The article by Zhao S et al (1) in this issue of Arthritis and Rheumatism gives further insight into the influence of certain microRNAs (miRNAs) on the promoter methylation status regulated by DNA methyltransferases (DNMTs). In recent years, there has been an increased interest in the role of epigenetic modifications of DNA and pathogenetic mechanisms of human diseases such as Systemic Lupus Erythematosus (SLE) (2). Epigenetics is linked to stable and potentially heritable changes in gene expression that do not entail a change in the DNA sequence. DNA methylation and histone modifications are the two major changes that contribute to the epigenome of a cell. The first mechanism usually occurs in the context of CpG dinucleotides at the 5' position of cytosine in the promoter region, leading to functional consequences such as transcriptional repression. In fact, some tissue-specific genes are silenced by promoter methylation (2). Post-translational modifications that occur in histones make up a second group of epigenetic modifications. Both DNA methylation and histone modifications are coupled through different machineries, including DNMTs as well as histone modifying enzymes in multiprotein complexes (2). Three main types of DNMTs are involved in genomic DNA methylation: DNMT1, DNMT3A and DNMT3B. Whereas DNMT1 preferentially replicates existing methylation patterns and maintains DNA methylation, DNMT3A and DNMT3B are responsible for establishing new DNA methylation markers therefore referred to de novo DNA methyltransferase (3).
A remarkable example of disease in which epigenetic-DNA methylation abnormalities and patterns of inheritance are extremely complex is SLE, characterized by the production of a variety of autoantibodies against nuclear and cytoplasmic components associated with inflammation and injury of multiple organs. The high incidence of twin pairs in which only one of the siblings has developed SLE supports the notion that environmental factors and their involvement in epigenetic modifications could affect the onset of the disease. The influence on SLE onset could occur at several levels. The epigenetic deregulation of genes can contribute to, or increase, the activation of apoptosis. Moreover, it may lead to an exacerbated activation of T and B cells (2). In fact, the DNA extracted from T cells of SLE patients is hypomethylated compared with the DNA from normal T cells (3). The mechanisms by which hypomethylated T cells induce SLE are not well understood. Additional evidence of the role of global methylation changes in the development of SLE comes from studies with DNA demethylating drugs, such as 5-azacytidine, procainamide and hydralazine (2). In all cases, exposing T cells to demethylating drugs results in the demethylation-dependent induction of lupus-like disease (2).
MiRNAs are small, non-coding RNAs, usually 21–23 nucleotides long, which mediate post-transcriptional silencing of target genes (4). MiRNAs usually bind to partially complementary sites in the 3' untranslated region (UTR) of target mRNAs, and efficient mRNA targeting requires continuous base-pairing of miRNA nucleotides 2 to 8, the so called “seed sequence” (4). In this way, miRNAs can regulate target gene expression by either translational inhibition, mRNA degradation or both (3, 4). Dysregulation of miRNAs by several mechanisms has been described in various disease states, including SLE (5, 6). Only recent works have suggested that miRNAs can regulate DNA methylation by targeting the DNA methylation machinery in SLE (3) (Figure 1, panel A). The discovery of the association between miRNAs and methylation regulation provides an insight into the role of miRNAs in lupus CD4+ T cell hypomethylation and the pathogenesis of SLE.
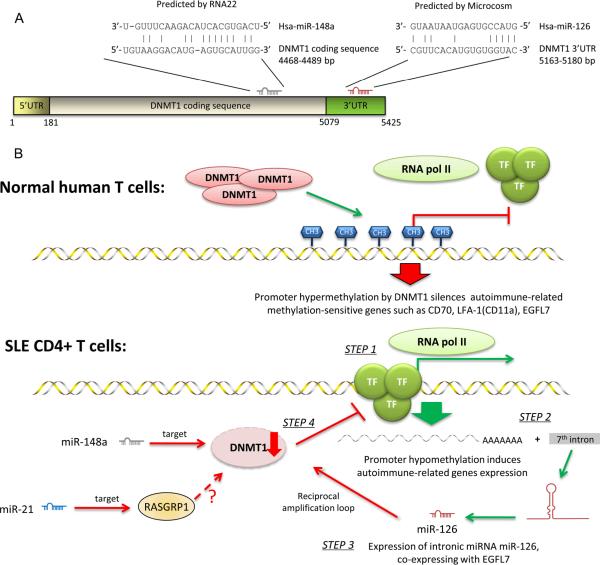
FIGURE 1.
MiR-21, miR-126, and miR-148a directly or indirectly regulate DNMT1 in SLE CD4+ T cells.
A. MiR-148a and miR-126 directly regulate DNMT1 validated experimentally. In particular, miR-148a binds to the DNMT1 coding sequence, as predicted by the program RNA22, while miR-126 binds to the 3'-UTR region of DNMT1, as predicted by the program Microcosm.
