Abstract
To assess effects of carbon nanoparticle (CNP) exposure on renal epithelial cells, fullerenes (C60), single-walled carbon nanotubes (SWNT), and multi-walled carbon nanotubes (MWNT) were incubated with a confluent renal epithelial line for 48 h. At low concentrations, CNP-treated cells exhibited significant decreases in transepithelial electrical resistance (TEER) but no changes in hormone-stimulated ion transport or CNP-induced toxicity or stress responses as measured by lactate dehydrogenase or cytokine release. The changes in TEER, manifested as an inverse relationship with CNP concentration, were mirrored by an inverse correlation between dose and changes in protein expression. Lower, more physiologically relevant, concentrations of CNP have the most profound effects on barrier cell function and protein expression. These results indicate an impact of CNPs on renal epithelial cells at concentrations lower than have been previously studied and suggest caution with regard to increasing CNP levels entering the food chain due to increasing environmental pollution.
Keywords: Carbon nanotubes, fullerene, cortical collecting duct, dielectric spectroscopy, proteomics, transepithelial resistance
Introduction
Carbon nanoparticles (CNPs) are important components of the rapidly expanding nanotechnology field. Due to their size and unique electrical, mechanical, and thermal properties, they have found widespread application in electronic, aerospace, medical, agricultural, pharmaceutical, and other industries. Consequently, mass production and widespread application of nanoparticles continues to rise and, along with it, the likelihood of occupational and environmental exposure (Maynard et al. 2004; Borm et al. 2006; Lam et al. 2006) and potential for exposurerelated inflammation, human illness, and dysfunction. Depending on the manufacturing process, CNPs are released to the air and water and ultimately contaminate soil and food products (Reijnders 2006).
Despite recent efforts to characterize potential health hazards, the overall understanding of the biological effects of CNP exposure is far from complete. Three of the most common types are single-walled carbon nanotubes (SWNT), multi-walled carbon nanotubes (MWNT), and fullerenes (C60). SWNT consist of covalently bound carbon atoms arranged in a long, thin tube-like structure with a diameter of approximately 1.4 nm (Jia et al. 2005). MWCNT have a similar structure, but multiple layers of graphene sheets are concentrically rolled up for their formation with a diameter in the range of 10–50 nm (Jia et al. 2005). C60, also known as fullerenes or ‘buckyballs’, typically consist of 60 carbon atoms covalently linked together to form a spherical molecule.
Recent research has revealed diverse effects of CNPs on biological systems. One study indicated that SWNT and MWNT inhibit growth by apoptosis and loss of cell adhesion (Cui et al. 2005), while other studies suggest that carbon nanotubes seem to increase the growth of mesenchymal cells, cause fibrogenesis, and granuloma formation (Donaldson et al. 2006). We have previously shown that MWNT alter expression of genes for cellular transport, metabolism, cell cycle regulation, and stress response (Witzmann and Monteiro-Riviere 2006). MWNT are of special interest because of their structural similarity to asbestos (Poland et al. 2008). Early experiments with SWNT have shown them to be cytotoxic, and they have been shown to bind to ion channels (Park et al. 2003). Various types of nanoparticles are endocytosed and can alter the cytoskeletal organization (Gupta and Gupta 2005).
Assuming the possibility of systemic availability of nanoparticles via lung, skin, or gastric/intestinal absorption, secondary renal exposure is a distinct likelihood (Chen et al. 2006), particularly if the CNP exposure is chronic. Lacerda et al. (2008) have shown that mice injected with purified non-functionalized MWNTs contain both aggregated and individual carbon nanotubes in the glomerular capillaries. In elegant electron microscopy images, the authors show that aggregated nanoparticles appear to remain in the capillaries while individual nanotubes pass into Bowman’s space by crossing the endothelial fenestrations in a longitudinal conformation. From Bowman’s capsule they traverse the nephron and enter the urinary bladder.
Our current study examines the effect of carbon nanoparticles on the function of the epithelial cells lining the renal nephron. The model used in this study is the mouse principal cell type of the kidney cortical collecting duct, clone 4 (mpkCCDcl4) cell line. mpkCCDcl4 cells grow to form a confluent monolayer that simulates the barrier epithelial function and hormone responsiveness found in vivo in renal distal tubule and collecting ducts (Bens et al. 1999). Principal cells are of particular interest because they are responsible for much of the hormonally-regulated ion transport in the kidney. If the CNP exposure alters the hormonal responsiveness of these cells, salt homeostasis could be modulated, resulting in changes in blood pressure. If the barrier function of the intact epithelium is compromised, the resulting equilibration between the filtrate and the renal interstitium can have serious consequences ranging from an inability to concentrate urine to complete renal failure.
Experiments were conducted to determine functional, structural, and proteomic changes induced by application of CNPs to the renal barrier epithelial cells. Electrophysiological studies were used to determine the effect of CNPs on transepithelial electrical resistance (TEER), a measure of barrier integrity, and hormone responsiveness. Quantitative proteomic studies were conducted to correlate the observed structural and functional studies with CNP-induced changes in the expressed cellular proteome.
Methods
Materials
CNPs were purchased from SES Research (Houston, TX, USA) and used with no further purification. As reported by the manufacturer, C60 (#600-9980) was 99.95+%, ultra-pure and vacuum oven-dried; SWNT (#900-1301) (long) were purified single-walled nanotubes with an outer diameter <2 nm, length ranging from 5–15 μtivm, purity >90%CNT (>50% SWNT), ash <2% wt and amorphous carbon <5% wt; and purified MWNT (# 900-1203) with an outer diameter of 40–60 nm, length ranging from 5–15 μm, >95% nanotubes vs. amorphous carbon (<2%), and ash content <0.2%.
ADH ([Arg8]-Vasopressin), amiloride, transferrin, sodium selenide, and triiodothyronine were purchased from Sigma Chemical Co. (St Louis, MO, USA). DMEM/F12 tissue culture media, Glutamax, penicillin, streptomycin were purchased from Invitrogen (Carlsbad, CA, USA); fetal bovine serum came from ICN Biochemicals, Inc (Irvine CA); and ciprofloxacin from Mediatech Inc. (Herndon, VA, USA). Permeable tissue culture supports in six-well format were obtained from Costar-Corning (Acton, MA, USA). Mouse monoclonal antibody to proliferating cell nuclear antigen (PCNA) was obtained from Sigma Chemical Co. and goat anti-mouse Alexafluor red 594 was obtained from Invitrogen-Molecular Probes (Eugene, OR, USA).
CNP preparation
SWNT, MWNT, and C60 were diluted in fetal bovine serum to 5 mg/ml. A control was prepared by treating with fetal bovine serum in the absence of added nanoparticles. Using a Branson Sonifier 450, samples were sonicated at a duty cycle of 30% and an output control of 3 for 20 sec. Before sonication of each sample, the probe of the Branson Sonifier 450 was cleaned with ethanol and coated for 10 sec with normal serum. After sonication, the samples were sterilized via autoclave and diluted to a final concentration of 2% FBS-CNP in media. The final experimental concentrations of CNP varied over 6 orders of magnitude. In the manuscript we have expressed the concentrations as concentration per cm2 of epithelial cell layer. For conversion, X μg/cm2 = 2.5X μg/ml.
Background experiments performed using untreated and autoclaved serum in matched cultures showed that adding autoclaved serum to the confluent cultures for 48 h did not alter the TEER or the hormonal responses of the cell line as compared to untreated serum.
Cell culture
mpkCCDcl4 cells were grown in a humidified chamber at 37°C and 5% CO2. The cell line was maintained in plastic culture flasks and, for CNP exposures, cells were seeded onto Transwell filters or six-well tissue culture plates. The media was replaced thrice weekly and consisted of Dulbecco’s modified Eagle’s medium (DMEM): Ham’s F12 basal media supplemented with 2% fetal bovine serum, 1 mM Glutamax, 25 U/ml penicillin, 25 mg/ml streptomycin, 12 mg/l ciprofloxacin, 5 mg/l transferrin, 20 μg/l sodium selenite, and 10−7 M triiodothyronine. Cultures were used for exposures only after achieving a confluent monolayer of cells.
