NA, Lefebvre PA. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr. Biol. 2003;13:1145–1149. doi: 10.1016/s0960-9822(03)00415-9. [DOI] [PubMed] [Google Scholar]
- Berthold P, Tsunoda SP, Ernst OP, Mages W, Gradmann D, Hegemann P. Channelrhodopsin-1 initiates phototaxis and photophobic responses in Chlamydomonas by immediate light-induced depolarization. Plant Cell. 2008;20:1665–1677. doi: 10.1105/tpc.108.057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. Organelle positioning and cell polarity. Nat. Rev. Mol. Cell Biol. 2008;9:874–886. doi: 10.1038/nrm2524. [DOI] [PubMed] [Google Scholar]
- Boyd JS, Mittelmeier TM, Lamb MR, Dieckmann CL. Thioredoxin-family protein EYE2 and ser/thr kinase EYE3 play interdependent roles in eyespot assembly. Mol. Biol. Cell. 2011;22:1420–1428. doi: 10.1091/mbc.E10-11-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S, Simons K. Apical, basal, and lateral cues for epithelial polarization. Cell. 1995;82:5–8. doi: 10.1016/0092-8674(95)90045-4. [DOI] [PubMed] [Google Scholar]
- Ehler LL, Dutcher SK. Pharmacological and genetic evidence for a role of rootlet and phycoplast microtubules in the positioning and assembly of cleavage furrows in Chlamydomonas reinhardtii. Cell Motil. Cytoskeleton. 1998;40:193–207. doi: 10.1002/(SICI)1097-0169(1998)40:2<193::AID-CM8>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Feldman JL, Geimer S, Marshall WF. The mother centriole plays an instructive role in defining cell geometry. PLoS Biology. 2007;5:1284–1297. doi: 10.1371/journal.pbio.0050149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimer S, Melkonian M. The ultrastructure of the Chlamydomonas reinhardtii basal apparatus: identification of an early marker of radial asymmetry inherent in the basal body. J. Cell Sci. 2004;117:2663–2674. doi: 10.1242/jcs.01120. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Weiss RL. Interrelationships between microtubules, a striated fiber, and the gametic mating structure of Chlamydomonas reinhardtii. J. Cell Biol. 1978;76:430–438. doi: 10.1083/jcb.76.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press; San Diego, California: 1989. p. 780. [DOI] [PubMed] [Google Scholar]
- Holmes JA, Dutcher SK. Cellular asymmetry in Chlamydomonas reinhardtii. J. Cell Sci. 1989;40:273–285. doi: 10.1242/jcs.94.2.273. [DOI] [PubMed] [Google Scholar]
- Horst CJ, Fishkind DJ, Pazour GJ, Witman GB. An insertional mutant of Chlamydomonas reinhardtii with defective microtubule positioning. Cell Motil. Cytoskeleton. 1999;44:143–154. doi: 10.1002/(SICI)1097-0169(199910)44:2<143::AID-CM6>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Lamb MR, Dutcher SK, Worley CK, Dieckmann CL. Eyespot-assembly mutants in Chlamydomonas reinhardtii. Genetics. 1999;153:721–729. doi: 10.1093/genetics/153.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. Cytoplasmic microtubules containing acetylated α-tubulin in Chlamydomonas reinhardtii: spatial arrangement and properties. J. Cell Biol. 1986;103:13–22. doi: 10.1083/jcb.103.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVittie A. Flagellum mutants of Chlamydomonas reinhardtii. J. Gen. Microbiol. 1972;71:525–540. doi: 10.1099/00221287-71-3-525. [DOI] [PubMed] [Google Scholar]
- Melkonian M. Flagellar apparatus ultrastructure in relation to green algal classification. In: Irvine DEG, John DM, editors. Systematics of the Green Algae. Academic Press; London: 1984. pp. 75–120. [Google Scholar]
- Melkonian M, Robenek H. Eyespot membranes of Chlamydomonas reinhardtii: a freeze-fracture study. J. Ultrastruct. Res. 1980;72:90–102. doi: 10.1016/s0022-5320(80)90138-0. [DOI] [PubMed] [Google Scholar]
- Mittelmeier TM, Berthold P, Danon A, Lamb MR, Levitan A, Rice M, Dieckmann CL. C2 domain protein MIN1 promotes eyespot organization in Chlamydomonas reinhardtii. Euk. Cell. 2008;7:2100–2112. doi: 10.1128/EC.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelmeier TM, Boyd JS, Lamb MR, Dieckmann CL. Asymmetric properties of the Chlamydomonas reinhardtii cytoskeleton direct rhodopsin photoreceptor localization. J. Cell Biol. 2011;193:741–753. doi: 10.1083/jcb.201009131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moestrup Ø . On the phylogenetic validity of the flagellar apparatus in green algae and other chlorophyll A and B containing plants. Biosystems. 1978;10:117–144. doi: 10.1016/0303-2647(78)90035-7. [DOI] [PubMed] [Google Scholar]
- Preble AM, Giddings TH, Dutcher SK. Extragenic bypass suppressors of mutations in the essential gene BLD2 promote assembly of basal bodies with abnormal microtubules in Chlamydomonas reinhardtii. Genetics. 2001;157:163–181. doi: 10.1093/genetics/157.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringo DL. Flagellar motion and fine structure of flagellar apparatus in Chlamydomonas. J. Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüffer U, Nultsch W. Flagellar photoresponses of Chlamydomonas cells held on micropipettes-2: change in flagellar beat pattern. Cell Motil. Cytoskeleton. 1991;18:269–278. [Google Scholar]
- Schmidt M, et al. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. The Plant Cell. 2006;18:1908–1930. doi: 10.1105/tpc.106.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L-W, Wilson NF, Lefebvre PA. LF2 encodes a CDK-related kinase that regulates flagellar length and assembly in Chlamydomonas. J. Cell Biol. 2007;176:819–829. doi: 10.1083/jcb.200610022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne PL. The effects of colchicine on cellular organization in Chlamydomonas. II. Ultrastructure. Amer. J. Bot. 1967;54:564–577. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.