B. DNMT1 is directly or indirectly regulated in SLE CD4+ T cells and it triggers the over-expression of autoimmune-associated methylation-sensitive genes involved in SLE. In normal human T cells, gene hypermethylation controlled by DNMT1 blocks the transcription activity of RNA pol II, thus silencing autoimmune-related methylation-sensitive genes such as CD70, LFA-1(CD11a) and EGFL7. In SLE CD4+ T cells, promoter hypomethylation allows binding of transcriptional factors (TF) and recruiting of RNA pol II (step 1). This leads to the overexpression of autoimmune-related genes, including EGFL7 (step 2). Interestingly, miR-126 is encoded in the 7th intron of EGFL7. Thus the expression of EGFL7 leads to elevated levels of miR-126, which downregulates the production of DNMT1 in a reciprocal negative feedback (amplification) loop (step 3). Low level of DNMT1 leads to the hypomethylation status which maintains the aberrant autoimmune response (step 4). Other miRNAs contributing to decreased DNMT1 activity are miR-148a, through direct inhibition, and miR-21 which indirectly regulates DNMT1, by targeting RASGRP1 in the Ras-MAPK pathway, and thus also influencing DNMT1 protein levels.
DNMT1=DNA methyltransferase 1; TF= transcription factor; RNA pol II= RNA polymerase II; RASGRP1= RAS guanyl-releasing protein 1; bp= base pairs.
Pan W et al (3) recently identified two miRNAs, miR-21 and miR-148a, to be upregulated in CD4+ T cells in both lupus patients and MRL/lpr mice. Moreover, both miRNAs downregulate the protein level of the enzyme DNMT1, one of the major components in the demethylation of DNA, thus resulting in hypomethylation status in CD4+ T cells. In particular, miR-21 indirectly downregulates DNMT1 by targeting its upstream regulator, RASGRP1, while miR-148a directly downregulates DNMT1 by targeting the protein coding region of its transcript (Figure 1, panel B). The final result is the derepression of autoimmune-associated methylation-sensitive genes in CD4+T cells, such as CD70 and LFA-1(CD11a). These investigators were also able to induce the potential alleviation of hypomethylation in CD4+ T cells from patients with lupus by transfection with miR-21 and miR-148a inhibitors.
The article published in this issue by Zhao S et al (1) further expands the role of miRNAs and epigenetic changes in SLE. The novel finding is that, among 11 miRNAs found to be increased or decreased in SLE CD4+ T cells, miR-126 was significantly overexpressed and its upregulation was inversely correlated with DNMT1 protein levels (Figure 1, panel B). These investigators were then able to demonstrate that miR-126 can directly inhibit DNMT1 translation by interacting with its 3'UTR region, leading to a significant reduction in the DNMT1 protein levels (Figure 1, panel B). Through this mechanism, overexpression of miR-126 causes demethylation and upregulation of genes encoding for LFA-1(CD11a) and CD70, two autoimmune-related proteins, which are directly proportional to disease activity. The inhibition of miR-126 in SLE CD4+ T cells has opposite effects. The miR-126 host gene EGFL7 was also overexpressed in SLE CD4+ T cells, in a hypomethylation-dependent manner.
Another interesting point of the work by Zhao S et al (1) is that these investigators focused their attention not only in the influence of the DNA methylation machinery on lupus CD4+ T cells, but also on the co-stimulation between active T cells and B cells, leading to IgG overproduction. They were also able to show that knocking-down miR-126 in SLE CD4+ T cells reduced their autoimmune activity and their stimulatory effect on IgG production in the co-cultured B cells.
The main differences in the reports by Pan et al (3) and Zhao et al (1) focusing on the role of miRNA overexpression affecting the DNA methylation mechanism in SLE T cells are shown in Table 1. Pan W et al (3) and Zhao et al (1) detected different miRNAs involved in the DNA methylation machinery. One possible explanation is that Pan W et al (3) performed the microarray analysis of miRNAs in splenic CD4+ T cells and B cells isolated from MRL/lpr mice and normal control mice, but not in samples from SLE patients. On the contrary, in the present study, Zhao S et al (1) perform this analysis from CD4+ T cells of SLE patients and healthy controls. However, even though the initial microarray analyses were performed on different substrates, both of them proceed with the miRNA analysis on SLE patients, as shown in Table 1. Moreover, it is well known that SLE is a very heterogeneous disease, so it is possible that the different altered miRNAs detected in the two works can be associated with the selection of patients, disease subsets, autoantibody expression, immunosuppressive therapy, and phases of disease activity. Ethnic background is usually another important aspect to consider when studying SLE patients, but it does not seem to play a role in this case because both groups of SLE patients are from Chinese population.
TABLE 1.