Electrophysiology
Electrophysiological techniques were used to monitor TEER as well as to observe changes in ion flux across the cellular monolayers in response to hormonal stimulation. Cells were grown to confluency over a period of 13 days on Transwell filters with CNP treatment in the last 48 h at concentrations indicated in the Figures. The filters were excised, mounted in a Ussing chamber, and connected to a DVC-1000 Voltage/Current Clamp (World Precision Instruments) with voltage and current electrodes on either side of the membrane as described in detail previously (Shane et al. 2006). The spontaneous transepithelial potential difference was clamped to zero, and the resultant short-circuit current (SCC) was monitored continuously. The cells were bathed in serum-free medium maintained at 37°C via water-jacketed buffer chambers on either side of the filter. Medium was circulated and kept at constant pH using a 5% CO2/95% O2 gas lift. TEER was recorded every 200 sec throughout each experiment by applying a 2 mV pulse and the resulting deflection in the SCC was measured and used to calculate the TEER by Ohm’s law. After the basal current stabilized, anti-diuretic hormone (ADH, vasopressin; 100 mU/ml) was added to the serosal bathing medium, and 30 min after the addition of ADH, amiloride (10−5 M), a specific blocker of the epithelial Na+ channel, was added to the apical bathing medium. Groupwise comparisons of TEER were conducted using one-way ANOVA and all pair wise multiple comparison procedures used the Holm-Sidak method, at P < 0.05.
Histochemistry
Confluent cellular monolayers were washed with Hanks Balanced Salt Solution (HBSS), fixed in 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100 in DI water. After rinsing with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer, cells were treated for 8 min with hot citric acid buffer, washed with HEPES and blocked with Normal Goat Serum diluted [1:10] in Blocking Buffer (NGSBB; 0.75 g Blocking Reagent/150 ml 1× Tris Buffered Saline with Tween-20 (TBST)) for 2 h at room temperature. Mouse monoclonal PCNA antibody in NGSBB (1:3000) was added to the cells overnight at 4°C. Cells were washed with HEPES buffer, and goat anti-mouse Alexafluor Red 594 secondary antibody in Blocking Buffer (1:1000) was added to the cells for 2 h at room temperature. Cells were washed with HEPES buffer and treated with DAPI. Cells were visualized using a Nikon Eclipse TE2000-U Microscope fitted with a Nikon Digital Camera (DXM1200F).
Cytokine assay
The expression pattern of eight cytokines; IL-1a (interleukin-1a), IL-1b (interleukin-1b), G-CSF (granulocyte-colony stimulating factor), GM-CSF (granulocyte macrophage-colony stimulating factor), MCP-1 (monocyte chemotactic protein-1), MIP-1a (macrophage inflammatory protein-1 alpha), SCF (stem cell factor), and RANTES (Regulated on Activation, Normal T Expressed and Secreted), was profiled using a kit, Mouse Inflammation ELISA Strip for Profiling 8 Cytokines (Signosis, Sunnyvale CA, USA). Apical and basolateral media from mpkCCDcl4 cell cultures were assayed in triplicate according to the manufacturer’s protocol. Briefly, 100 μl of media were added to wells coated with a primary antibody against a specific cytokine and incubated for 1 h at RT. Medium was aspirated from wells followed by three washes with 200 μl of assay wash buffer. 100 μl of biotin-labeled antibody mixture was added to each well and incubated for 1 h at RT. Wells were aspirated then washed three times with 200 μl of assay wash buffer. Streptavidin-HRP conjugate (100 μl) was added to each well and incubated for 45 min at RT followed by the aspiration and wash procedure. 100 μl of substrate was added and incubated for 10 min; subsequently 50 μl of stop solution was applied to each well. A microplate reader determined the optical density of each well at 450 nm. The concentrations of the inflammatory cytokines were directly proportional to the color intensity of the samples.
LDH assay
Media was collected from both apical and basolateral sides of exposed cells for electrophysiological studies and assayed for lactate dehydrogenase (LDH). The LDH assay was performed using Promega CytoTox 96 non-radioactive cytotoxicity assay kit (Promega, Madison, WI, USA). This is a colorimetric assay that quantitatively measures lactate dehydrogenase, a stable cytosolic enzyme released upon cell lysis. The released LDH oxidizes lactate to pyruvate which promotes conversion of tetrazolium salt INT to formazan, a water soluble molecule with absorbance at 490 nm. The amount of LDH released is proportional to the number of cells damaged or lysed. As a positive control for toxicity, we used cadmium dioxide (CdO2) (Sigma-Aldrich, St Louis, MO, USA), known for its cytotoxic properties. The CdO2 was prepared exactly like the other nanoparticles. The cells were exposed to CdO2 for 48 h and media collected from both apical and basolateral sides.
Scanning electron microscopy (SEM) and energy dispersive X-ray spectroscopy (EDS)
SEM and EDS data were collected on a LEO 1530 VP Scanning Electron Microscope equipped with an energy-dispersive X-ray analyzer. A few drops of SWNT and MWNT dispersions in cell culture media were placed on silicon wafers and allowed to air-dry. The silicon wafers were then mounted on aluminum stubs for SEM and EDS analysis.
Particle size analysis and dielectric spectroscopy of carbon nanotubes in media
The samples (the culture medium blank and CNPs dispersed in the medium at two different concentrations) for particle size analysis and dielectric spectroscopy were prepared at IUPUI. Particle size distribution of the samples was measured at 26°C using dynamic light scattering (Beckman Coulter N4-Plus submicron particle size analyzer) operating at 90° detector angle). Dielectric spectroscopy was carried out at NJIT with a Novacontrol BDS-80 broadband dielectric spectrometer, using a liquid sample cell with the size of 11.04 × 6.6 mm (diameter × thickness). The bias voltage was set at 1 volt, and the dielectric spectra were collected at room temperature in the frequency range 1–105 Hz.
Atomic force microscopy
To study CNP agglomeration on cell surfaces, morphology of these surfaces was evaluated with tappingmode atomic force microscopy (AFM) using a Digital Instrument Nanoscope II.
Proteomics
Proteins from duplicate samples were analyzed by label-free quantitative mass spectrometry (LFQMS) (Higgs et al. 2008; Wang et al. 2008). Cultured cell proteins were extracted in situ using a lysis buffer containing 8 M urea and 10 mM dithiothreitol (DTT) and protein concentrations were determined by Bradford assay (Bradford 1976). The same lysis buffer was used as the background reference for the protein assay and for BSA protein standards. The resulting protein extracts were reduced and alkylated with triethylphosphine and iodoethanol (Hale et al. 2004). Protein mixtures were digested with trypsin and filtered through 0.45 μm spin filters before being applied to the high-performance liquid chromatography (HPLC) system. Prior to tryptic digestion, each sample was spiked with chicken lysozyme as an internal reference to assess technical variation.
Tryptic peptides (20 μg) were injected randomly onto a Surveyor HPLC system (Thermo-Finnigan) with a C18 microbore column (Zorbax 300SB-C18, 1 mm × 65 cm). Peptides were eluted with a linear gradient from 5–45% acetonitrile developed over 120 min at a flow rate of 50 μl/min and the effluent was electro-sprayed into the LTQ mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). Data were collected in the ‘Triple Play’ (MS scan, Zoom scan, and MS/MS scan) mode. The acquired data were filtered and analyzed by a proprietary algorithm (Higgs et al. 2005) licensed to Monarch Lifesciences LLC, Indianapolis IN. Database searches against the International Protein Index (IPI) mouse database and the non-redundant Mus musculus database (NCBI) were carried out using the X!Tandem (Craig and Beavis 2004) and SEQUEST (Eng et al. 1994) algorithms.
Only those proteins identified with >90% confidence were evaluated quantitatively as described previously (Higgs et al. 2005, 2008). Briefly, when raw files were acquired from the LTQ mass spectrometer, all extracted ion chromatograms (XICs) were aligned by retention time. After alignment, area under the curve (AUC) for each individually aligned peak from each sample was measured, normalized, and compared for relative abundance. Significant differences in protein expression across dose groups were determined by ANOVA. To eliminate technical bias, randomization of order of measurement and ‘quantile normalization’ was used (Bolstad et al. 2003) and data normalized using a log2 scale.
Pathway analysis
To assist with interpretation of the numerous protein expression effects, we analyzed the differentially expressed protein data using Ingenuity Pathways Analysis (IPA) tools (Ingenuity Systems, Mountain View, CA; http://www.ingenuity.com/index.html), a web-delivered application that enables the discovery, visualization, and exploration of molecular interaction networks and canonical pathways in protein expression data (see Tables VII and VIII). Lists of differentially expressed proteins were uploaded into the IPA system and proteins that were associated with a specific functional network or canonical pathway in the Ingenuity Pathways Knowledge Base were considered for evaluation. The IPA computes a score for each network according to the fit of the user’s set of proteins of interest. The score is derived from a p value and indicates the likelihood that the selected proteins are categorized in a network due to random effects.
Table VII.
Functional networks in mpkCCDcl4 cells impacted by CNP exposure (P < 0.001).
| CNP | Functional network |
|---|---|
| C60 0.004 ug/cm2 (34▲; 27▼) | 1) Gene Expression, Cell Death, Energy Production (13▲; 4▼) 2) Cancer, Cellular Growth & Proliferation, Cell Cycle (6▲; 6▼) |
| C60 0.04 ug/cm2 (11▲; 5▼) | No significant relationship to networks |
| C60 40 ug/cm2 (4▲; 1▼) | No significant relationship to networks |
| SWNT 0.004 ug/cm2 (57▲; 59▼) | 1) Protein Synthesis, Gene Expression, Cellular Growth & Proliferation (4▲; 14▼) 2) Carbohydrate Metabolism, Cancer, Genetic Disorder (13▲; 4▼) 3) Cancer, Cellular Movement, Connective Tissue Development & Function (8▲; 5▼) |
| SWNT 0.04 ug/cm2 (14▲; 15▼) | 1) Cancer, Cell Death, Cellular Growth & Proliferation (7▲; 6▼) |
| SWNT 40 ug/cm2 (7▲; 5▼) | No significant relationship to networks |
| MWNT 0.004 ug/cm2 (45▲; 60▼) | 1) Cancer, Cell Cycle, Cell Death (7▲; 8▼) 2) Inflammatory Disease, Skeletal & Muscular Disorders (9▲; 7▼) 3) Neurological Disease, Connective Tissue Disorders (10▲; 4▼) 4) Endocrine Disorders, Cellular Movement, Metabolic Disease (9▲; 7▼) |
| MWNT 0.04 ug/cm2 (10▲; 7▼) | 1) DNA Replication, Recombination & Repair, Cancer, Genetic Disorder (3▲; 2▼) |
| MWNT 40 ug/cm2 (3▲; 7▼) | No significant relationship to networks |
Numbers in parentheses indicate the number of proteins whose abundance increased ▲ or decreased ▼. Protein alterations in the left column refer to those altered at each exposure (corresponding to Table III). Protein alterations appearing in the right column are only those corresponding to the specific functional networks determined by IPA, represented by the altered proteins (i.e., other proteins had no significant relationship to other networks). Those networks appearing in bold are those that are common to one or more CNPs.
Table VIII.
Canonical pathways in mpkCCDcl4 cells impacted by CNP exposure (only ≥ 3 proteins) (P < 0.001).
| CNP | Canonical pathway |
|---|---|
| C60 0.004 μg/cm2 | Fatty Acid Elongation in Mitochondria ▲ Glycolysis/Gluconeogenesis ▲▼ Mitochondrial Dysfunction ▲ Methane Metabolism ▲ Phenylalanine, Tyrosine and Tryptophan Biosynthesis ▲ |
| C60 0.04 μg/cm2 | No multiprotein pathways |
| C60 40 μg/cm2 | No multiprotein pathways |
| SWNT 0.004 μg/cm2 | Glycolysis/Gluconeogenesis ▲ Pentose Phosphate Pathway ▲ Galactose Metabolism ▲ Fructose and Mannose Metabolism ▲ Methane Metabolism ▼ |
| SWNT 0.04 μg/cm2 | Tight Junction Signaling ▲ NRF2-mediated Oxidative Stress Response ▲ |
| SWNT 40 μg/cm2 | Xenobiotic Metabolism Signaling ▼ NRF2-mediated Oxidative Stress Response ▼ |
| MWNT 0.004 μg/cm2 | Glycolysis/Gluconeogenesis ▲ Methane Metabolism ▲ One Carbon Pool by Folate ▲ Glycine, Serine and Threonine Metabolism ▲ Cellular Effects of Sildenafil ▲▼ |
| MWNT 0.04 μg/cm2 | Lysine Degradation ▼ |
| MWNT 40 μg/cm2 | No multiprotein pathways |
Arrows indicate whether proteins in those pathways increased ▲ or decreased ▼, or both ▲▼; Shaded areas emphasize pathways common to all three CNPs.
Results
Characterization of nanoparticles in aqueous media
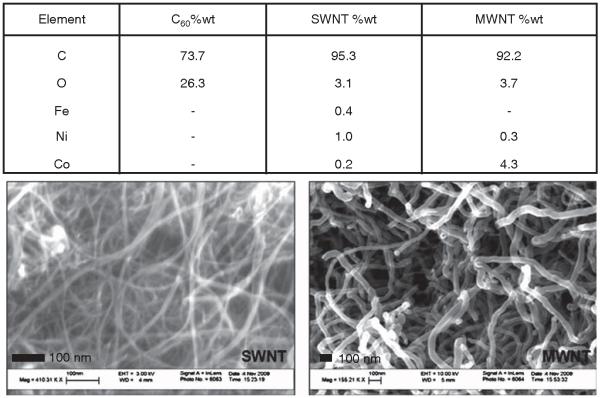
The nanomaterials used in these studies were ‘as-manufactured’ particles of the types that are likely to occur as environmental or workplace contaminants. An analysis of the three types of particles indicate that the fullerenes are virtually free of contaminating metals while the carbon nanotubes have measureable levels of iron, nickel and/or cobalt which are likely contaminates from the manufacturing process (Figure 1). SEM of SWNT and MWNT indicated that these materials are relatively free of amorphous carbon.
Figure 1.
Elemental analysis of the carbon nanoparticles determined by energy dispersive X-ray spectroscopy and SEM of SWNT and MWNT.
As-manufactured, unrefined nanotubes are sparingly dispersible in aqueous media. The highest concentrations of nanotubes used in these experiments, 40 and 4 μg/cm2 (100 and 10 μg/ml), show substantial agglomeration and precipitation onto the apical surface of the cellular monolayers. Figure 2 shows the particles that remain on the surface of the cells after removal of the media. Lower concentrations show no visible aggregates.
Figure 2.
Nanotube agglomeration in the tissue culture media. Multi-wall and single-wall nanotubes were prepared in serum and diluted into tissue culture media as described in Methods. The solutions, at the concentrations indicated, were added to six-well Transwell chambers and incubated for 48 h at 37°C. The media was removed. The Figure shows the agglomerated particles that were deposited on the apical side of the cellular monolayers under these incubation conditions.
Atomic force microscopy
In agreement with the visual observations (Figure 2), AFM demonstrated that at high dose (40 μg/cm2) exposures, roughness factor (Ra) is high, suggesting that large agglomerates are associated with the cell surface (Table I). These measurements represent the general filter surface and not the large black CNT particles seen in Figure 2. At a lower dose (0.4 μg/cm2), Ra is very low (near blank levels), suggesting that the particles are more dispersed and may, therefore, have greater potential for cellular interaction and biological activity. Agglomeration at cell surface would serve as a barrier which would prevent the dispersed CNPS from interacting with the cells.
Table I.
Results of AFM-Roughness (Ra) values on cell surface.
| CNP | Ra value |
|---|---|
| Blank | 147.5 |
| SWNT 0.4 μg/cm2 | 113.5 |
| MWNT 0.4 μg/cm2 | 160.1 |
| C60 40 μg/cm2 | 622.2 |
| SWNT 40 μg/cm2 | 432.2 |
| MWNT 40 μg/cm2 | 214.0 |
Particle size analysis and dielectric spectroscopy of CNTs in media
Results of particle size analysis of CNPs (C60, SWNT, and MWNT) dispersed in the culture media are listed in Table II. The particle size is expressed as ‘mean diameter’. The increase in particle size of fullerene dispersion was negligible when the concentration was increased by an order of magnitude, indicating little aggregation of fullerene in the culture medium. On the other hand, the increase in particle size with increasing CNT concentration is an indication of nanotube aggregation at higher concentration. Note that, at higher concentration of 0.04 μg/cm2, although the mean diameter of the MWNT dispersion was very close to that of the SWNT dispersion, the much higher standard deviation (STD) was observed for the former (7.1 vs. 1.0). This indicates that some larger agglomerates were formed in the case of the MWNT.
Table II.