Main differences in the two recent reports focusing on the role of miRNA overexpression affecting the DNA methylation mechanism in SLE T cells.
| Pan W et al (3) | Zhao S et al (1) | |
|---|---|---|
| Overexpressed miRNAs | MiR-21, miR-148a | MiR-126 |
| Cell substrate used for microarray analysis | Splenic CD4+ T cells and B cells isolated from MRL/lpr mice and controls | CD4+ T cells of SLE patients and healthy controls, no mouse model |
| Number of SLE patients studied for miRNA expression | 36 (33 female, 3 male) | 30 (all female) |
| SLE patients' information | Complete (male/female ratio, age, disease duration, SLEDAI score, anti-dsDNA, lupus nephritis, use of steroids and secondary agents) | Limited (age, gender, SLEDAI score, medications) |
| Overexpressed methylation-sensitive gene targets | LFA-1(CD11a), CD70 | LFA-1(CD11a), CD70, EGFL7 |
| Cell types examined for miRNA overexpression | CD4+ T cells | CD4+ T cells and B cells |
One possible limitation in the work by Zhao S et al (1) is the lack of information on SLE patients, in terms of clinical features and immunosuppressive therapy. Patients were recruited from in-patient ward and dermatology department, and it is not clear whether these SLE patients had predominantly dermatological manifestations, which could be a reason for the different miRNA expression in the two studies. Moreover, Pan W et al (3) provide more detailed information on SLE patients, including disease duration, anti-dsDNA titer, number of patients affected by lupus nephritis, detection of proteinuria, and use of immunosuppressive therapy.
Both studies analyze the influence of DNA methylation on the expression of genes that are linked to T cell autoreactivity. The CD70 and LFA-1(CD11a) genes are the targets studied in both reports, as they are demethylated genes overexpressed post-treatment with hypomethylating agents in CD4+ T cells (3). Zhao S et al (1) further studied a new target, the miR-126 host gene EGFL7, which is also overexpressed in SLE CD4+ T cells. MiR-126 upregulation is associated with a reduction of EGFL7 promoter methylation and thus also with T cell autoreactivity in SLE, leading to an amplification cycle that further contributes to the disease.
Zhao S et al (1) study the regulation of miR-126 on DNMT1 at the 3' UTR level through the use of miRBase information. However, this prediction was not confirmed by other algorithms, such as TargetScan or PicTar, commonly used in these bioinformatic analyses. This may be particularly important because, in the present work, the 5' interaction of miR-126 and the 3' UTR of DNMT1 only involve 4 bases. It is generally known that the seed sequence is expected to be 7–8 bases for most strong miRNA-mRNA interactions. Thus, it could be possible that the proposed interaction is weak and the in vivo biological relevance needs to be further examined (Figure 1, panel A).
It is also clear that miRNAs can regulate the immune response through pathways independent from the DNA methylation machinery (7). In fact, recent reports have shown that miR-146a is a negative regulator of the Interferon (IFN) pathway in lupus patients, and that underexpression of miR-125 contributes to the elevated expression of the pro-inflammatory cytokine RANTES in lupus (5–7).
A number of analytical techniques are now available for studying epigenetic modifications in a genome-wide manner. The systematic use of these genomic techniques will serve to provide a full profile of epigenetic deregulation in SLE. Unlike genetic alterations, which are permanent, epigenetic alterations are reversible. This opens the possibility of using epigenetic drugs to reverse the pattern of epigenetic alterations to relieve the phenotype. To date, histone deacetylase (HDAC) inhibitors such as suberoylanilide hydroxamic acid (SAHA) and trichostatin A (TSA) have proved to be useful for relieving SLE disease in mice (2). The effects of TSA on human T cells are predominantly immunosuppressive and reminiscent of the signaling aberrations that have been described in patients with SLE (2). Current evidence indicates that most of the genes that exhibit aberrant patterns of DNA methylation are hypomethylated, although gene-gene-specific hypermethylation cannot be ruled out (2). Therefore, the detailed analysis of DNA methylation at the gene level will serve to evaluate how useful histone deacetylase inhibitors and DNA demethylating drugs could be. It is not clear whether the increased expression of specific miRNAs is an indirect effect rather than the cause of SLE, and also this point needs further investigation in future studies.
Acknowledgments
This work was supported in part by a grant from the Lupus Research Institute and the National Institutes of Health grant AI47859. P.R.D.G. was supported by NIH training grant T32 DE007200.
Footnotes
Conflict of interests: The authors have no competing interests.
REFERENCES
- 1.Zhao S, Wang Y, Liang Y, Zhao M, Long H, Ding S, et al. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2010 doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]
- 2.Ballestar E, Esteller M, Richardson BC. The epigenetic face of systemic lupus erythematosus. J Immunol. 2006;176:7143–7. doi: 10.4049/jimmunol.176.12.7143. [DOI] [PubMed] [Google Scholar]
- 3.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–81. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 4.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 5.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–75. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X, Tang Y, Qu B, Cui H, Wang S, Wang L, et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010 doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]
- 7.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–94. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]