Results of particle size analysis of CNTs in cell culture media.
| CNPs | Mean diameter (nm) | STD |
|---|---|---|
| C60 0.004 μg/cm2 | 230 | 0.7 |
| C60 0.04 μg/cm2 | 233 | 0.6 |
| SWNT 0.004 μg/cm2 | 218 | 0.1 |
| SWNT 0.04 μg/cm2 | 260 | 0.6 |
| MWNT 0.004 μg/cm2 | 224 | 1.0 |
| MWNT 0.04 μg/cm2 | 263 | 7.1 |
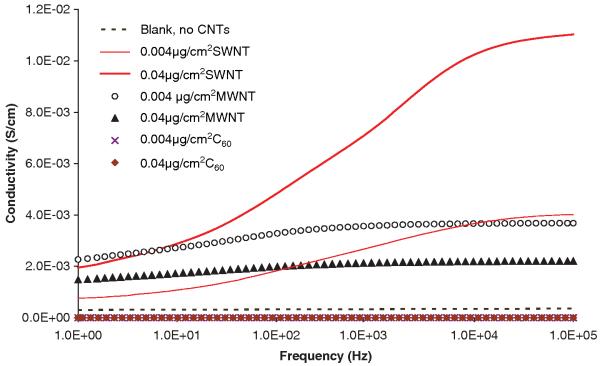
We applied dielectric spectroscopy (DS) for the investigation on the dispersion of CNPs in the cell culture medium. DS has been used extensively to characterize colloidal suspensions. The measurements were carried out using a liquid sample cell at a bias voltage of 1 volt. Dielectric spectra were collected in the frequency range of 1–105 Hz. Frequency dependence of AC conductivity of C60, SWNTs or MWNTs dispersed in the medium at two different concentrations (0.004 μg/cm2 and 0.04 μg/cm2) is shown in Figure 3. Frequency dependence of AC conductivity of the medium blank is included for comparison. The conductivity of the medium blank and the C60 dispersions are relatively low and underwent little change over the entire frequency range. An increase in the conductivity with increasing frequency is evident for all media with dispersed CNTs. This could be attributed to the effect of interfacial polarization due to surface charge on CNT particles. The difference in the conductivity of the medium matrix and the conductivity of the CNTs caused interfacial polarization resulting in the motion of the surface charge which is trapped at the interface of components.
Figure 3.
Frequency dependency of AC conductivity for a culture medium (dashed line), and the media with 0.004 μg/cm2 (thin solid line) or 0.04 μg/cm2 (thick solid line) of SWNT, the media with 0.004 μg/cm2 (open circle) or 0.04 μg/cm2 (filled triangle) of MWNT, and the media with 0.004 μg/cm2 (cross) or 0.04 μg/cm2 (filled diamond) of C60.
In the case of SWNT, conductivity increased with an increase in the concentration of SWNT. For example, at 9.8 kHz, AC conductivity was increased from 3.62 × 10−4 S/cm for the medium blank to 3.66 × 10−3 S/cm at 0.004 μg/cm2 SWNT concentration and further to 1.02 × 10−2 S/cm at 0.04 μg/cm2 SWNT concentration. The conductivity reached a plateau at higher frequency because the polarization process can no longer follow the change in the applied electrical field. Increased conductivity with concentration indicates finely dispersed SWNTs even at higher concentrations.
As for MWNT, at 0.004 μg/cm2 MWNT concentration, conductivity was comparable to that of the medium with SWNT at the same concentration level at higher frequency range but reached a plateau at lower frequency. The difference between the MWNT and SWNT dispersed in the medium is that the conductivity decreased with the increase in MWNT concentration, contrary to the trend shown in the case of SWNT dispersion. One possible reason is that, compared to SWNT, the MWNTs tend to form large agglomerates at higher concentration. Higher agglomeration in MWNTs resulted in the reduction of effective conduction pathways. In general, the dielectric spectroscopy data provide an insight into agglomeration pattern in the biological suspensions, which is expected to affect CNP-cell interactions at both physical and biological levels. It is interesting to observe that the SWNT and MWNT behaved so differently. This is consistent with the DLS data, where the formation of larger agglomerates led to a significantly higher variability in the particle size distribution.
Cellular effects of nanoparticle exposure
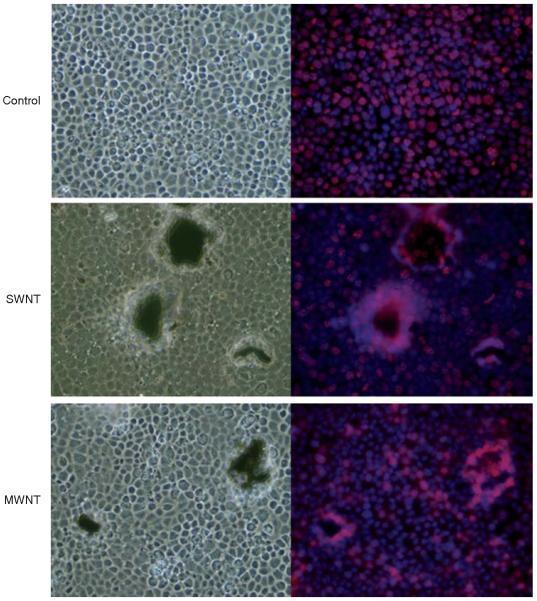
PCNA staining was used to visualize cellular proliferative responses (Figure 4). At high concentrations, where agglomeration of particles is pronounced, in the immediate vicinity of the agglomerated nanotubes, there is an increase in cellular proliferation. This effect was never seen in C60 exposures, nor was it observed at SWNT or MWNT concentrations ≤0.4 μg/cm2.
Figure 4.
Effects of CNP exposure in the mpkCCDcl4 cell line. Cells were seeded in 24-well plates and exposed to 40 μg/cm2 of SWNT or MWNT for 48 h. The cells were fixed and stained with proliferating cell nuclear antigen (PCNA) to indicate the nuclei or proliferating cells and DAPI to stain all cell nuclei (red and blue respectively in the merged images on the right). The brightfield images (left-hand panels) clearly show the agglomerated nanoparticles as irregular black spots. The panels on the right indicate the increased cellular proliferation (PCNA; red) in the areas surrounding the nanoparticles.
While the agglomerated particles induce specific effects on the cellular proliferation, we hypothesized that more subtle functional effects may be exerted at lower concentrations where the particles were not agglomerated and could, potentially, enter the cells or permeate the cellular junctions as individual particles. To mimic the in vivo situation of a healthy epithelium, the cells were exposed to nanoparticles only after they had achieved at high-resistance monolayer (13 days). The effects of 48 h incubation with various nanoparticles on the transepithelial resistance (TEER) of the mpkCCDcl4 renal cell line were measured using standard Ussing-type electrophysiological techniques. 48 h incubation with multi-walled nanotubes decreases the TEER of the renal cells with equal efficacy over a 6 order of magnitude dose response range. The single-walled nanotubes and fullerenes, on the other hand, show an inverse dose response relationship with the lower concentrations showing a significant decrease in TEER while the higher concentrations are without effect (Figure 5).
Figure 5.

Effect of nanoparticle incubation on the TEER in the mpkCCDcl4 cell line. Cells were grown to confluence on Transwell permeable supports and incubated for 48 h with media containing the indicated concentrations of nanoparticles provided as concentration per cm2 diagonally under each bar. After the incubation, the Transwells were removed and placed in Ussing chambers and allowed to stabilize for 0.5–1 h before measuring the TEER. The bars represent the means ± SEM. * indicates experiments where the mean values were significantly different than the mean control value. The number of replicate experiments performed at each concentration is indicated as the values in the bars.
After measurement of the TEER, the cells were exposed to antidiuretic hormone (ADH; vasopressin). Figure 6 illustrates the response to ADH in control and nanoparticle-exposed cellular monolayers. The mpkCCDcl4 cells respond to three hormones that stimulate ion transport in renal principal cells – aldosterone, insulin and ADH (Shane et al. 2006). We have chosen to study the effects of ADH because this peptide hormone elicits a multi-phasic response consisting of both Cl− secretion via the cystic fibrosis transmembrane regulator (CFTR) and Na+ absorption via the epithelial Na+ channel (ENaC). Aldosterone and insulin both cause an increase in ENaC-mediated Na+ absorption with no detectable change in Cl− secretion. Thus, studying the response to ADH maximizes the ability to detect potential nanoparticle effects on ion transporters. Figure 6 contains the data from control, 40 μg/cm2 (highest dose) and 0.4 ng/cm2 (lowest dose). Nanoparticle exposure did not alter either the Cl− secretory or the Na+ absorption response to ADH. The experiments were repeated at every concentration shown in Figure 5 and all doses were without effect (data not shown).
Figure 6.
Response of mpkCCDcl4 renal cells to ADH in control and nanotube-exposed cultures. Cells were grown in Transwell supports until they formed a confluent monolayer. The media was then replaced with either control or nanoparticle containing media and the incubation was continued for 48 h. The cells were mounted in Ussing chambers and the basal SCC was allowed to stabilize. ADH was added to the serosal bathing media at time zero. The panels on the left denote the entire time course of the response of the mpkCCDcl4 cells to ADH. The hormone causes an immediate response that consists of a CFTR-mediated Cl− secretory phase (~ 0–1 min) followed by an ENaC-mediated increase in Na+ flux (~ 5–30 min). Amiloride, a specific inhibitor of ENaC, was used to terminate the experiment at time t = 30 min. Panels on the right show the first 2 min which is the Cl− secretory phase. This Figure contains the data from control, 40 μg/cm2 (highest dose) and 0.4 ng/cm2 (lowest dose).
Cytokine profiles and LDH measurements (data not shown) revealed that none of the CNP exposures, at any dose, altered their levels, indicating the absence of any irritation or cell damage. Regarding cell viability, none of the changes in TEER reached levels that represent a decrease in cell viability. The observed decreases in TEER are indicative of a healthy and high resistance, intact epithelium. Therefore, no other measures of viability were deemed necessary.
Proteomic effects of nanoparticle exposure
Carbon nanoparticle treatment induced significant alterations in protein expression as measured by label-free quantitative mass spectrometry. Across all samples analyzed, an average of 1,878 proteins were identified, quantified, and compared. As Table III indicates, all CNPs caused differences in protein expression compared to controls and the number of proteins whose expression was significantly altered declined with increasing dose, with C60 having the least effect overall.
Table III.
Number of proteins altered (P ≤ 0.01) by CNP exposure.
| CNP dose (μg/cm2) |
|||
|---|---|---|---|
| CNP | 0.004 | 0.04 | 40 |
| C60 | 61 | 16 | 5 |
| SWNT | 116 | 29 | 12 |
| MWNT | 105 | 17 | 10 |
A complete list of the detected proteins and their pertinent information can be found in the supplementary material, available online. When all doses and CNP-types were compared, no differentially expressed proteins common to all exposure conditions were found. This was also true across all three doses within individual CNPs. However, at the two lowest doses, 0.04 and 0.004 μg/cm2, expression of a number of proteins was affected by the respective CNP, and these are listed in Table IV.
Table IV.
mpkCCDcl4 cell proteins altered in both lower dose exposures (0.004 and 0.04 μg/cm2) in response to specific CNP.
| CNP | Gene ID | Protein name |
|---|---|---|
| C60 | CPPED1 | calcineurin-like phosphoesterase domain containing 1 ▲,▼ |
| SWNT | ACBD3 | acyl-Coenzyme A binding domain containing 3 ▲,▲ |
| GAPDHS | glyceraldehyde-3-phosphate dehydrogenase ▲,▲ | |
| H2AFX | histone family, member X ▲,▲ | |
| MIF | macrophage migration inhibitory factor (glycosylation-inhibiting factor) ▲,▲ | |
| RPL17 | ribosomal protein L17 ▼,▼ | |
| RPL27 | ribosomal protein L27 ▼,▼ | |
| EG237361 | ribosomal protein L27a ▼,▼ | |
| RPS8 | ribosomal protein S8 ▼,▼ | |
| GM5121 | ribosomal protein S8 isoform 1 ▼,▼ | |
| RPS17 | ribosomal protein S17 ▼,▼ | |
| SECTM1B | secreted and transmembrane 1B ▼,▼ | |
| MWNT | H2AFJ | H2A histone family, member J ▲,▼ |
| H2AFX | H2A histone family, member X ▲,▼ | |
| H2AFY | H2A histone family, member Y ▲,▼ | |
| HIST2H2AC | histone cluster 2, H2ac ▲,▼ | |
| HADH | hydroxyacyl-Coenzyme A dehydrogenase ▼,▲ | |
| SHMT2 | serine hydroxymethyltransferase 2 (mitochondrial) ▼,▲ |
In each row, the first arrow refers to change associated with the 0.004 μg/cm2 exposure; the second arrow refers to change associated with the 0.04 μg/cm2 exposure.
At the lowest dose, the expression of seven proteins was altered by all three CNPs (Table V), while only histone cluster 2, H2ac was altered by all three CNPs at the 0.04 μg/cm2 dose, and none at 40 μg/cm2.
Table V.
mpkCCDcl4 cell proteins altered by all CNPs (C60, SWNT, MWNT) at the lowest dose (0.004 μg/cm2).
| Gene ID | Protein name |
|---|---|
| HIST1H2BF | histone cluster 1, H2bf ▲,▲,▼ |
| HNRNPU | heterogeneous nuclear ribonucleoprotein U ▼,▼,▼ |
| LHX9 | LIM homeobox 9 ▼,▼,▼ |
| MYBBP1A | MYB binding protein (P160) 1a ▼,▼,▼ |
| PDE2A | phosphodiesterase 2A, cGMP-stimulated ▲,▲,▲ |
| PIWIL1 | piwi-like 1 (Drosophila) ▼,▲,▼ |
| TMPO | thymopoietin ▼,▼,▼ |
In each row, the first arrow refers to the effect of C60; the second arrow refers to the effect of SWNT; and the third arrow refers to the effect of MWNT.
When comparing the effect of the two nanotubes, 26 proteins were similarly affected by exposure to SWNT and MWNT at the 0.004 μg/cm2 dose. These are listed in Table VI. At the 0.04 μg/cm2, only members J and X of the H2A histone family and histone cluster 2 were altered by both SWNT and MWNTs, while at 40 μg/cm2, catenin, beta like 1 (CTNNBL1) was upregulated.
Table VI.
mpkCCDcl4 cell proteins altered by both SWNT and MWNT exposures at the lowest dose (0.004 μg/cm2).
| Gene ID | Protein name |
|---|---|
| ACBD3 | acyl-Coenzyme A binding domain containing 3 ▲,▼ |
| AHNAK | AHNAK nucleoprotein ▼,▼ |
| DNAHC9 | dynein, axonemal, heavy chain 9 ▲,▼ |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase ▲,▼ |
| H2AFX | H2A histone family, member X ▲,▼ |
| H2AFY | H2A histone family, member Y ▲,▼ |
| HIST1H2BF | histone cluster 1, H2bf ▲,▼ |
| HNRNPU | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) ▼,▼ |
| KCNN3 | K+ intermediate/small conductance Ca++-activated channel, subfamily N, member 3 ▲,▼ |
| LHX9 | LIM homeobox 9 ▼,▼ |
| ABCG3 | similar to ATP-binding cassette transporter ▲,▼ |
| MAT2B | methionine adenosyltransferase II, beta ▼,▲ |
| MIF | macrophage migration inhibitory factor (glycosylation-inhibiting factor) ▲,▼ |
| MYBBP1A | MYB binding protein (P160) 1a ▼,▼ |
| OIP5 | Opa interacting protein 5 ▲,▼ |
| PDE2A | phosphodiesterase 2A, cGMP-stimulated ▲,▲ |
| PFN1 | profilin 1 ▼,▲ |
| PIWIL1 | piwi-like 1 (Drosophila) ▲,▼ |
| PPIL1 | peptidylprolyl isomerase (cyclophilin)-like 1 ▼,▼ |
| RDH5 | retinol dehydrogenase 5 (11-cis/9-cis) ▲,▼ |
| RPS5 | ribosomal protein S5 ▼,▼ |
| SHMT1 | serine hydroxymethyltransferase 1 (soluble) ▼,▲ |
| SHMT2 | serine hydroxymethyltransferase 2 (mitochondrial) ▼,▲ |
| SLC39A10 | solute carrier family 39 (zinc transporter), member 10 ▲,▼ |
| TLN1 | talin 1 ▼,▲ |
| TMPO | thymopoietin ▼,▼ |
In each row, the first arrow refers to the effect of SWNT; the second arrow refers to the effect of MWNT.
To interpret the biological significance of the affected proteins, Ingenuity Pathways Analysis was used to categorize proteins differentially expressed by mpkCCDcl4 cells in response to each dose and CNP exposure, and those 254 proteins altered across all exposures. Pathway analysis technology enables the mapping of protein expression data into relevant molecular interaction networks and canonical pathways based on functional annotations and known molecular interactions. Tables VII and VIII list the most significant (P ≤ 0.001) functional networks and canonical pathways involving proteins altered by the CNP exposures. Table VII lists the various CNP exposures and corresponding number of up- and down-regulated proteins (corresponding to totals listed in Table III) in one column and, in a second column, the functional networks corresponding to the number of proteins (included parenthetically) from the first column that were statistically assigned to IPA networks. Table VIII lists the various CNP exposures along with canonical pathways identified by IPA that are impacted by each exposure. Only those pathways in which three or more differentially expressed proteins were determined statistically (P < 0.001) to be members are included. Interpretation of these results is presented in the Discussion section.
To compare the proteomic results with multiple previous studies, it is also informative to highlight proteins that did not change in response to CNP exposure. The general absence of toxicity is exemplified by the observation that numerous proteins typically upregulated by ROS or cellular toxicity were unaffected at all doses. The following glutathione-S-transferases were identified and quantified: A3, mu-1, mu-2, mu-4, mu-5, mu-7, mu-7, omega-1, P1, P2, and theta-3. With the exception of GST mu-4, which was up-regulated (10.1%) by SWNT at 0.004 μg/cm2, and GST omega-1 which was up-regulated slightly by fullerenes (10.7%) at 0.04 μg/cm2 and down-regulated by SWNT (10.6%) at 0.004 μg/cm2, all others were normally expressed, as were glutathione reductase and formylglutathione lyase (glyoxalase I). Similar observations were made for both superoxide dismutases Mn and Cu-Zn and stress proteins. Seventeen proteins associated with the peroxiredoxin family of antioxidant enzymes and with peroxisomes were unaffected at all exposures, as were four members of the cytochrome P450 family. Seventy-eight kinases were profiled and none with toxicity-related signaling function were affected by the exposures.
Discussion
Raw, non-functionalized nanoparticles such as the materials used in this study form the basis for the synthesis of other functionalized particles and composite materials that are useful in both industry and medicine. As such, it is the non-functionalized forms that will be manufactured in large quantities and are, therefore, the most likely forms that will contribute to environmental contamination as well as work-place exposures. As shown in the Results these particles are very sparingly dispersible, even after sonication in a protein-rich solution such as serum. The results predict that there may be two very different types of effects of the nanoparticles on cellular response – those manifested in response to the presence of large agglomerants of particles that adhere to the surface of the epithelial cells (micro effects) and those manifested by exposure to individual particles (nanoeffects). At concentrations above 1 μg/ml (0.4 μg/cm2), both single- and multi-walled nanotubes form visible aggregates. These observations are corroborated by the dielectric spectroscopy results and AFM analysis.
In a previous study examining the effect of MWNT on human epidermal keratinocytes (Witzmann and Monteiro-Riviere 2006), the authors observed significant alterations in protein expression, particularly in membrane-related proteins, such as those involved with organization of membrane domains and/or - membrane-cytoskeleton linkages, certain exocytotic and endocytotic transport steps, cytoskeletal integrity, and related signaling functions. A subsequent experiment in which mpkCCDcl4 cells were exposed to very high concentrations of CNPs (200 μg/cm2) (Amos et al. 2008) demonstrated significant declines in TEER but minor proteomic alterations. However, those proteins that were affected were related to junctional and cell adhesion functions. This leads us to hypothesize that MWNT and other related CNPs might alter barrier function in these and other epithelial cells that play such a protective role.
Studies have suggested that carbon nanoparticles may have carcinogenic properties (Murr et al. 2005). MWNT have been specifically implicated due to their structural similarity to chrysotile asbestos that is widely accepted to cause carcinogenic responses in humans (Kane and Hurt 2008; Poland et al. 2008). After exposure to high concentrations of nanotubes, we observed changes in cells surrounding agglomerations of SWNT and MWNT. Cells seemed to exhibit proliferating nuclei as indicated by PCNA staining. Under control conditions, once confluence is reached, cells no longer actively divide. The results with the nanotubes indicate that SWNT and MWNT agglomerations cause cells to replicate abnormally, suggesting that hyperplasia is occurring. Despite elevated PCNA staining at agglomerant foci, PCNA expression was detected but remained unchanged at all doses analyzed using quantitative mass spectrometry.
The mpkCCDcl4 studies were conducted to show the effects of relatively short-term exposure to nanoparticles. High resistance cell lines most closely mimic the in vivo situation if potential effectors are added to the cultures after the cells have achieved a confluent monolayer, thus limiting the timing of the experiments to several days. However, the effects seen in this model could portend additional, more serious, outcomes with longer-term exposures.
In some organs, nanoparticles may remain embedded in the tissues for long periods of time. MWNT administered intratracheally persisted in the lung tissue for more than 60 days (Muller et al. 2005). In the rodent models and in cultured lung cells, high levels of MWNT caused inflammatory and fibrotic reactions as well as evidence of mutagenesis (Muller et al. 2005, 2008). In contrast, a single oropharyngeal aspiration of SWNT caused no inflammatory response 1 or 21 days post-exposure. However, at 21 days, the investigators did observe small interstitial fibrotic lesions in the alveolar regions, particularly near groups of macrophages containing micron-sized aggregates of the nanotubes (Mangum et al. 2006).
While studies employing high concentrations of CNPs are useful for predicting effects, concentrations above 1 μg/ml are unlikely to occur in vivo except during topical exposure of keratinocytes and perhaps in lung epithelia in an industrial setting. Thus, the current studies also explored the effects of very low concentrations of nanoparticles on cellular function and protein expression in renal cells. To our knowledge these are the first studies to comprehensively examine concentrations in the low ng/ml (ng/cm2) range.
TEER is a measure of monolayer integrity and is also a very sensitive measure of cellular viability. As cellular viability decreases, TEER falls precipitously. In the experiments shown in Figure 5, TEER was significantly decreased after treatment with all concentrations of MWNT from 40 μg/cm2 down to 0.4 ng/cm2. Interestingly, after incubation with fullerenes or SWNT, resistances were lowered only by exposure to concentrations below 0.4 μg/cm2.
It is important to note that none of the changes in TEER reached levels that represent a decrease in cell viability. Control monolayers had an average TEER of 1867 Ω · cm2. A decrease to values between 750 and 1000 Ω · cm2 is still considered a high resistance, intact epithelium. The changes in resistance indicate more subtle changes within the cells, and perhaps between them. Examples of cellular alterations which could be manifested in the magnitude of the changes observed would be minor modifications of the cytoskeleton which is a major component in determining the impermeability of the junctional complexes or changes in the composition of the cellular membrane which would be sufficient to alter permeability. We have not tested concentrations below those listed in Figure 5 so cannot yet comment on the lowest concentration that causes this change in transepithelial resistance.
To further investigate nanotube-induced changes in cellular function, the responses to external stimuli were assessed using the method of short circuit current (SCC), a measure of net ion transport. Interestingly, there were no alterations in the cellular response to ADH, a hormone known to regulate both Na+ and Cl− in the principal cells. Thus, this cellular function is maintained in the presence of nanoparticle exposure.
The proteomic analyses have highlighted several important aspects of the responses to nanoparticle exposure. The first observation is that, in general, the effects on protein expression correspond to the results of the transepithelial resistance in that they are, interestingly, inversely related to dose. Both the number of altered proteins and the fold-changes increase as dose declines. No one has previously studied carbon nanoparticle doses this low so there are no parallels in the published literature. Size measurements suggest that at high doses where there is significant agglomeration, particles act at a microscale, while at the lower doses, CNPs may act at a nanoscale level where little if any agglomeration is observed.
Second, none of the protein groups reflect a ‘traditional’ set of toxicity-related proteins. Thus, at least in the mpkCCDcl4 cells we are not observing a ‘toxic effect’ in the classic sense. This finding is in agreement with the lack of cell death in the TEER and LDH measurements and lack of substantial stress responses as measured by cytokine secretion. In fact, stress responses proteins such as those upregulated by ROS were notably unchanged by CNP exposure.
Third, there is little overlap among the three nanoparticle types in terms of the types of proteins, molecular interaction networks and pathways, suggesting the physico-chemically distinct particles act differently at the biochemical and molecular level, with respect to effects on the proteome. While the nanomaterials decreased TEER in the mpkCCDcl4 cells, the protein alterations observed do not provide evidence of mechanism in this regard. There is only one exception where SWNT at 0.04 μg/cm2 upregulated tight junction signaling proteins (Table VII). However, we have only conducted the cytoskeletal and proliferating nuclei labeling experiments at high concentrations, where the carbon forms can be located for analysis of surrounding cells, while the proteomic changes are observed predominately at the lower concentrations. Therefore additional experimentation will be necessary to fully explain the correlation, if any, between the observed changes in cellular proliferation and actin cytoskeleton structure and the proteomic changes that have been observed.
In general, the proteomic results demonstrate a complex and diverse biological response to CNP exposure, at the lowest concentrations ever studied. Furthermore, as already noted, this response is inversely related to dose, is CNP-specific, and occurs in the absence of overt irritation, inflammation, or toxicity. The inverse relationship between CNP exposure concentration and protein expression is not surprising when considered in the context of particle size. At 40 and 4 μg/cm2, significant agglomeration is evident while visible particles are absent below that level. Physico-chemical characterization evidence supports this.
When one examines mpkCCDcl4 cell proteins whose expression was altered by CNP exposure, the 40 μg/cm2 had little effect. At P < 0.01 one would expect ~ 19 proteins to be altered by chance alone, thus one can conclude this exposure was insignificant relative to protein expression, despite the decline in TEER observed in MWNT. However, at exposures several orders of magnitude lower, CNP-mediated protein expression was significantly greater and these alterations parallel significant declines in TEER. This is likely the result of CNP agglomerants (at 40 μg/cm2) exerting focal effects that may be significant in those foci, but are rendered largely undetectable when all cells on the transwell are examined. As mentioned earlier, this accounts for focal elevations of PCNA staining, but no significant change overall in PCNA as measured by label-free quantitative mass spectrometry.
As Table IV indicates, the lowest two SWNT exposures studied by proteomics resulted in a decreased expression of six ribosomal proteins. Neither C60 nor MWNT exerted such an effect. Ribosomal proteins are integral components of basic cellular machinery involved in protein synthesis and individual ribosomal proteins are known play a role in regulating cell growth, transformation and death (Warner and McIntosh 2009). It has been suggested that both the differentiation state and the proliferative status of the cells affect the expression of ribosomal protein mRNAs (Bevort and Leffers 2000) and both L and S-type ribosomal protein gene expression have been shown to decline in nephrotoxicity (Leussink et al. 2003).
Of the proteins altered by MWNT low-dose exposure, conflicting responses were observed. The four histone family proteins were up-regulated by the 0.004 μg/cm2 exposure, while the same proteins were significantly down-regulated by the higher 0.04 μg/cm2 exposure. One of these, H2AFX, is a major component of the nucleosome core structure that comprises 10–15% of total cellular H2A family proteins in mammalian cells, is critical to genomic stability and DNA repair (Fernandez-Capetillo et al. 2004), and play an important role in the regulation of gene expression and cellular proliferation (Svotelis et al. 2009).The conflicting effects of MWNT on these members of the histone H2A family at these exposures are difficult to interpret but they suggest significant MWNT effects on nuclear function of these barrier epithelia.
To aid in interpretation of the complex proteomic results, we used Ingenuity Pathways Analysis. As mentioned above, protein expression was relatively unaffected by the 40 μg/cm2 exposure and no statistically significant relationships to functional networks were detected.
One of the effects observed in all three CNP exposures only at the 0.004 μg/cm2 dose was a general up-regulation of glycolysis/gluconeogenesis pathways. As Table VIII indicates, expressions of proteins in this pathway were generally increased. These included enolase 2, enolase 3, and lactate dehydrogenase A (C60 exposure); aldolase A, enolase 1, glyceraldehyde-3-phosphate dehydrogenase, glucose phosphate isomerase, lactate dehydrogenase A, phosphofructokinase, phosphoglycerate kinase, phosphoglucomutase, pyruvate kinase and triosephosphate isomerase (SWNT exposure); and aldolase B, enolase 2, enolase 3, phosphoglycerate kinase, phosphoglycerate mutase, and pyruvate kinase (MWNT exposure). The only proteins associated with this pathway that were down-regulated were hexokinase 1 (C60 exposure); alcohol dehydrogenase 4 (SWNT exposure); and glyceraldehyde-3-phosphate dehydrogenase (MWNT exposure).
Gou et al. (2008) reported a dose-dependent adsorption and depletion of nutrients from RPMI cell culture medium by purified SWNTs that contained 10% Fe. HepG2 cells cultured in these depleted media showed significantly reduced viability that was restored by replenishment of folate. However, those effects occurred at doses of 10 μg/ml (corresponding to 25 μg/cm2) and above, higher than the doses used in the present study. Alternatively, one can view the increased glycolysis/gluconeogenesis pathway as a generalized response to stress. Systemically, such phenomena are usually manifested in a fight or flight response. However, the stimuli for a more subtle, analogous change at a tissue or molecular level are not well documented.
These altered components of the glycolysis/gluconeogenesis pathway are also reflected in the Functional Networks presented in Table VII. For instance, in low-dose C60 exposure, the predominant interaction network affected is a network of gene expression, cell death, and energy production. This network received a significance score of 32 (with 35 being the maximum), the highest in this exposure analysis (thus the proteins have a 1 in 1032 probability of having been assigned to this network by chance). The network contains the following differentially expressed (13 up-regulated and four down-regulated) proteins (by gene symbol): acetyl-Coenzyme A acyltransferase 2, ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, ATP synthase, H+ transporting, mitochondrial F1 complex, beta, ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit, DEAD (Asp-Glu-Ala-Asp) box polypeptide 5, enolase 2 (gamma, neuronal), enolase 3 (beta, muscle), GTPase activating protein (SH3 domain) binding protein 2, glutathione transferase zeta 1, hexokinase 1, high-mobility group box 2, high-mobility group box 1, hydroxysteroid (17-beta) dehydrogenase 10, lactate dehydrogenase A, microtubule-associated protein, RP/EB family, member 1, phospholipase C, gamma 1, and ubiquitin-conjugating enzyme E2L 3, including members of the glycolysis/gluconeogenesis pathway. This is also observed to an even greater extent in the low-dose SWNT exposure (Table VIII), where Carbohydrate Metabolism is part of the second-rated network (significance score of 20) along with Cancer and Genetic Disorder, and contains the following differentially expressed proteins: acyl-Coenzyme A binding domain containing 3, actin, alpha 1, aldolase A, fructose-bisphosphate, acidic (leucine-rich) nuclear phosphoprotein 32 family, member A, HLA-B associated transcript 1, ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R), enolase 1, (alpha), glyceraldehyde-3-phosphate dehydrogenase, glucose phosphate isomerase, heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A), lactate dehydrogenase A, myosin, heavy chain 9, non-muscle, nucleoside diphosphate kinase, phosphoglycerate kinase 1, ribosomal protein S5, talin 1, and triosephosphate isomerase 1. Again, glycolytic/gluconeogenic proteins are interacting broadly in this functional network and are implicated in the biological effects of CNPs in this regard. As mentioned earlier, such effects are absent at exposures where agglomeration is evident. The functional networks and pathways impacted by low-dose CNP exposure are thus likely to be nanoscale effects.
Other interaction networks and pathways presented in Tables VII and VIII are established in fundamentally the same manner as that described above. Only the specific protein components differ. For instance, fullerene exposure at 0.004 μg/cm2 is associated with a predominant up-regulation in the expression of proteins in cell cycle, gene expression and cell death functional networks. These, and several other functional networks, appear in bold print in Table VII to emphasize the fact that these network components are similarly affected to some degree by all three CNPs studied, at the lowest two doses. Studies of bronchoalveolar cells in mice have demonstrated increased apoptosis and GI arrest after 14 and 28 days of C60 instillation (Park et al. 2010). That same study also showed that the expression of the tissue damagerelated MMPs, Timp and Slpi gene, and the expression of oxidative stress related SOD gene were increased until day 28 after a single instillation. Protein changes related to cell death have also been reported by SWNT exposures by Cui in HEK293 cells (Cui et al. 2005), along with down-regulated cellular growth and proliferation. These changes were accompanied by a decrease of markers associated with G1 to S transition (e.g., cdk2, cdk4, and cyclin A, E, and D3) as well as markers associated with S, G2, and M phase and down-regulation of expression of adhesion proteins (laminin, fibronectin, cadherin FAK and collagen IV). The relevance of the changes we have observed to those alterations seen by others in various tissues will require additional studies.
Although we have no evidence of oxidative stress, as ROS-related protein alterations are otherwise absent from our data as is any other evidence of cytotoxic effect, the pathway analysis reported in Table VIII suggests the SWNT-mediated up-regulation of NRF-2 mediated oxidative stress response at 0.04 μg/cm2 (in contrast to the decrease in this response at 40 μg/cm2), is consistent with an overall trend observed in our study, that the CNPs have greater biological effect at lower as opposed to higher exposure levels.
Other pathways impacted by CNP exposure at very low levels include alteration in functional networks involved in inflammatory diseases and regulation of amino acid metabolism. Changes in the expression of components of various metabolic pathways may be particle specific as well as dose- and exposure time-dependent. These aspects require further investigation.
Conclusion
The present study has shown that CNP induced significant alterations in renal collecting duct cell function, and protein expression. At high doses, CNP suspensions cause an increased nuclear proliferation. At low, more physiologically relevant concentrations, the CNPs cause changes in epithelial barrier function as well as changes in protein expression that remain, for the moment, documented but difficult to characterize. The observed changes are subtle, not related to overt toxicity, and likely represent cellular alterations that would have physiological effects over a prolonged time-course.
Supplementary Material
Footnotes
Supplementary material available online
Complete list of the detected proteins and their pertinent information.
Declaration of interest: These studies were supported by a grant from NIGMS (R01GM085218), NIEHS (RC2ES018810), and the IUPUI Undergraduate Research Opportunities Program. The authors gratefully acknowledge the technical support of Heather Ringham and Susana Ado Ntim, and sincerely thank Dr Alain Vandewalle (INSERM U478, Paris, France) for providing the mpkCCDcl4 cell line. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Amos AD, Witzmann FA, Chernoff EA, Hong D, Lai X, Ringham HN, Blazer-Yost BL. Changes in cell function and protein expression of mouse renal principal cells, mpkCCD, after carbon nanoparticle (CNP) exposure. FASEB J. 2008;22:942. [Google Scholar]
- Bens M, Vallet V, Cluzeaud F, Pascual-Letallec L, Kahn A, Rafestin-Oblin ME, Rossier BC, Vandewalle A. Corticosteroid-dependent sodium transport in a novel immortalized mouse collecting duct principal cell line. J Am Soc Nephrol. 1999;10:923–934. doi: 10.1681/ASN.V105923. [DOI] [PubMed] [Google Scholar]
- Bevort M, Leffers H. Down regulation of ribosomal protein mRNAs during neuronal differentiation of human NTERA2 cells. Differentiation. 2000;66:81–92. doi: 10.1046/j.1432-0436.2000.660203.x. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Borm PJ, Robbins D, Haubold S, Kuhlbusch T, Fissan H, Donaldson K, Schins R, Stone V, Kreyling W, Lademann J, Krutmann J, Warheit D, Oberdörster E. The potential risks of nanomaterials: A review carried out for ECETOC. Part Fibre Toxicol. 2006;3:11. doi: 10.1186/1743-8977-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen Z, Meng H, Xing G, Chen C, Zhao Y, Jia G, Wang T, Yuan H, Ye C, Zhao F. Acute toxicological effects of copper nanoparticles in vivo. Toxicol Lett. 2006;163:109–120. doi: 10.1016/j.toxlet.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- Cui DX, Tian FR, Ozkan CS, Wang M, Gao HJ. Effect of single wall carbon nanotubes on human HEK293 cells. Toxicol Lett. 2005;155:73–85. doi: 10.1016/j.toxlet.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Donaldson K, Aitken R, Tran L, Stone V, Duffin R, Forrest G, Alexander A. Carbon nanotubes: A review of their properties in relation to pulmonary toxicology and workplace safety. Toxicol Sci. 2006;92:5–22. doi: 10.1093/toxsci/kfj130. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: The histone guardian of the genome. DNA Repair. 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Guo L, Von Dem Bussche A, Buechner M, Yan A, Kane AB, Hurt RH. Adsorption of essential micronutrients by carbon nanotubes and the implications for nanotoxicity testing. Small. 2008;4:721–727. doi: 10.1002/smll.200700754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta AK, Gupta M. Cytotoxicity suppression and cellular uptake enhancement of surface modified magnetic nanoparticles. Biomaterials. 2005;26:1565–1573. doi: 10.1016/j.biomaterials.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Hale JE, Butler JP, Gelfanova V, You JS, Knierman MD. A simplified procedure for the reduction and alkylation of cysteine residues in proteins prior to proteolytic digestion and mass spectral analysis. Anal Biochem. 2004;333:174–181. doi: 10.1016/j.ab.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Higgs RE, Knierman MD, Gelfanova V, Butler JP, Hale JE. Comprehensive label-free method for the relative quantification of proteins from biological samples. J Proteome Res. 2005;4:1442–1450. doi: 10.1021/pr050109b. [DOI] [PubMed] [Google Scholar]
- Higgs RE, Knierman MD, Gelfanova V, Butler JP, Hale JE. Label-free LC-MS method for the identification of biomarkers. Methods Mol Biol. 2008;428:209–230. doi: 10.1007/978-1-59745-117-8_12. [DOI] [PubMed] [Google Scholar]
- Jia G, Wang H, Yan L, Wang X, Pei R, Yan T, Zhao Y, Guo X. Cytotoxicity of carbon nanomaterials: Single-wall nanotube, multi-wall nanotube, and fullerene. Environ Sci Technol. 2005;39:1378–1383. doi: 10.1021/es048729l. [DOI] [PubMed] [Google Scholar]
- Kane AB, Hurt RH. Nanotoxicology: The asbestos analogy revisited. Nat Nanotechnol. 2008;3:378–379. doi: 10.1038/nnano.2008.182. [DOI] [PubMed] [Google Scholar]
- Lacerda L, Herrero MA, Venner K, Bianco A, Prato M, Kostarelos K. Carbon-nanotube shape and individualization critical for renal excretion. Small. 2008;4:1130–1132. doi: 10.1002/smll.200800323. [DOI] [PubMed] [Google Scholar]
- Lam CW, James JT, McCluskey R, Arepalli S, Hunter RL. A review of carbon nanotube toxicity and assessment of potential occupational and environmental health risks. Crit Rev Toxicol. 2006;36:189–217. doi: 10.1080/10408440600570233. [DOI] [PubMed] [Google Scholar]
- Leussink BT, Baelde HJ, den Berg TMB-v, de Heer E, van der Voet GB, Slikkerveer A, Bruijn JA, de Wolff FA. Renal epithelial gene expression profile and bismuth-induced resistance against cisplatin nephrotoxicity. Human Experim Toxicol. 2003;22:535–540. doi: 10.1191/0960327103ht393oa. [DOI] [PubMed] [Google Scholar]
- Mangum JB, Turpin EA, Antao-Menezes A, Cesta MF, Bermudez E, Bonner JC. Single-Walled Carbon Nanotube (SWCNT)-induced interstitial fibrosis in the lungs of rats is associated with increased levels of PDGF mRNA and the formation of unique intercellular carbon structures that bridge alveolar macrophages in situ. Part Fibre Toxicol. 2006;3:15. doi: 10.1186/1743-8977-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard AD, Baron PA, Foley M, Shvedova AA, Kisin ER, Castranova V. Exposure to carbon nanotube material: Aerosol release during the handling of unrefined single-walled carbon nanotube material. J Toxicol Environ Health A. 2004;67:87–107. doi: 10.1080/15287390490253688. [DOI] [PubMed] [Google Scholar]
- Muller J, Decordier I, Hoet PH, Lombaert N, Thomassen L, Huaux F, Lison D, Kirsch-Volders M. Clastogenic and aneugenic effects of multi-wall carbon nanotubes in epithelial cells. Carcinogenesis. 2008;29:427–433. doi: 10.1093/carcin/bgm243. [DOI] [PubMed] [Google Scholar]
- Muller J, Huaux F, Moreau N, Misson P, Heilier J-F, Delos M, Arras M, Fonseca A, Nagy JB, Lison D. Respiratory toxicity of multi-wall carbon nanotubes. Toxicol Appl Pharmacol. 2005;207:221–231. doi: 10.1016/j.taap.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Murr LE, Garza KM, Soto KF, Carrasco A, Powell TG, Ramirez DA, Guerrero PA, Lopez DA, Venzor J., 3rd Cytotoxicity assessment of some carbon nanotubes and related carbon nanoparticle aggregates and the implications for anthropogenic carbon nanotube aggregates in the environment. Int J Environ Res Public Health. 2005;2:31–42. doi: 10.3390/ijerph2005010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E-J, Kim H, Kim Y, Yi J, Choi K, Park K. Carbon fullerenes (C60s) can induce inflammatory responses in the lung of mice. Toxicol Applied Pharmacol. 2010;244:226–233. doi: 10.1016/j.taap.2009.12.036. [DOI] [PubMed] [Google Scholar]
- Park KH, Chhowalla M, Iqbal Z, Sesti F. Single-walled carbon nanotubes are a new class of ion channel blockers. J Biol Chem. 2003;278:50212–50216. doi: 10.1074/jbc.M310216200. [DOI] [PubMed] [Google Scholar]
- Poland CA, Duffin R, Kinloch I, Maynard A, Wallace WAH, Seaton A, Stone V, Brown S, MacNee W, Donaldson K. Carbon nanotubes introduced into the abdominal cavity of mice show asbestos-like pathogenicity in a pilot study. Nat Nano. 2008;3:423–428. doi: 10.1038/nnano.2008.111. [DOI] [PubMed] [Google Scholar]
- Reijnders L. Cleaner nanotechnology and hazard reduction of manufactured nanoparticles. J Cleaner Product. 2006;14:124–133. [Google Scholar]
- Shane MA, Nofziger C, Blazer-Yost BL. Hormonal regulation of the epithelial Na+ channel: From amphibians to mammals. Gen Comp Endocrinol. 2006;147:85–92. doi: 10.1016/j.ygcen.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Svotelis A, Gevry N, Gaudreau L. Regulation of gene expression and cellular proliferation by histone H2A.Z. Biochem Cell Biol. 2009;87:179–188. doi: 10.1139/O08-138. [DOI] [PubMed] [Google Scholar]
- Wang M, You J, Bemis KG, Tegeler TJ, Brown DPG. Label-free mass spectrometry-based protein quantification technologies in proteomic analysis. Brief Funct Genomic Proteomic. 2008;7:329–339. doi: 10.1093/bfgp/eln031. [DOI] [PubMed] [Google Scholar]
- Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Molec Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzmann FA, Monteiro-Riviere NA. Multi-walled carbon nanotube exposure alters protein expression in human keratinocytes. Nanomedicine. 2006;2:158–168. doi: 10.1016/j.nano.2006.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.